Abstract
The maize, cut-and-paste transposon Ac/Ds is mobile in Saccharomyces cerevisiae, and DNA sequences of repair products provide strong genetic evidence that hairpin intermediates form in host DNA during this transposition, similar to those formed for V(D)J coding joints in vertebrates. Both DNA strands must be broken for Ac/Ds to excise, suggesting that double-strand break (DSB) repair pathways should be involved in repair of excision sites. In the absence of homologous template, as expected, Ac excisions are repaired by nonhomologous end joining (NHEJ) that can involve microhomologies close to the broken ends. However, unlike repair of endonuclease-induced DSBs, repair of Ac excisions in the presence of homologous template occurs by gene conversion only about half the time, the remainder being NHEJ events. Analysis of transposition in mutant yeast suggests roles for the Mre11/Rad50 complex, SAE2, NEJ1, and the Ku complex in repair of excision sites. Separation-of-function alleles of MRE11 suggest that its endonuclease function is more important in this repair than either its exonuclease or Rad50-binding properties. In addition, the interstrand cross-link repair gene PSO2 plays a role in end joining hairpin ends that is not seen in repair of linearized plasmids and may be involved in positioning transposase cleavage at the transposon ends.
The homologous recombination pathway is the primary means to repair double-strand breaks (DSBs) in Saccharomyces cerevisiae (reviewed in reference 74). Gene conversion, a conservative mechanism, and single-strand annealing (SSA), a nonconservative mechanism, each require extensive homology between the broken ends and the repair template (19, 34, 54). However, when homologous recombination is blocked (e.g., in a rad52 mutant), nonhomologous end joining (NHEJ) is also shown to be involved in yeast DSB repair (36, 42, 51). The details of NHEJ are clearest in yeast, and at least eight yeast genes are required directly for efficient and relatively accurate repair by NHEJ: YKU70, YKU80, DNL4, LIF1, NEJ1, RAD50, MRE11, and XRS2. Extensive searches have shown that there are unlikely to be any other major NHEJ genes in yeast (53, 80). A better understanding of NHEJ is of general interest, as NHEJ is the dominant DSB repair mechanism in animals and, based on the difficulties in achieving homologous gene replacement, NHEJ is thought to be the overwhelmingly predominant DSB repair mechanism in plants (6, 26, 33).
Plant transposable elements provide a valuable resource for studying DNA breakage and rejoining events in plants. These elements have to break both strands of the DNA in order to excise from one site and move to another, although the nature of those breaks and any host factor involvement remain somewhat unclear (for reviews, see references 6, 25, 37, 76, and 78). For example, it is likely that at least some plant transposases (TPases), such as those of hAT superfamily elements like Ac/Ds and Tam3, initiate sequential single-stranded cuts similar to those described in immunoglobulin gene rearrangement, rather than cutting both strands simultaneously (reviewed in references 23 and 37). While TPase is thought to bend and align the two transposon ends prior to excision (61) and possibly recruit a transposition target site (15, 32), it is not yet clear what role, if any, plant TPases might play in repair of excision sites. In addition, few host factors that are involved in the transposition process per se are known (65, 69).
The repair of plant transposon excision sites is primarily through NHEJ, which usually creates sequence alterations, or transposon footprints, at the excision sites (7, 14, 64, 67). As NHEJ is also the predominant means for repair of DSBs in plants, it is useful to compare directly the roles of these genes in repairing DSBs and repairing transposon excision sites.
Manipulating these genes in a meaningful way in plants is difficult. However, yeast cells provide an excellent system for making this comparison. The maize Ac/Ds transposons have been modified to transpose in yeast cells (79), allowing us to assay the repair of excision sites in a variety of yeast mutant backgrounds. DNA hairpins appear to form in the host DNA during transposition, and NHEJ is involved in the repair of these excision sites in haploid cells, often creating long palindromic sequences when the hairpins reopen. Both autonomous Ac elements and nonautonomous Ds elements (provided with TPase in trans) can excise from yeast plasmids and yeast chromosomes. Similar repair products arise in all these cases as long as the sequence flanking the transposon is held constant (J. Yu and C. F. Weil, unpublished data).
The mechanism used to repair DNA breaks in yeast is controlled by a variety of factors. These include the availability of homologous repair templates, phase of the cell cycle, whether the cells are haploid or diploid, the presence of other breaks in the genome, complementarity of the broken ends, and overall growth conditions (10a, 41, 48, 51, 56). A common occurrence in a yeast cell with a broken DNA molecule and an available, homologous repair template is that either recombination or gene conversion is used to repair the broken DNA, a feature that has made gene replacement in yeast a widespread technique. However, if there is no homologous template available, then nonhomologous ends can be rejoined by three other mechanisms (42, 56). One is SSA, a deletion-prone, microhomology-dependent mechanism rejoining the two DNA ends via short (≥10-bp) direct repeats, deleting one of the two direct repeats and the sequences between them (18, 34a, 44). SSA is dependent on the function of RAD52 and RAD59 and appears to be suppressed by HPR5/SRS2, increasing in hpr5/srs2 mutants (12, 60, 70, 80). Alternatively, NHEJ can occur precisely by simple religation, in which two DNA ends are ligated together without the loss of nucleotide sequences (41, 50, 71). Finally, NHEJ can occur imprecisely, resolving through very short (<10 bp) microhomologies and creating deletions; however, imprecise NHEJ shows different gene requirements from those of SSA (36, 47, 51, 81, 82).
A more thorough understanding of the repair events in yeast cells following Ac/Ds transposon excision is necessary before gene function in that repair can be analyzed. In our previous studies, repair of the excision sites occurred by NHEJ; however, these experiments were carried out in the absence of any homologous repair templates. Here we describe repair of those sites in the presence of available, homologous repair templates and find that >50% of the repair events still occur by end joining resolved using microhomologies. We then show that mutants lacking genes involved in NHEJ show a reduced ability to repair Ac/Ds excisions, suggesting that these genes are important for the repair. In particular, MRE11 separation-of-function alleles suggest a specific role for this gene in processing Ac excision sites.
MATERIALS AND METHODS
Ac/Ds constructs.
Plasmids pWL117 (containing a mini-Ds), pWL80 (containing a GALIS-driven Ac TPase cDNA fused to the Gal4p nuclear localization signal), pWL67A (with a mini-Ds in the HpaI site of ADE2), pWL88 (pRS402 [4] with a unique XhoI site introduced into the 5′ untranslated region [UTR] of ADE2), pWL89A (a BglII fragment of pWL88 containing the entire ADE2 gene plus an XhoI site cloned into YCpLac33 [24]), and pWL90A (with a mini-Ds in the 5′ UTR of ADE2) have all been described previously (79).
Plasmid pWL205 was constructed by removing a 2-kb BstXI-KpnI CEN4/ARS fragment from pWL89A, blunting the BstXI end with T4 DNA polymerase, and ligating the fragment into Ecl136II/KpnI-cut pWL67A. We synthesized an autonomous Ac element by inserting a PvuII fragment of pWL80 containing TPase into the blunted XhoI site of the mini-Ds on pWL117, creating pWL137A. HpaI and BglII restriction sites were then introduced on both sides of the transposon by site-directed mutagenesis to make pWL155. We constructed a second, autonomous Ac element on plasmid pWL211 by insertion of the PvuII TPase fragment of pWL80 into the blunted XhoI site of mini-Ds on plasmid pWL205. The TPase on pWL211 parallels the orientation of the Ds. The PvuII fragment of pWL80 containing the TPase was also inserted into the blunted XhoI site of pWL67A to create pWL204. Plasmid pWL200 was made by insertion of a HpaI fragment from pWL155 into the HpaI site of the ADE2 open reading frame on plasmid pWL89A. pWL174 was constructed by insertion of a PvuII fragment of a hyperactive TPase mutant, D459A (unpublished data) (a generous gift from R. Kunze), into the blunted XhoI site of pWL90A. The TPase on pWL174 parallels the orientation of ADE2. pWL216 was made by ligating the PvuII TPase fragment of pWL80 into EcoRV/Ecl136II-cut pGEV-HIS (22), which contains a HIS3 marker.
Site-directed mutagenesis of the flanking sequences of Ds.
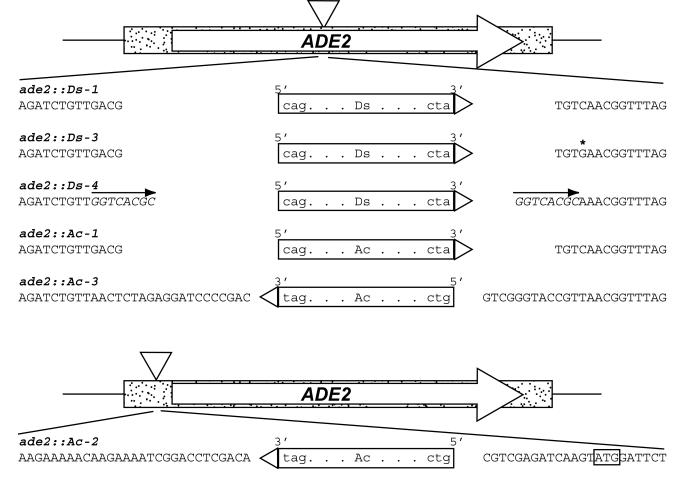
PCR primers were as follows: P1, 5′-GGAAATGATTCCGGAAGCTTTGGAAGTACTGAAGG-3′; P2, 5′-AGTCCGGAACTCTAGCAGGCGCATAACATAAGTCACAAATATTGTCCTTGTGGATAGTCTCTACAATTGGGTAAGAAAACACTAACGTCAACAGACCGTTCACGATAGGGATGAAAACGGTCGG-3′; P3, 5′-ATTCCGGAAGCTTTGGAAGTACTGAAGGATCGTCCTTTGTACGCCGAAAAATGGGCACCATTTACTAAAGAATTAGCAGTCATGATTGTGAGATCTGTTGGTCACGCCAGGGATGAAAGTAGGATGG-3′; and P4, 5′-AGTCCGGAACTCTAGCAGGCGCATAACATAAGTCACAAATATTGTCCTTGTGGATAGTCTCTACAATTGGGTAAGAAAACACTAAACCGTTTGCGTGACCTAGGGATGAAAACGGTCGGT-3′. Primers P1 and P2 were used to site-direct a 1-bp change into the DNA flanking Ds in pWL205, and the amplified fragment was digested with BspEI and used to replace the original BspEI fragment of pWL205 to create pWL252, carrying the ade2::Ds-3 allele (Fig. 1). Similarly, for construction of pWL264, the BspEI fragment of a PCR fragment amplified from pWL205 by using primers P3 and P4 was used to replace the original BspEI fragment of pWL205. By doing so, the 8-bp direct repeat of the maize wx-m7 allele (64) was placed adjacent to Ds, creating ade2::Ds-4 (Fig. 1).
FIG. 1.
Ac/Ds constructs with their flanking sequences used in this study. The ADE2 open reading frame is indicated by a stippled box. ade2::Ds-3, ade2::Ds-4, and ade2::Ac-1 are derived from ade2::Ds-1 (see Materials and Methods). Transposon insertions are represented as triangles. The site-directed single-base change in ade2::Ds-3 is denoted by an asterisk. The 8-bp direct repeat of wx-m7 in ade2::Ds-4 is indicated by heavy black arrows. In ade2::Ac-2, Ac is inserted in the 5′ UTR of ADE2 instead of in the middle of the gene; the ATG start codon is boxed. Ac/Ds transposons are indicated as boxes; the orientation of Ds (5′ and 3′ ends) is based on the transcription direction of wild-type Ac in maize. Italicized bases are those that do not belong to the original open reading frame of ADE2 and have been added by cloning.
Yeast strains.
Yeast strains used in these experiments are shown in Table 1. pWL204 was transformed into BY4723, selecting Ura+ transformants on synthetic complete glucose (SC) medium lacking uracil. The transformants were streaked on 5-fluoroorotic acid (5-FOA) to select red, Ura− derivatives, indicating disruption of ADE2 and loss of URA3. A red 5-FOAr colony that had been verified to contain ade2::Ac by PCR amplification was designated JYY24.
TABLE 1.
S. cerevisiae strains
| Strain | Parent strain/plasmid(s) | Genotype | Source or reference |
|---|---|---|---|
| BY4723 | MATaura3Δ0 his3Δ0 | 4 | |
| BY4725 | MATα ade2::hisG ura3Δ0 | 4 | |
| CWY1 | BY4723 | MATaura3Δ0 his3Δ0 ade2::Ds-1 | 79 |
| JYY1 | CWY1 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1 | This study |
| JYY2 | JYY1/pWL200 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1/CEN6/ARS4 URA3 ade2::Ac-3 | This study |
| JYY13 | JYY1/pWL211 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1/CEN6/ARS4 URA3 ade2::Ac-1 | This study |
| JYY16 | JYY1/pWL174 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1/CEN6/ARS4 URA3 ade2::Ac-2 | This study |
| JYY24 | MATaura3Δ0 his3Δ0 ade2::Ac-1 | This study | |
| JYY26 | BY4725/pWL174 | MATα ade2::HisG ura3Δ0/CEN6 ARS4 URA3 ade2::Ac-2 | This study |
| JYY37 | CWY1/pWL216 | MATaura3Δ0 his3Δ0 ade2::Ds-1/CEN6 ARS4 HIS3 TPase | This study |
| JYY41 | JYY1/pWL205/pWL216 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1/CEN6 ARS4 URA3 ade2::Ds-1/CEN6 ARS4 HIS3 TPase | This study |
| JYY54 | JYY1/pWL252/pWL216 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1/CEN6 ARS4 URA3 ade2::Ds-3/CEN6 ARS4 HIS3 TPase | This study |
| JYY60 | JYY1/pWL264/pWL216 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1/CEN6 ARS4 URA3 ade2::Ds-4/CEN6 ARS4 HIS3 TPase | This study |
| MY031 | JYY1 | MATaura3Δ0 his3 Δ0 ade2ΔHpaI-PflM1 trp1::hisG | This study |
| MY032 | MY031 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1 trp1::hisG mre11::hisG | This study |
| MY033 | MY031 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1 trp1::hisG sae2::kanMX4 | This study |
| MY034 | MY031 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1 trp1::hisG nej1::kanMX4 | This study |
| MY037 | MY031 | MATaura3Δ0 his3Δ0 ade2ΔHpaI-PflM1 trp1::hisG pso2::kanMX4 | This study |
A partial deletion of ADE2 was made in BY4723 to get JYY1. To this end, the BglII-BclI fragment containing CEN/ARS was removed from pWL89A to get pWL198. Then the HpaI-PflM1 fragment of ADE2 was deleted from pWL198 to create pWL201. pWL201 was transformed into BY4723 to select Ura+ transformants. Selection on 5-FOA was carried out to identify Ura− Ade− derivatives carrying ade2-ΔHpaI/PflM1 on chromosome XV. The deletion was then confirmed by PCR.
The strain MY031 was constructed by knocking out the TRP1 gene of JYY1 by the hisG cassette replacement technique (1). Strains MY032, MY033, MY034, and MY037 were then constructed from MY031 by replacing the MRE11, SAE2, NEJ1, and PSO2 open reading frames, respectively, with the kanMX4 cassette (77).
In vivo plasmid rejoining assay.
Plasmid pWL89A was cut with HpaI, and the linearized fragment was gel purified, recut with HpaI, and gel purified again to remove any uncut plasmid. This fragment was transformed into JYY1 cells and selected on both SC-Ura and SC-Ura-Ade plates. White colonies recovered from the transformation were restreaked onto SC-Ura-Ade, and total DNA was extracted from them by using a Y-DER yeast DNA extraction kit (Pierce). The extracted DNA was used to amplify end-joining repair products with primers 5′-TCGTCTTGAAGTCGAGGACTTTGGCA-3′ and 5′-AACGGAGTCCGGAACTCTAGCAGGCGCA-3′. Those PCR fragments of correct size that could still be cut by HpaI were scored as simple-ligation end joining.
Characterization of independent Ac/Ds footprints.
A single colony was streaked on SC medium containing galactose as the sole carbon source (SCGal)-Ura-Ade for Ade+ revertants. To ensure that the recovered footprints were independent, only one revertant was picked among all the revertants derived from each original Ade− colony. The revertants were purified to homogeneity, and a single colony was grown in SC-Ade liquid culture. DNA was extracted and used to amplify Ac/Ds excision sites by PCR as described previously (79). The gel-purified PCR products were then sequenced directly.
RESULTS
Experimental system.
Ac/Ds transposons can excise and reinsert in yeast cells given a TPase source under the transcriptional control of the yeast GAL1S promoter and CYC1 terminator sequences (79). Transposons inserted into the yeast ADE2 open reading frame can be used to monitor excision events in much the same way that excision markers are utilized in plants. Ade+ revertants on galactose medium are then purified, and their DNA is analyzed for excision and reinsertion events (Fig. 1). While this system depends on the restoration of the open reading frame to recover ADE2 function, revertants are typically observed with a frequency of 5 × 10−4 (thus, an overall predicted excision rate of ∼1.5 × 10−3), allowing recovery of a wide variety of excision events. Insertions in the 5′ UTR of ADE2 were also used to analyze Ac excisions in the presence of a homologous repair template. However, while these insertions do not require reading frame restoration to become Ade+, they result in a leaky Ade− phenotype due to an outward reading promoter in the transposon ends (Yu and Weil, unpublished). Revertants can be selected against the background growth after 3 to 7 days.
End-joining repair of Ac/Ds excision sites in yeast is primarily microhomology dependent.
Analysis of 62 independent footprints from ade2::Ac-1 or ade2::Ds-1 in the strains JYY13 (with Ac on a plasmid), JYY24 (Ac in chromosome XV), and JYY41 (Ds on a plasmid) suggest that 95.2% of them are resolved through microhomology, resulting in deletions (Fig. 2). Sequences of 1, 2, and 3 bp serve as the microhomology with frequencies of 40.3, 25.8, and 29.0%, respectively. In these strains, homologous repair is prevented by lack of a homologous repair template. The short length of the apparent microhomologies argues that imprecise NHEJ is the mechanism rejoining these ends, as opposed to SSA (34a). Similarly, analysis of 110 additional footprints derived from the ade2::Ds-3, ade2::Ds-4, ade2::Ac-2, and ade2::Ac-3 alleles showed that these could also be resolved through short direct repeats (91.9% of the total 172) (Fig. 2, 3, and 4). Overall, the direct repeats that serve as microhomologies range from 1 to 6 bp. These results are consistent with previous studies, in which ∼88% of end-joining events following either HO-induced or dicentric chromosome breaks had homology of the terminal bases in the deletion (36).
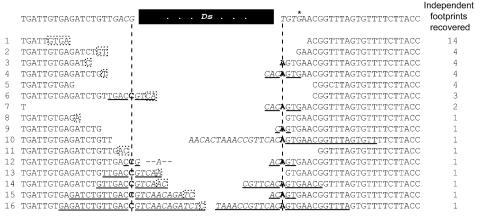
FIG. 2.
DNA sequences of independent footprints recovered from ade2::Ac-1 and ade2::Ds-1. (A) All of these sequences are collected from the strains JYY13 (Ac on plasmid), JYY24 (Ac on chromosome), and JYY41 (Ds on plasmid). The 62 independent transposon footprints correspond to 25 distinct types. Palindromes of sequences are underlined, with the added “P” nucleotides in italics and the altered base at the axis of each palindrome in boldface (see reference 37). Bases representing putative microhomologies are shown in dashed boxes; these bases would also be present in the sequence of the opposite end and then removed when the footprint is resolved. (B) An example of how microhomology between the P nucleotides present on one side of the footprint could serve as microhomology for resolution of the footprint. The arrow represents the opening of the hairpin on the right (the one on the left has already been opened), the boldface letter represents the base around which the palindrome has its axis, the microhomology is indicated in the dashed boxes, and the heavy, dashed lines represent DNA synthesis.
FIG. 3.
DNA sequences of independent footprints recovered from the ade2::Ds-3 allele in strain JYY54. ade2::Ds-3 was derived from ade2::Ds-1 by site-directed mutagenesis, with a single base change, C→G (asterisk). Representation is as in Fig. 2. The 44 footprints correspond to 16 distinct types.
In plants, one predominant Ac/Ds footprint arises, determined by flanking sequence and likely the most frequent repair product (64, 67). Similar predominant products have been reported for Ascot excision in the fungus Ascobolus immersus (10). A predominant Ac/Ds footprint also arises in yeast, apparently through a combination of the length and the distance from the broken DNA ends of the microhomology (see Discussion). Of the 62 independent footprints from ade2::Ac-1 or ade2::Ds-1, one footprint is found 22 times (33.5%) and represents the predominant footprint for this particular sequence context (Fig. 2). The probability of recovering independently derived, identical footprints in which both ends are processed identically, or even processed coincidentally to produce identical sequences, is extremely small. The predominant Ade+ footprint from the ade2::Ac-1 and ade2::Ds-1 alleles, which are flanked by the same host DNA sequences, is determined by a 3-bp, TGA microhomology. One copy of TGA is present upstream of and close to the Ac/Ds excision site. The other is downstream of the excision site and is part of a palindrome that forms when the DNA hairpin is opened. In contrast, among minor footprints shown in Fig. 2 only one (shown as footprint 3) uses a 3-bp microhomology and the rest form through microhomologies of either 1 or 2 bp. Although the microhomology is also TGA for this footprint, the two copies of TGA are further apart.
In yeast, the predominant footprints are the same and occur with similar frequencies for Ac as for Ds for any given flanking sequence, whether transposition is from a plasmid or from a chromosome (Table 2). The ade2::Ac-1, ade2::Ac-2, and ade2::Ac-3 alleles, each with different flanking sequences (Fig. 1), all have different predominant footprints (data not shown). Different flanking sequences would likely lead to availability of different microhomologies and, therefore, determine the formation of different predominant footprints and minor footprints.
TABLE 2.
Frequency of predominant footprints of ade2::Ac-1 and ade2::Ds-1 on chromosomes or plasmids
| Strain | Element and location | No. of total footprints sequenced | No. of predominant footprints | Frequency of predominant footprints (%) |
|---|---|---|---|---|
| JYY13 | Ac on plasmid | 21 | 7 | 33.3 |
| JYY24 | Ac on chromosome | 22 | 7 | 31.8 |
| JYY41 | Ds on plasmid | 19 | 8 | 42.1 |
To test this hypothesis further, we altered the downstream TCA of ade2::Ds-1 to TGA by site-directed mutagenesis to produce the ade2::Ds-3 allele (Fig. 3). This 1-bp change destroyed the microhomology that forms the predominant Ade+ footprint for ade2::Ds-1 and created a new direct repeat of 4 bp (GTGA) flanking the excision site at a different position in ade2::Ds-3. As expected, the ade2::Ds-3 allele showed a new predominant Ade+ footprint (31.8%, n = 44), formed through this 4-bp microhomology. The predominant footprint observed in the progenitor construct was no longer observed. In addition, no other Ade+ footprints formed from this allele utilize microhomologies as long as 4 bp.
Microhomology and “+0” excisions.
In plants, insertions, deletions, or substitutions associated with Ac/Ds excisions usually occur in the 1 or 2 bp closest to the element (64, 67). The majority of footprints typically do not restore reading frame, and precise, or “+0,” excisions, which restore the DNA to the same sequence that was present before transposon insertion, are rare.
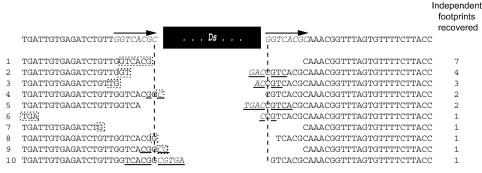
None of the footprints shown in Fig. 2 and 3 are recovered from alleles flanked by the characteristic 8-bp target site duplication that typically accompanies insertion of hAT family transposons like Ac/Ds (reviewed in reference 37). Each was created by Ac/Ds cloning rather than transposition. We therefore mutagenized the DNA flanking Ds in ade2::Ds-1 to add the 8-bp target duplication found flanking Ac in the maize wx-m7 allele (64), creating the ade2::Ds-4 allele (Fig. 1). Among independent, Ade+ revertants of ade2::Ds-4 (admittedly, a biased sample selected for restoration of ADE2 reading frame), +0 footprints were the largest single group (7 of 23) (Fig. 4). In both Arabidopsis thaliana and maize, using this same wx-m7 sequence context in PCR assays that are not biased by phenotype, we observe similar proportions of +0 events among those footprints that restore reading frame, though they do not predominate overall (64). These +0 footprints could form through a 6-bp microhomology (the middle part of the 8-bp target site duplication), although this microhomology would be available only in the subset of excisions where both DNA hairpins open >8 bp away from the tip of the hairpin. Further characterization is required, but it is intriguing that +0 footprints do not occur with similar frequency in those alleles examined to date that lack an 8-bp target site duplication.
FIG. 4.
DNA sequences of independent footprints recovered from ade2::Ds-4 in strain JYY60. Other representations are as in Fig. 2. The 23 footprints correspond to 10 distinct types.
Choice of repair pathway.
Microhomology searching during repair of Ac excision sites would require that additional processing of the host DNA hairpins into nonhomologous, single-stranded tails occur before the ends are ligated. Boulton and Jackson (2, 3) reported that blunt-ended, SmaI-cut pBTM116 was repaired using microhomology-driven end joining, resulting in deletions. Oettinger and coworkers have recently observed similar results (M. Oettinger, personal communication). The interpretation of their results was that the blunt ends were resected to produce single-stranded overhangs. This processing is also consistent with the recombinogenic nature of blunt-ended DNAs transformed into yeast, which further suggests that the blunt ends are processed to have single-stranded 3′ ends. If 3′ overhangs are also created during Ac excision site repair, homologous sequences might be subject to strand invasion by those 3′ ends, and Ac excision might stimulate recombination.
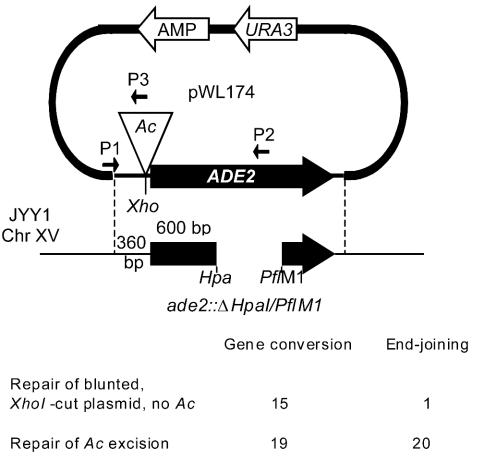
In our previous experiments, yeast cells were forced to use NHEJ to repair Ac excisions because no homologous repair template was available. We therefore wanted to assay repair of excision sites in cells that could carry out both homologous repair and NHEJ to ask which pathway was used (Fig. 5). As a control, we first tested repair of a blunt-ended, linear DNA using the same constructs that we proposed to test in our transposition assay. The plasmid pWL89A cut with XhoI and treated with T4 DNA polymerase leaves blunt ends at the same site as does the transposon insertion in ade2::Ac-2; this XhoI site is not present in the chromosomal copy of ADE2 (79). We transformed purified XhoI-linearized and blunted pWL89A, carrying an ADE2 gene with no Ac, into JYY1 cells carrying the ade2::ΔHpaI-PflM1 allele, as described in Materials and Methods. Of transformants selected initially for Ura+ and then screened for ADE2 function, 15 of 16 (93.7%) were Ade−, and all these contained gene conversions of the ade2::ΔHpaI-PflM1 allele from the chromosome to the plasmid (see Discussion). The same pWL89A plasmid cut with HpaI leaves blunt DSB ends in the middle of the ADE2 gene at the same site as does the Ac insertion in ade2::Ac-1. With the use of HpaI-linearized pWL89A in JYY1, one side of the DNA break is homologous to the repair template while the other is not, due to the ade2::ΔHpaI-PflM1 allele in the chromosome. These too were predominantly Ade− (∼98%, n = 600), and 60 of 60 Ade− plasmids analyzed contained gene conversions of the ade2::ΔHpaI-PflM1 allele from the chromosome to the plasmid. We had a 95% chance of seeing at least one of any other type of event, had it occurred with a frequency of 5% or greater [n = −ln(1 − P)/μ] but did not. Among Ade+ plasmids recovered, the ADE2 gene could be recut with HpaI in 15 of 16 independent events, suggesting that this subclass was rejoined by simple, blunt-end ligation without resection.
FIG. 5.
Repair choice experiment. Small arrows indicate PCR primers. The region of homology between the plasmid and the chromosome is indicated by dashed lines.
The Ac-containing plasmid pWL174 was then transformed into strains carrying a chromosomal ADE2 locus with a deletion of the internal HpaI-PflM1 fragment in its 3′ end (Fig. 1). This arrangement provided ∼360 bp 5′ and ∼600 bp 3′ to the Ac insertion site (34) and allowed us to distinguish end joining from homologous repair events because of sequence polymorphisms immediately flanking the Ac added while cloning the transposon into ADE2 (see reference 79). Restoration of completely wild-type sequence to any functional ADE2 gene recovered could come only from either gene conversion between the plasmid and the chromosomal ADE2 gene or double-crossover events that would also transfer Ac reciprocally to the chromosomal ADE2, creating an allele with both the HpaI-PflM1 deletion and the transposon insertion.
Unexpectedly, 20 of 39 Ade+ repair products that we recovered following transposition were formed by end joining, even though a repair template with extensive homology on either side of the excision site was available (Fig. 5). Overall, we recovered Ade+ colonies with a similar frequency in this set of experiments (∼10−4) as in experiments where homologous repair templates were not available. Fully wild-type ADE2 genes were recovered in 19 of the 39 independent events, presumably the result of gene conversion events. While we cannot conclusively associate these gene conversions with transposition and our sample size was relatively small, we did not observe them among a similar number of cells plated on glucose medium where transposition was not induced.
Roles for host factors in transposition and repair of Ac excision sites.
We assayed the repair of excision sites in cells deficient in various NHEJ-associated genes. Deletion of the yku70, yku80, rad50, mre11, xrs2, sae2, or pso2 loci resulted in a striking decrease in Ade+ revertants of the ade2::Ac-1 allele compared with isogenic, repair-proficient cells (Fig. 6A). In each case, complementation of all these mutations by transformation with plasmids carrying the appropriate wild-type genes restored Ade+ reversion to nonmutant levels (data not shown). Most of these deletion alleles completely abolish reversion of the ade2::Ac-1 allele. However, deletion of the PSO2 gene still allows ∼10% of the wild-type reversion frequency, and the transposon footprints from these revertants are intriguing.
FIG. 6.
(A) Ac excision in strains carrying DNA repair mutations. Bars indicate reversion of the ade2::Ac-1 allele on plasmid pWL210 to Ade+. Values were averaged over four independent experiments, error bars indicate standard deviations. (B) Ac footprints obtained in pso2Δ mutant yeast. Footprints 1 to 24 are as in Fig. 2. Footprints 25 to 32 contain sequences from the Ac transposon, indicated in white letters on a black background.
Half of the footprint sequences recovered from three independent experiments with the pso2Δ mutant are similar to those seen in nonmutant controls (Fig. 6B). Of particular interest, however, 8 of the 32 types of footprints recovered from the pso2Δ mutant retain part of one or both ends of the transposon. This class of footprint has been quite rare in all our previous experiments (Fig. 2 to 4) (79), arising only one time each in 2 of over 150 separate experiments with repair-proficient strains (∼200 different footprint types overall [C. F. Weil, unpublished data]). Microhomology appears to be involved in resolving only two of these eight footprints. In one of these (sequence 25, Fig. 6B), both ends of the transposon remain at the site and resolution appears to be through a TACCGACC repeat present near both transposon ends, similar to a mechanism that forms deleted elements in plants and other eukaryotes (31, 83). In the other case (sequence 31, Fig. 6B), one side of the footprint appears to have formed and resolved a hairpin structure in host DNA; however, it is not yet clear whether DNA hairpins form within transposon sequences. In four cases, cleavage at one end appears to be precise in its removal of only the base adjacent to the transposon while the other end retains transposon sequence. In one instance, a hairpin appears to have formed in host DNA and resolved to create a short palindrome on one side while leaving Ac sequence on the other. We note that microhomology appears to be involved in the resolution of this particular footprint but not in the others, where host DNA on one side is joined to residual transposon sequence on the other. The mechanism forming footprints that include part of the transposon remains unclear but is likely to involve TPase miscutting, as they do not arise when the TPase is not induced (data not shown). The pso2Δ mutation does not significantly affect NHEJ of HO endonuclease-cleaved chromosomal DNA or of restriction endonuclease-linearized plasmids transformed into yeast (data not shown).
Ac excision and Mre11.
The apparent involvement of NHEJ genes further prompted us to investigate the involvement of the MRE11 gene in more detail. Mre11p is involved in a wide variety of cellular functions (reviewed in references 11, 59, and 84) and has been shown to have 5′-to-3′ exonuclease, 3′-to-5′ exonuclease, endonuclease, DNA-binding, and Rad50p-binding activities. Its exonuclease function has been implicated in the microhomology-dependent repair of nonhomologous ends (58), and its endonuclease function has been implicated in the opening of DNA hairpins in vitro (45, 57). We were interested in the possibility that it might be involved in opening the DNA hairpins created in host DNA during Ac transposition. This function is typically carried out by the Artemis gene product in V(D)J rearrangement (66), and although PSO2 bears some protein sequence similarity to Artemis, functional homologs of Artemis have not yet been identified either in yeast or in plants (see Discussion).
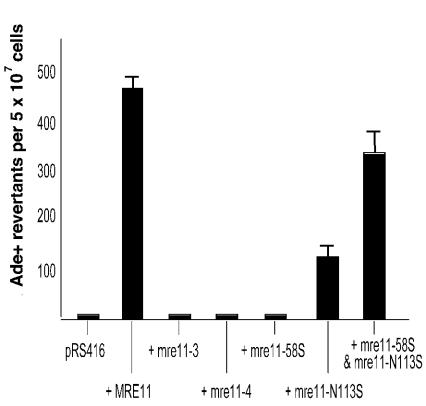
JYY1 cells with a completely deleted mre11 locus (MY032) were transformed with the plasmid pWL210, containing an Ac element inserted into the 5′ UTR of the ADE2 gene. No revertants were recovered from plates on which the TPase was induced or from the same strain transformed with a second, empty vector (Fig. 7). Transformation was restored by transforming in a second plasmid containing the wild-type MRE11 gene. Similar experiments were then carried out to add each of several separation-of-function mutant alleles, mre11-3, mre11-4, mre11-11, mre11-58S, and mre11-N113S, on plasmids (40; Bressan et al., 1998) (Fig. 7). These alleles have alterations in phosphodiesterase motifs located in the N-terminal nuclease domain of the protein. The mutations differentially eliminate the exonuclease and/or Rad50p-binding capabilities of Mre11p and cause differences in the ability of the cell to prevent mitotic chromosome loss, maintain telomere length, and respond to radiation-induced breaks (40). Most of the mutations behaved identically to the mre11Δ allele, and no Ade+ colonies were recovered. Only mre11-N113S restored any ability to repair the Ac excision site and then only to ∼35% of wild-type levels. Interestingly, when the mre11-58S and mre11-N113S alleles were added together, the reversion rate increased to nearly 70% of the wild-type rate. A similar complementation was seen in the suppression of methyl methanesulfonate sensitivity and in end joining of restriction endonuclease-cleaved plasmids by these two mutants in the N terminus of the Mre11 protein (40) (see Discussion).
FIG. 7.
Ac excision in strains carrying separation-of-function mutations of mre11. An mre11Δ0 mutant strain was transformed with either empty pRS416 vector or pRS416 with the indicated insertion. Bars indicate reversion of the ade2::Ac-1 allele on plasmid pWL210 to Ade+. Values were averaged over four independent experiments; error bars indicate standard deviations.
DISCUSSION
In this paper, we present evidence to show that yeast cells primarily use a microhomology-dependent end-joining repair pathway to repair Ac/Ds transposon excision sites. Based on palindromes observed at rejoined footprints, these excision sites are thought to involve DNA hairpin intermediates in the rejoining DNA, similar to those seen in V(D)J rearrangement of vertebrate immunoglobulin genes (37, 79). Also similar is the apparent involvement of gene products that carry out NHEJ. Data suggest that yeast can use NHEJ to repair Ac excision sites even in the presence of homologous repair templates, although it is not yet clear whether this is a result of bias towards NHEJ or a reflection of when in the cell cycle excision occurs or of the fact that TPase remains associated with one or both DSB ends. One source of bias towards NHEJ may be that nonhomologies must be removed from DSB ends in yeast before strand invasion can prime DNA synthesis needed for gene conversion (9, 55), and Ac excision events may leave heterologous sequences that are long enough to inhibit homologous repair relative to NHEJ. A plasmid carrying a 5-bp homology at the broken ends and lacking Ac is repaired primarily through gene conversion, suggesting either that the heterologies created during Ac excision events are longer or that additional factors may be involved.
As in plants, a predominant repair product of Ac/Ds excision is formed in yeast, and the microhomology used for the predominant footprint is longer and closer to the excision site than those used for minor footprints. Manipulation of these microhomologies results in predictable changes in the predominant footprint. While our data are more consistent with imprecise NHEJ than they are with SSA, our results parallel those of Sugawara et al. (70), who found similar relationships among homology length, proximity to the DNA break, and frequency with which sequences were used to resolve SSA events. RAD52, RAD59, RAD1, RAD10, and MSH2 are important genes for SSA, and studies to determine whether deleting any of these genes similarly affects repair of Ac excision sites are in progress. For example, Rad1/10 and Msh2/3 would also be needed to remove heterologies of >20 nucleotides from DSB ends for homologous recombination, and we would predict that mutations in these genes could further bias repair of Ac excisions towards end joining.
Most Ac/Ds footprints in yeast involve deletions of 5 to 20 bp or additions of palindromic sequences of 1 to >20 bases. In contrast, footprints recovered from plants typically show much smaller additions or deletions, particularly in maize (64, 67, 79). The reasons for these differences in NHEJ remain unknown. Now that we have compared the same Ac TPase source, cis-acting transposon sequences, and even the same flanking sequence context (that of maize wx-m7) in yeast and plants, the explanation probably lies in the host factors involved.
Candidates for these host factors are genes involved in NHEJ repair of broken DNA. YKU70, YKU80, RAD50, MRE11, and XRS2, genes known to be involved in end joining, are all required for reversion of ade2 mutations caused by Ac/Ds insertions. We have not yet tested deletions of other important NHEJ genes, such as DNL4 or LIF1 (28), but predict that these deletions should also show defects in repairing Ac excision sites. Our results further predict that a genome-wide screen for mutations affecting Ac transposition and repair of Ac excision sites, similar to that of Wilson (80), would be highly informative, particularly in finding additional candidate genes for further study in plants.
As an example, the PSO2 gene is not typically associated with NHEJ in yeast, being involved instead in DNA damage recognition of interstrand cross-links such as those induced by psoralen under UV light (5, 38). Pso2p is also similar in sequence to a family of interstrand cross-link repair proteins (1A [NP_055696], 1B [NP_073747.1], and 1C [CAC37570.1]) identified in human, mouse, and rat, which also include the Artemis protein, which opens the DNA hairpins formed during vertebrate immunoglobulin gene [V(D)J] rearrangement (46). While the similarity between Pso2p and cross-link repair protein 1A is much stronger than the similarity with Artemis (BLASTP scores of 2e-40 versus 8e-07, respectively), Pso2p and all three mammalian proteins share a beta-lactamase fold (52). If PSO2 is, in fact, a yeast homolog for Artemis, then our data suggest that Pso2p cannot be the only factor opening DNA hairpins in yeast cells, because genetic evidence for hairpin formation and opening persists even in pso2Δ mutants. Alternatively, Pso2 may recognize a DNA hairpin structure as similar to covalent, interstrand cross-linking between bases and bind it. The increase in TPase miscutting in the absence of PSO2, leaving transposon sequences behind, could indicate that this binding might help position the TPase to cut the second transposon end correctly. Two genes highly similar to PSO2 (At2g45700 and At3g26680) are found in Arabidopsis, and it will be interesting to determine their functions and whether they play any role in Ac transposition.
Transposon excision and recombination.
Ac/Ds transposon excisions have been proposed as a source of DSBs to stimulate somatic recombination events in plants to frequencies as high as 10−4 (82a). However, only intrachromosomal recombination has been demonstrated in these systems, and the observed stimulation could simply be a consequence of TPase bringing homologous sequences near one another as the transposon ends synapse. In addition, the stimulation does not extend to meiotic recombination (13), although meiosis is a somewhat specialized context. We suggest that in yeast and, quite possibly, in plants the host DNA ends are either sequestered in hairpin structures, protected by proteins, or both throughout the transposition process and that, as a result, only abortive transposition events would provide DSBs that might become recombination substrates.
Single-stranded 3′ ends at excision sites are an important part of the idea that transposition can stimulate recombination. The use of microhomology in repairing excision sites supports the idea that single-stranded DNA ends are a regular part of repairing transposon excisions, and this idea is consistent with the rearrangements long known to accompany transposition. How often those single-stranded DNA tails can initiate strand invasion remains an open question. This question becomes still more intriguing considering that transposon excision sites are still preferentially repaired by end joining even in the presence of homologous repair templates. Recent data suggest that the availability of substrates dictates which repair pathway is most likely to be used (21). The relative lack of strand invasion, crossover formation, or synthesis-dependent strand annealing further supports the idea that any 3′ overhangs that form during transposition are sequestered away from homologous recombination. This idea would also predict that DNA DSB signaling mechanisms might not be triggered during typical transposition events.
Transposition and host factors.
Although there are some differences, V(D)J recombination in vertebrates is a strong model mechanistically for Ac/Ds transposition, at least in yeast (49, 75). Our data suggest that cellular proteins may play similar roles in both situations, particularly those binding early in the process of forming V(D)J coding joints (analogous to repair of Ac excision sites).
For example, one striking similarity between these systems is the direct involvement of Ku proteins (62). After Rag1/Rag2 cuts are introduced at recombination signal sequences in vertebrate lymphoid cells, Ku proteins bind the broken ends (29). Ku proteins also appear to be involved in repair of Ac/Ds excisions (Fig. 6A), and we suggest that they may act early in the process, before any microhomology searching begins. It seems unlikely that microhomology searching would initiate before the DNA hairpins are opened and processed, and Ku mutations are epistatic to hpr5/srs2 helicase mutations in cells repairing DSBs on linearized plasmids (27). The involvement of yku70 and yku80 in repair of Ac excisions is consistent with the results of Boulton and Jackson (3), who found that end-joining repair in a yku70 mutant was accompanied by formation of deletions. Similar results are observed in mammalian cells (reviewed in references 16, 43, and 72). Presumably, the broken DNA ends are more vulnerable to degradation in Ku-deficient mutants (17, 21, 39). Preliminary data also suggest that, for Ac excision sites in an AtKu80 mutant of Arabidopsis, deletions often extend up to 200 bp from the initial DNA break, >10-fold farther than typical results in nonmutant plants. (A. Saballos, K. Marshall, J. Friesner, A. Britt, and C. F. Weil, unpublished data). Our ADE2 screen would not detect such deletions because they would remove part of the ADE2 coding sequence and fail to restore ADE2 function (see Discussion). PCR screens for Ac excision events in cultures of yku80 mutant yeast, looking for a decrease in the size of the amplified excision product instead of for Ade+ revertants, have been inconclusive thus far.
Involvement of the Mre11/Rad50/Xrs2 complex is less straightforward. Mutations in any one of these proteins sharply inhibit successful end-joining repair of Ac excisions in yeast (Fig. 6 and 7). Similarly, mutations of Mre11, Rad50, or Nbs1 in mammalian cells disrupt V(D)J coding joint formation (23). The Mre11 complex may help bridge broken ends and align them and could serve a similar function in repair of Ac excisions (30). Strikingly, deletion of SAE2 has no effect on NHEJ although it does affect repair of Ac excisions; this suggests that the MRX(S) complex has both a role in NHEJ and another role in the Ac excision and repair process. A potential role in transposition could be in opening the DNA hairpin ends or in processing the ends into single-strand tails. Our data suggest that some functions of Mre11p can be ruled out as important. For example, neither the mre11-3 nor the mre11-4 mutant supports repair of Ac excisions, even though one binds Rad50p and the other does not, indicating that the Rad50-binding property of Mre11p is not sufficient for repairing Ac excisions and that the MRX complex likely serves more than just a structural role. Similarly, both mre11-3 and mre11-N113S support the exonucleolytic resection of HO endonuclease-induced DSBs, yet only mre11-N113S supports the repair of Ac excisions, suggesting that the exonuclease function(s) of Mre11p may not be important in repairing Ac excision sites. However, the endonuclease activity of the mre11-N113S and mre11-58S alleles remains untested, and this may be the activity of Mre11p that is important for opening hairpins (45). Hairpin opening in V(D)J recombination appears to be carried out in vivo by the Artemis protein together with a DNA-dependent protein kinase (DNA-PK) (46); however, no functional homologs of either Artemis or the catalytic subunit of DNA-PK known have been demonstrated yet in yeast or in plants. Furthermore, human Mre11p has been shown to have hairpin-opening activity in vitro (57). It remains formally possible that the Ac TPase will also be involved in the reopening of these hairpins.
The requirement for Nej1p (Fig. 6) is likely to come from its role in localizing Lif1/XRCC4p to the nucleus to interact with ligase IV, the major ligase in NHEJ (20, 35, 73). NEJ1 is sharply downregulated in a/α diploid cells, with a parallel decrease in NHEJ, and it will be interesting to see what effect this may have on repair of Ac excisions in diploid cells.
Lobachev et al. (45) describe a role for the SAE2 gene in repair of hairpins induced by palindromic copies of an Alu sequence in yeast. Consistent with their observations, we also observe a role for SAE2 in repairing Ac excisions (Fig. 6). How SAE2 is involved at either sort of hairpin remains unclear, but one suggestion has been that it may regulate the nuclease activity of the MRX complex (63). An intriguing possibility is that Sae2p might stimulate MRX exonuclease activity but only after MRX endonucleolytic cleavage.
Implications for transposition.
Most Ac/Ds footprints from yeast and from maize, Arabidopsis, and other plants (e.g., references 26, 64, and67) are explained by a variation of the “hairpin model” first proposed by Coen et al. (8) for the Antirrhinum majus transposon Tam3. Recent observations of V(D)J-related transposition, and even reverse transposition (68), as well as in vitro studies of the TPase encoded by the related hAT family transposon Hermes (N. Craig, personal communication), have further supported the formation of DNA hairpins and parallels between V(D)J and transposition of hAT transposons.
As it might apply to Ac, the present model proposes a single-stranded cut made by the TPase 1 base from the transposon on each end (reviewed in reference 37). Whether these cuts occur before or after the transposon ends synapse (83a) or whether they occur simultaneously remains an interesting question, particularly with regard to the timing of transposon movement, target site acquisition, and the potential role of Pso2p in aligning the transposon ends correctly. For example, TPase cleavage and hairpin formation in the DNA flanking one end of the transposon might occur and be recognized by Pso2p, which would then help position the TPase cleavage at the other end of the transposon. Once both transposon ends are free, the transposon could attack its next insertion target with these nucleophiles. At the excision site, hairpin opening, processing, and rejoining follow. We suggest that this hairpin processing may take place with the transposition complex still present, possibly preventing the use of the broken ends in homologous repair. Cellular repair proteins such as Ku and the Mre11 complex are likely to be parts of that complex.
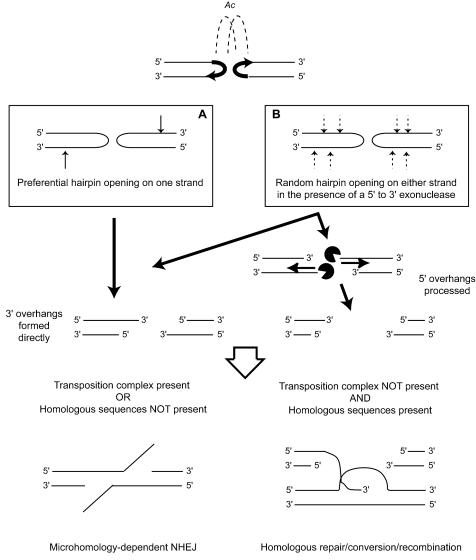
Our results suggest two possible models for how hairpins might be opening. In the first model, whatever endonuclease opens the hairpin would preferentially cut the strand that the TPase did not cut initially (Fig. 8A). Strand preference in hairpin opening could create 3′ single-stranded tails that could then undergo microhomology searching or homologous repair, if it is available. This model explains why the base adjacent to the element is always either changed to its complement or deleted. Such a strand preference would be most easily explained if TPase itself were either opening the hairpin ends or helping to position the factor that does.
FIG. 8.
Models for end-joining repair of Ac/Ds excision sites (see text for details). At the top of the figure, the ends of the transposon (dashed lines) have synapsed, TPase cleavage has occurred, and DNA hairpins have formed in the host DNA flanking the transposon. (A) Model showing preferential hairpin opening on the strand that has been attacked to form the DNA hairpin. (B) An alternative model in which hairpins are opened randomly on either strand but subjected to processing by a 5′-to-3′ exonuclease activity. 3′ ends are left as such, and 5′ overhangs are processed into 3′ overhangs that are then resolved through either microhomology-dependent end joining or homologous repair.
However, we cannot tell whether hairpin opening forms 5′ overhangs or 3′ overhangs in those half-footprints where the base closest to the transposon has been deleted (∼50% of the different half-footprints that we observed, both overall and for each allele individually). Thus, we cannot rule out a second model in which the DNA hairpin can be opened on either strand, and hairpin opening occurs in the presence of 5′-to-3′ exonuclease activity (Fig. 8B). Hairpins opened to create 3′ overhangs would undergo microhomology searching, while 5′ overhangs would be subject to further processing before ligation, perhaps enough to remove the base adjacent to the transposon before rejoining occurs, probably through microhomology. The lack of Ade+ revertants in mre11Δ, rad50Δ, or xrs2Δ mutant cells suggests that one candidate for this 5′-to-3′ exonuclease could be the Mre11/Rad50/Xrs2 complex. Interestingly, microhomology appears to be less of a factor in the repair of Ac excision sites in plants. Only about half of the footprints are even consistent with the use of possible microhomologies, and these are typically only 1 or 2 bp. Ironically, some of the clearest examples of deletions occurring through microhomologies that have been reported in plants come from examining formation of defective transposons (31, 83). Interestingly, plants do not appear to contain either a Rad52p or a Rad59p homolog, two gene products thought to mediate SSA in yeast (12, 70). While plant-specific factors are not required for Ac transposition (79), differences between the repair of Ac excision sites in yeast and that in plants should still prove informative with regard to specific repair factors that may be involved. In addition, the apparent formation of DNA hairpin intermediates in a system that lacks a DNA-PKcs and may lack an Artemis protein should provide an interesting perspective on alternative ways in which cells handle these structures.
Acknowledgments
We thank Avi Levy, Maria Valencia, Margie Oettinger, and Wolf Heyer for helpful discussions and Steve Scofield for critical reading of the manuscript.
This work was supported by USDA grant 98-35301-6740 and research grant no. US-3223-01C from BARD, the United States-Israel Binational Agriculture Research and Development Fund, to C.F.W. J.E.H. has been supported by DOE grant DOE 01ER63229 and NIH grant GM20056.
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulton, S. J., and S. P. Jackson. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24:4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulton, S. J., and S. P. Jackson. 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15:5093-5103. [PMC free article] [PubMed] [Google Scholar]
- 4.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 5.Brendel, M., and J. A. Henriques. 2001. The pso mutants of Saccharomyces cerevisiae comprise two groups: one deficient in DNA repair and another with altered mutagen metabolism. Mutat. Res. 489:79-96. [DOI] [PubMed] [Google Scholar]
- 5a.Bressan, D. A., H. A. Olivares, B. E. Nelms, and J. H. Petrini. 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britt, A. B. 1999. Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 4:20-25. [DOI] [PubMed] [Google Scholar]
- 7.Britt, A. B., and V. Walbot. 1991. Germinal and somatic products of Mu excision from the Bronze-1 gene of Zea mays. Mol. Gen. Genet. 227:267-276. [DOI] [PubMed] [Google Scholar]
- 8.Coen, E. S., T. P. Robbins, J. Almeida, A. Huon, and R. Carpenter. 1989. Consequences and mechanisms of transposition in Antirrhinum majus, p. 413-436. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 9.Colaiacovo, M. P., F. Paques, and J. E. Haber. 1999. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics 151:1409-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colot, V., V. Haedens, and J. L. Rossignol. 1998. Extensive, nonrandom diversity of excision footprints generated by Ds-like transposon Ascot-1 suggests new parallels with V(D)J recombination. Mol. Cell. Biol. 18:4337-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Cromie, G. A., J. C. Connelly, and D. R. Leach. 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell 8:1163-1174. [DOI] [PubMed] [Google Scholar]
- 11.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signaling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 12.Davis, A. P., and L. S. Symington. 2001. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159:515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dooner, H. K., and I. M. Martinez-Ferez. 1997. Germinal excisions of the maize transposon activator do not stimulate meiotic recombination or homology-dependent repair at the bz locus. Genetics 147:1923-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doseff, A., R. Martienssen, and V. Sundaresan. 1991. Somatic excision of the Mul transposable element of maize. Nucleic Acids Res. 19:579-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.English, J. J., K. Harrison, and J. Jones. 1995. Aberrant transpositions of maize double Ds-like elements usually involve Ds ends on sister chromatids. Plant Cell 7:1235-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Featherstone, C., and S. P. Jackson. 1999. Ku, a DNA repair protein with multiple cellular functions? Mutat. Res. 434:3-15. [DOI] [PubMed] [Google Scholar]
- 17.Feldmann, E., V. Schmiemann, W. Goedecke, S. Reichenberger, and P. Pfeiffer. 2000. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 28:2585-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fishman-Lobell, J., and J. E. Haber. 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258:480-484. [DOI] [PubMed] [Google Scholar]
- 19.Fishman-Lobell, J., N. Rudin, and J. E. Haber. 1992. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 12:1292-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank-Vaillant, M., and S. Marcand. 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 15:3005-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank-Vaillant, M., and S. Marcand. 2002. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell 10:1189-1199. [DOI] [PubMed] [Google Scholar]
- 22.Gao, C. Y., and J. L. Pinkham. 2000. Tightly regulated, beta-estradiol dose-dependent expression system for yeast. BioTechniques 29:1226-1231. [DOI] [PubMed] [Google Scholar]
- 23.Gellert, M. 2002. V(D)J recombination: Rag proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 24.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 25.Gorbunova, V., and A. A. Levy. 1999. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4:263-269. [DOI] [PubMed] [Google Scholar]
- 26.Gorbunova, V., and A. A. Levy. 2000. Analysis of extrachromosomal Ac/Ds transposable elements. Genetics 155:349-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hegde, V., and H. Klein. 2000. Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res. 28:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrmann, G., T. Lindahl, and P. Schar. 1998. Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 17:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopfner, K. P., C. D. Putnam, and J. A. Tainer. 2002. DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 12:115-122. [DOI] [PubMed] [Google Scholar]
- 30.Hopfner, K. P., L. Craig, G. Moncalian, R. A. Zinkel, T. Usui, B. A. Owen, A, Karcher, B. Henderson, J. L. Bodmer, C. T. McMurray, J. P. Carney, J. H. Petrini, and J. A. Tainer. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418:562-566. [DOI] [PubMed] [Google Scholar]
- 31.Hsia, A. P., and P. S. Schnable. 1996. DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. Genetics 142:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudson, A. D., R. Carpenter, and E. S. Coen. 1990. Phenotypic effects of short-range and aberrant transposition in Antirrhinum majus. Plant Mol. Biol. 14:835-844. [DOI] [PubMed] [Google Scholar]
- 33.Jackson, S. P. 2002. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23:687-696. [DOI] [PubMed] [Google Scholar]
- 34.Jinks-Robertson, S., M. Michelitch, and S. Ramcharan. 1993. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3937-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Karathanasis, E., and T. E. Wilson. 2002. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161:1015-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kegel, A., J. O. Sjostrand, and S. U. Astrom. 2001. Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr. Biol. 11:1611-1617. [DOI] [PubMed] [Google Scholar]
- 36.Kramer, K. M., J. A. Brock, K. Bloom, J. K. Moore, and J. E. Haber. 1994. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol. Cell. Biol. 14:1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunze, R., and C. F. Weil. 2002. The hAT and CACTA superfamilies of plant transposons, p. 565-610. In N. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 38.Lambert, S., S. J. Mason, L. J. Barber, J. A. Hartley, J. A. Pearce, A. M. Carr, and P. J. McHugh. 2003. Schizosaccharomyces pombe checkpoint response to DNA interstrand cross-links. Mol. Cell. Biol. 23:4728-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 40.Lee, S. E., D. A. Bressan, J. H. Petrini, and J. E. Haber. 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair 1:27-40. [DOI] [PubMed] [Google Scholar]
- 41.Lewis, L. K., J. M. Kirchner, and M. A. Resnick. 1998. Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA. Mol. Cell. Biol. 18:1891-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis, L. K., and M. A. Resnick. 2000. Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat. Res. 451:71-89. [DOI] [PubMed] [Google Scholar]
- 43.Liang, F., and M. Jasin. 1996. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 271:14405-14411. [DOI] [PubMed] [Google Scholar]
- 44.Lin, F. L., K. Sperle, and N. Sternberg. 1984. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol. Cell. Biol. 4:1020-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 46.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J. recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 47.Manthey, G. M., and A. M. Bailis. 2002. Multiple pathways promote short-sequence recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:5347-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martini, E., and S. Keeney. 2002. Sex and the single (double-strand) break. Mol. Cell 9:700-702. [DOI] [PubMed] [Google Scholar]
- 49.McBlane, F., D. van Gent, D. Ramsden, C. Romeo, C. Cuomo, M. Gellert, and M. Oettinger. 1995. Cleavage at a V(D)J. recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 50.Milne, G. T., S. Jin, K. B. Shannon, and D. T. Weaver. 1996. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4189-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moshous, D., I. Callebaut, R. de Chasseval, C. Poinsignon, I. Villey, A. Fischer, and J. P. de Villartay. 2003. The V(D)J recombination/DNA repair factor Artemis belongs to the metallo-beta-lactamase family and constitutes a critical developmental checkpoint of the lymphoid system. Ann. N. Y. Acad. Sci. 987:150-157. [DOI] [PubMed] [Google Scholar]
- 53.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2001. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science 294:2552-2556. [DOI] [PubMed] [Google Scholar]
- 54.PÂques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paques, F., G. F. Richard, and J. E. Haber. 2001. Expansions and contractions in 36-bp minisatellites by gene conversion in yeast. Genetics 158:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pastink, A., J. C. Eeken, and P. H. Lohman. 2001. Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res. 480-481:37-50. [DOI] [PubMed] [Google Scholar]
- 57.Paull, T. T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell 1:969-979. [DOI] [PubMed] [Google Scholar]
- 58.Paull, T. T., and M. Gellert. 2000. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl. Acad. Sci. USA 97:6409-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petrini, J. H. 2000. S-phase functions of the Mre11 complex. Cold Spring Harbor Symp. Quant. Biol. 65:405-411. [DOI] [PubMed] [Google Scholar]
- 60.Plessis, A., A. Perrin, J. E. Haber, and B. Dujon. 1992. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics 130:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raina, R., M. Schlappi, B. Karunanandaa, A. Elhofy, and N. Fedoroff. 1998. Concerted formation of macromolecular Suppressor-mutator transposition complexes. Proc. Natl. Acad. Sci. USA 95:8526-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramsden, D. A., and M. Gellert. 1998. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 17:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rattray, A. J., C. B. McGill, B. K. Shafer, and J. N. Strathern. 2001. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for. SAE2/COM1. Genetics 158:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rinehart, T. A., C. Dean, and C. F. Weil. 1997. Comparative analysis of non-random DNA repair following Ac transposon excision in maize and Arabidopsis. Plant J. 12:1419-1427. [DOI] [PubMed] [Google Scholar]
- 65.Rio, D. C., and G. M. Rubin. 1988. Identification and purification of a Drosophila protein that binds to the terminal 31-base-pair inverted repeats of the P transposable element. Proc. Natl. Acad. Sci. USA 85:8929-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rooney, S., F. W. Alt, D. Lombard, S. Whitlow, M. Eckersdorff, J. Fleming, S. Fugmann, D. O. Ferguson, D. G. Schatz, and J. Sekiguchi. 2003. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 197:543-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scott, L., D. LaFoe, and C. F. Weil. 1996. Adjacent sequences influence DNA repair accompanying transposon excision in maize. Genetics 142:237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shih, I. H., M. Melek, N. D. Jayaratne, and M. Gellert. 2002. Inverse transposition by the RAG1 and RAG2 proteins: role reversal of donor and target DNA. EMBO J. 21:6625-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staveley, B. E., T. R. Heslip, R. B. Hodgetts, and J. B. Bell. 1995. Protected P-element termini suggest a role for inverted-repeat-binding protein in transposase-induced gap repair in Drosophila melanogaster. Genetics 139:1321-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugawara, N., G. Ira, and J. E. Haber. 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsukamoto, Y., J. Kato, and H. Ikeda. 1996. Effects of mutations of RAD50, RAD51, RAD52, and related genes on illegitimate recombination in Saccharomyces cerevisiae. Genetics 142:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tuteja, R., and N. Tuteja. 2000. Ku autoantigen: a multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 35:1-33. [DOI] [PubMed] [Google Scholar]
- 73.Valencia, M., M. Bentele, M. B. Vaze, G. Herrmann, E. Kraus, S. E. Lee, P. Schar, and J. E. Haber. 2001. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414:666-669. [DOI] [PubMed] [Google Scholar]
- 74.van den Bosch, M., P. H. Lohman, and A. Pastink. 2002. DNA double-strand break repair by homologous recombination. Biol. Chem. 383:873-892. [DOI] [PubMed] [Google Scholar]
- 75.van Gent, D. C., J. F. McBlane, D. A. Ramsden, M. J. Sadofsky, J. E. Hesse, and M. Gellert. 1995. Initiation of V(D)J recombination in a cell-free system. Cell 81:925-934. [DOI] [PubMed] [Google Scholar]
- 76.Vergunst, A. C., and P. J. J. Hooykaas. 1999. Recombination in the plant genome and its application in biotechnology. Crit. Rev. Plant Sci. 18:1-31. [Google Scholar]
- 77.Wach, A., A. Brachat, R. Pohlmann, and P. Phillipsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 78.Walbot, V., and G. N. Rudenko. 2002. MuDR/Mu transposable elements of maize, p. 533-564. In N. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 79.Weil, C. F., and R. Kunze. 2000. Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nat. Genet. 26:187-190. [DOI] [PubMed] [Google Scholar]
- 80.Wilson, T. E. 2002. A genomics-based screen for yeast mutants with an altered recombination/end-joining repair ratio. Genetics 162:677-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilson, T. E., U. Grawunder, and M. R. Lieber. 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388:495-498. [DOI] [PubMed] [Google Scholar]
- 82.Wilson, T. E., and M. R. Lieber. 1999. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J. Biol. Chem. 274:23599-23609. [DOI] [PubMed] [Google Scholar]
- 82a.Xiao, Y. L., and T. Peterson. 2000. Intrachromosomal homologous recombination in Arabidopsis induced by a maize transposon. Mol. Gen. Genet. 263:22-29. [DOI] [PubMed] [Google Scholar]
- 83.Yan, X., I. M. Martinez-Ferez, S. Kavchok, and H. K. Dooner. 1999. Origination of Ds elements from Ac elements in maize: evidence for rare repair synthesis at the site of Ac excision. Genetics 152:1733-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83a.Yu, K., and M. R. Lieber. 2000. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol. 20:7914-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zickler, D., and N. Kleckner. 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33:603-754. [DOI] [PubMed] [Google Scholar]