Abstract
The Rho family GTPases Rac1, RhoA, and Cdc42 function as molecular switches that transduce intracellular signals regulating gene expression and cell proliferation as well as cell migration. p19Arf and p53, on the other hand, are tumor suppressors that act both independently and sequentially to regulate cell proliferation. To investigate the functional interaction and cooperativeness of Rho GTPases with the p19Arf-p53 pathway, we examined the contribution of Rho GTPases to the gene transcription and cell proliferation unleashed by deletion of p19Arf or p53 in primary mouse embryo fibroblasts. We found that (i) p19Arf or p53 deficiency led to a significant increase in PI 3-kinase activity, which in turn upregulated RhoA and Rac1 activities; (ii) deletion of p19Arf or p53 led to an increase in cell growth rate that was in part dependent on RhoA, Rac1, and Cdc42 activities; (iii) p19Arf or p53 deficiency caused an enhancement of the growth-related transcription factor NF-κB and cyclin D1 activities that are partly dependent on RhoA or Cdc42 but not on Rac1; (iv) forced expression of the activating mutants of Rac1, RhoA, or Cdc42 caused a hyperproliferative phenotype of the p19Arf−/− and p53−/− cells and promoted transformation of both cells; (v) RhoA appeared to contribute to p53-regulated cell proliferation by modulating cell cycle machinery, while hyperactivation of RhoA further suppressed a p53-independent apoptotic signal; and (vi) multiple pathways regulated by RhoA, including that of Rho-kinase, were required for RhoA to fully promote the transformation of p53−/− cells. Taken together, these results provide strong evidence indicating that signals through the Rho family GTPases can both contribute to cell growth regulation by p19Arf and p53 and cooperate with p19Arf or p53 deficiency to promote primary cell transformation.
Rho family small GTPases are molecular switches that transduce diverse intracellular signals leading to cell proliferation, gene induction, and survival as well as cytoskeleton remodeling (7, 46). Many mitogenic signals, including those from growth factor receptors and integrins, can promote the exchange of GDP for GTP on Rho GTPases (56), enabling them to interact with an array of effector targets to elicit specific cellular effects (4). Accumulating evidence has implicated Rho GTPases in many aspects of tumor development (5, 37). RhoA, Rac1, and Cdc42 are proto-oncogene products themselves that when hyperactivated can transform fibroblast cells (31-33). Activation of these Rho proteins can stimulate transcriptional activation of some of the critical genes involved in cell growth regulation, such as nuclear factor κB and cyclin D1 and leads to cell cycle progression (49-52). These Rho family members are required for Ras transformation (3, 17, 23, 58), and their deregulation correlates with poor cancer prognosis in some cases (37). Moreover, the Rho GTPases appear to be intimately associated with morphological changes of tumor cells and have been linked to tumor cell migration and invasion through their ability to regulate actin cytoskeleton, cellular-extracellular matrix adhesion, and cell-cell communication (7, 15, 41).
The p53 cell cycle inhibitor and its regulators, including p19ARF, are well-established tumor suppressors that are components of a complex signaling network central to tumor suppression (13, 18, 43). Deletion or mutation in p53 or its regulators occurs in many tumor cases and correlates with the onset of a wide spectrum of cancers. p53 is a key transcription factor essential for the response to cellular stress from DNA damage, hypoxia, and oncogene activation. When activated, p53 can trigger cell cycle arrest or apoptosis (2, 18), whereas p19ARF may serve as a sensor to oncogenic insult to stabilize p53 by sequestering Mdm2, a negative regulator of p53 activity (43). The p19ARF-p53 tumor suppressor pathway therefore is thought to be primarily involved in monitoring proliferation signals to prevent cells from uncontrolled growth (43). For example, it has been well documented that excess of mitogenic signals can turn on Ras, which in turn transiently stimulates p53 activity to induce cell cycle arrest, apoptosis, or senescence (8, 22, 42).
With appreciation of a central role of the p53 pathway in tumor suppression and the critical involvement of Rho GTPases in cell cycle progression, it seems logical to envision a functional connection and/or cooperation between the p53 pathway and Rho GTPase-mediated signaling processes in tumorigenesis. In particular, we are interested in determining the contribution of Rho family members to cell behaviors in a genetic background bearing defects of p53 or its regulators that might better represent that of tumor cells. Previously, we have shown that the p19Arf-p53 pathway negatively modulates PI 3-kinase and Rho GTPase activities and regulates actin cytoskeleton and cell migration through the PI 3-kinase-Rac GTPase signaling module (12). To investigate the potential contribution of Rho GTPases to p19Arf- and p53-mediated cell growth control, in the present study we have further characterized the relationship between the p19Arf-p53 tumor suppressor pathway and Rho proteins in regulating cell proliferation and gene expression. The possible cooperative nature of the p19Arf-p53 pathway defect with hyperactive Rho GTPases in inducing cell hyperproliferation and transformation was examined in p19Arf- or p53-null mouse embryo fibroblasts (MEFs). Moreover, we have examined the contribution of distinct effector pathways emanating from active RhoA to the transformation of p53−/− cells. Our findings strongly indicate that the Rho family GTPases, Rac1, RhoA, and Cdc42, contribute to cell growth regulation through p19Arf and p53 and that mitogenic activation of the Rho proteins may further cooperate with p19Arf or p53 deficiency to promote cell transformation.
MATERIALS AND METHODS
DNA constructs.
The Rac1, RhoA, and Cdc42 dominant negative mutants (Rac1N17, RhoAN19, and Cdc42N17), fast-cycling mutants (Rac1L28, RhoAL30, and Cdc42L28), and constitutively active mutants (Rac1L61, RhoAL63, and Cdc42L61) and the effector domain mutants of RhoA in the constitutively active backbone (RhoAL63-V39, RhoAL63-T40, RhoAL63-L40, and RhoAL63-C42) were generated by site-directed mutagenesis based on oligonucleotide-mediated PCR (19). For retroviral expression, cDNAs encoding the dominant negative, fast-cycling, and constitutively active forms of Rac1, RhoA, and Cdc42, the effector domain mutants of RhoA, and ROCKI were ligated into the BamHI and EcoRI sites in frame with the nucleotides encoding a three-hemagglutinin (HA3) tag at the 5′ end of the retroviral vector MIEG3 that expresses enhanced green fluorescent protein bicistronically. The constructs expressing p19Arf and p53 were described previously (57).
Cell culture and retroviral induction.
Primary wild type, p53−/− and p19Arf−/− MEFs were kind gifts from Martine Roussel (St. Jude, Memphis, Tenn.) that were derived from mouse embryos of the C57BL/6 × 129/sv genetic background (57) and were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 2 mM glutamine, 0.1 mM nonessential amino acids, 55 μM β-mercaptoethanol, and 10 μg of gentamicin/ml. Recombinant retroviruses were produced using the Phoenix cell packaging system (11, 12). Primary MEFs were infected with the respective retroviruses and harvested 48 to 72 h postinfection. The enhanced green fluorescent protein-positive cells were isolated by fluorescence-activated cell sorting (FACS).
Immunoblotting.
Whole-cell lysates were prepared by extraction of the MEF cells by the lysis buffer containing 20 mM Tris-HCl (pH 7.6), 100 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 0.2% sodium deoxycholate, 2 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, and 0.5 mM dithiothreitol for 30 min. The nuclear proteins were purified by the method described before (14). Briefly, cells were washed in a hypotonic buffer (HB; 25 mM Tris-HCl [pH 7.6], 1 mM MgCl2, 5 mM KCl) and lysed in HB containing 0.25% Nonidet P-40, 2 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, and 10 μg of aprotinin/ml for 30 min. The lysates were centrifuged at 500 × g for 5 min. The nuclear pellet was washed with HB containing 2 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, and 10 μg of aprotinin/ml, resuspended in a solution containing 20 mM Tris-HCl (pH 8.0), 0.42 M NaCl, 1.5 mM MgCl2, and 25% glycerol, vortexed, and incubated at 4°C for 30 min. The extracts were centrifuged at 900 × g for 5 min, and the supernatants were taken as the nuclear protein lysates. Protein contents in the whole-cell lysates and nuclear lysates were normalized by the Bradford method. The lysates were separated by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. The ectopic expression of the dominant negative, the fast-cycling, or the constitutively active forms of Rac1, RhoA, and Cdc42 were probed by using an anti-HA antibody (Boehringer Mannheim). NF-κB and cyclin D1 from the nuclear extracts were probed by using the anti-NF-κB p65 and anti-cyclin D1 antibodies (Santa Cruz Biotechnology), respectively.
Endogenous Rho GTPase activity assay.
Glutathione S-transferase (GST)-PAK1, GST-Rhotekin, and GST-WASP, which contain Rac1, RhoA, and Cdc42 interactive domains of PAK1, Rhotekin, and WASP, respectively, were used to probe the endogenous Rac1-GTP, RhoA-GTP, and Cdc42-GTP activities by the affinity precipitation method as previously described (20).
PI 3-kinase assay.
The endogenous PI 3-kinase activities of Arf−/−, p53−/−, and Arf−/− or p53−/− cells reconstituted with Arf, p53, or Rho protein mutants, respectively, were assayed according to a described protocol (12). Briefly, the cells were lysed in buffer A, containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.3 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 1% Triton X-100, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 0.1 mM sodium orthoranadate, and 25 mM NaF. After centrifugation at 4°C for 30 min at 14,000 rpm, the protein contents in the supernatant of cell lysates were measured by the Bradford method. Equal amounts of protein were incubated with anti-p85 polyclonal antibody coupled to protein A-agarose (Upstate Biotech., Inc.) overnight or subjected to anti-p85 Western blot analysis. The immunoprecipitates were washed twice with buffer C, containing 20 mM Tris-HCl (pH 8.0) 100 mM NaCl, and 10 mM MgCl2 and washed once with a kinase assay buffer (20 mM Tris-HCl [pH 7.6], 10 mM MgCl2). Five micrograms of sonicated phosphatidylinositol (PI) together with [γ-32P]ATP (200 μCi/ml) in 45 μl of the kinase assay buffer was incubated with the washed beads at 25°C for 10 min. The reactions were terminated by the addition of 100 μl of 1 N HCl. The reaction products were extracted by 200 μl of CHCl3-MeOH (1:1) and resolved on a thin-layer chromatographic silica plate coated with potassium oxalate in a solvent containing CHCl3-MeOH-4 M NH4OH (9:7:2). The PI kinase reactions were analyzed by autoradiography. The anti-p85 of PI 3-kinase antibody was obtained from Upstate Biotechnology, Inc., and the PI 3-kinase inhibitor wortmannin was obtained from Sigma.
Cell proliferation assay.
Cell growth rates were measured by a [3H]thymidine incorporation assay. Cells were cultured in a medium containing 2% serum for the assays. The cell cultures were assayed at 0, 1, 2, 3, and 4 days after the addition of 1 μCi of [3H]-thymidine/ml to the medium followed by an incubation for 4 h at 37°C. The cells were harvested by trypsinization, and the [3H]-thymidine incorporated into the cells was quantified by scintillation counting.
Luciferase reporter assay.
To detect endogenous NF-κB and cyclin D1 gene expression, the luciferase reporter constructs fused with the promoter of NF-κB or cyclin D1 (Stratagene) that contain the promoter response elements of NF-κB and cyclin D1 were used to report transiently the relative activities of NF-κB and cyclin D1. Transient transfection of these reporter plasmids into primary MEFs was carried out using LipofectAMINE reagents (Invitrogen) according to the manufacturer's protocols. Twenty-four hours prior to harvesting, the cells were switched to a medium containing 0.5% serum. Analysis of luciferase and β-galactosidase activities of the transfected cells was performed by using a luciferase assay kit (Promega). Transfection efficiencies were routinely corrected by obtaining the ratio of the luciferase and the β-galactosidase activities observed in the same sample, as previously described (26).
Cell cycle progression and apoptosis analysis.
Cell cycle progression of the p53−/− MEFs was monitored by cell cycle marker staining (propidium iodide labeling and Cytofix/Cytoperm fixation) followed by flow cytometry (phosphatidylethanolamine-conjugated anti-BrdU antibody) to examine whether dominant negative or constitutively active RhoA could affect the G1/S phase and/or G2/M phase transition in a p53-independent manner (11). The effects of RhoA mutants on p53-independent apoptotic response were tested by apoptosis staining of the p53−/− MEFs after DNA damage induction by gamma irradiation (20 Gy). The apoptotic cell population was revealed by 7AAD and allophycocyanin-conjugated Annexin V staining followed by flow cytometry analysis on a FACSCaliber machine (11).
Cell transformation assay.
To determine the transforming activity of the p53−/− or p19Arf−/− MEF cells, 5,000 cells that stably express various fast-cycling or constitutively active mutants of Rac1, RhoA, or Cdc42 were combined with 5 × 104 parental p53−/− or p19Arf−/− cells. The cell cultures were fed every 2 days with fresh culture medium. Fourteen days postplating, foci were scored after fixation and staining of the cells on the plates.
RESULTS
p19Arf and p53 regulate PI 3-kinase, which in turn regulates Rho GTPase activities.
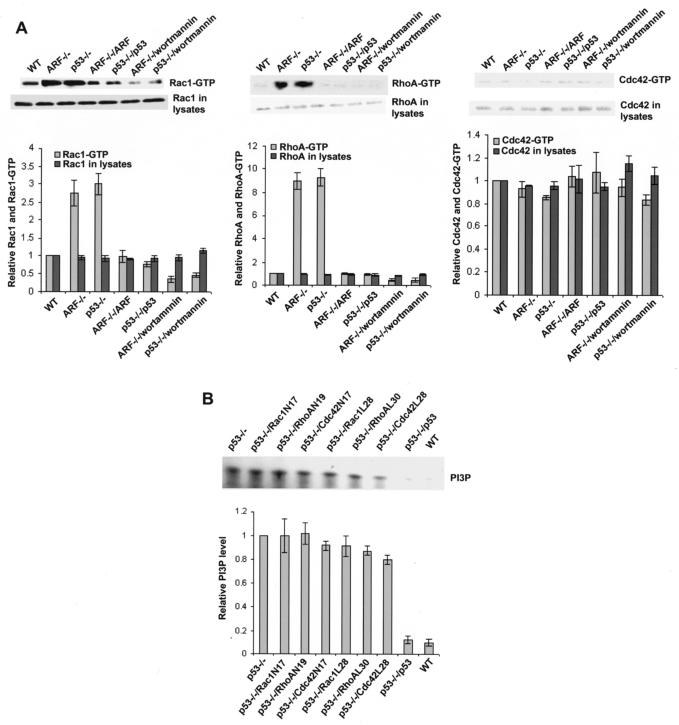
Previously, we found that the endogenous PI 3-kinase, RhoA, and Rac1 activities were elevated in the Arf−/− and p53−/− primary MEF cells and that the PI 3-kinase and Rac1 activities were required for a fast-migration phenotype of the Arf−/− and p53−/− cells. Reintroduction of the wild-type Arf or p53 gene into Arf−/− or p53−/− cells reversed the PI 3-kinase and Rho GTPase activities as well as the migration phenotype, indicating that p19Arf and p53 negatively regulate cell migration by suppression of PI 3-kinase and Rac1 activities (12). To further dissect the relationship between PI 3-kinase and Rho GTPases in the p19Arf−/− and p53−/− cells, we measured the Rac1, RhoA, and Cdc42 activities in the Arf−/− and p53−/− MEFs with or without the PI 3-kinase inhibitor (wortmannin) treatment. As shown in Fig. 1A, wortmannin (50 nM) treatment of the Arf−/− and p53−/− MEFs resulted in markedly decreased Rac1-GTP and RhoA-GTP species, to a level similar to that in the Arf- or p53-reconstituted cells or the wild-type cells (Fig. 1A). As we have observed previously, the Cdc42-GTP level was not significantly affected by the deletion of the Arf or p53 gene, nor was it changed upon wortmannin treatment or Arf or p53 reconstitution (Fig. 1A). On the other hand, the elevated endogenous PI 3-kinase activity in the p53−/− cells (compared with that of the wild-type MEFs) was not significantly altered by the expression of dominant negative Rac1N17, RhoAN19, or Cdc42N17 or by the fast-cycling Rac1L28, RhoAL30, or Cdc42L28 mutant, contrary to the effect of the reconstitution of wild-type p53 (Fig. 1B). Similarly, the dominant negative Rho protein mutants had no effect on the PI 3-kinase activity of the Arf−/− cells (data not shown). These results indicate that PI 3-kinase acts downstream of p19Arf or p53 but upstream of Rho proteins to regulate cell behaviors.
FIG. 1.
Activation of Rac1 and RhoA by the p19Arf or p53 defect is dependent on elevated PI 3-kinase activity. (A) The endogenous Rac1, RhoA, or Cdc42 activities in the Arf or p53 knockout and reconstituted cells with or without wortmannin (50 nM) treatment were assayed by using log phase cells that were serum starved for 12 h. The lysates were subject to GST-PAK1, GST-Rhotekin, or GST-WASP pull-down analysis. The amount of Rac1-GTP, RhoA-GTP, or Cdc42-GTP was detected by Western blotting of the respective glutathione-agarose coprecipitates with anti-Rac1, anti-RhoA, or anti-Cdc42 antibody and was normalized to that of Rac1, RhoA, or Cdc42 in wild-type (WT) MEFs. The results are shown as the means ± the standard deviations of three experiments. (B) The PI 3-kinase activities of the Arf or p53 knockout and reconstituted cells of the log phase cells were assayed. The cells were starved in a medium containing 0.5% serum for 12 h and harvested for anti-p85 immunoprecipitation. The PI 3-kinase activities in the immunoprecipitates were measured by an in vitro lipid kinase assay using exogenous PI as the substrate. The PI3P signals of various MEFs were normalized to those of the p53−/− MEFs in the quantification.
Rho GTPases contribute to the growth phenotype of p19Arf−/− and p53−/− cells.
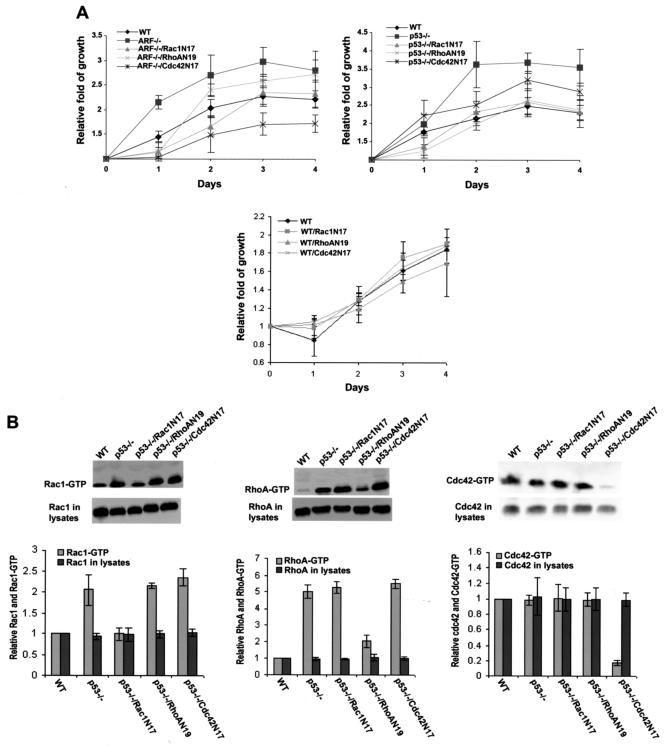
p19Arf and p53 are checkpoint molecules that upon overexpression can induce cell cycle arrest or apoptosis (43). Deletion of Arf or p53 resulted in a significant increase in cell growth rate and saturation density (Fig. 2). Since the Arf−/− and p53−/− cells contained elevated levels of active Rho GTPase species (Fig. 1A), we examined whether Rac1, RhoA, or Cdc42 might contribute to the proliferative phenotype of these cells. As shown in Fig. 2, the increased cell growth rates due to p19Arf or p53 deletion were partially inhibited by the dominant negative Rac1N17, RhoAN19, and Cdc42N17 mutants to various degrees under conditions at which the growth rate of wild-type MEFs was not significantly affected by these mutants. The expression levels of the dominant negative Rho mutants were comparable in these cells (12; data not shown), and the dominant negative inhibitory effects of Rac1N17, RhoAN19, and Cdc42N17 were specific toward the respective Rho proteins, as assayed by the effector domain pull-down assays (Fig. 2B). Although Cdc42 activity was not detectably upregulated by the Arf or p53 deficiency (Fig. 1A), the fact that Cdc42N17 could partially inhibit the growth phenotype of the Arf−/− and p53−/− cells suggests either that the effector probe (the p21-binding domain of WASP) used in the activity assay was not sensitive enough or that basal Cdc42 activity is required for the Arf or p53 defect-mediated growth. These results indicate that part of the growth stimulatory signals unleashed by p53 or p19Arf deletion are mediated through the Rho family members.
FIG. 2.
Rac1, RhoA, and Cdc42 contribute to the growth regulation of p19Arf- or p53-null cells. The dominant negative mutants of Rho proteins were expressed in the Arf−/− or p53−/− MEFs by retroviral induction, and the Rho mutant-expressing cells were isolated by FACS. (A) Five thousand cells/well of the indicated cells were plated in 1-ml culture medium containing 5% fetal bovine serum on 24-well plates. At the time points of day 0, 1, 2, 3, and 4, incorporation of [3H]thymidine into the cells was measured. Data are representative of three independent experiments and are expressed as the fold of growth relative to the respective value at day 0. Error bars represent the standard deviations of four repeats of one experiment. (B) Western blots of the endogenous Rac1, RhoA, and Cdc42 in the GTP-bound state were performed after the respective GST-effector domain pull-downs in the wild-type (WT) MEFs, p53-deficient MEFs, and p53-deficient MEFs expressing various dominant negative mutants of Rh proteins. The amounts of each Rho protein in the respective cell lysates and in the GTP-bound form were normalized to that of Rac1, RhoA, or Cdc42 in WT MEFs.
Involvement of Rho GTPases in NF-κB and cyclin D1 activation induced by p19Arf or p53 deficiency.
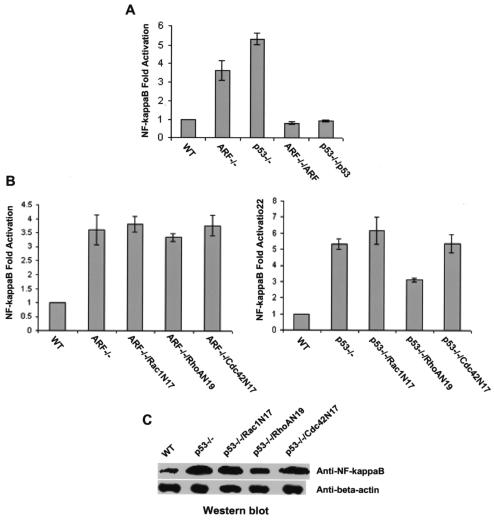
Transcription factor NF-κB has been shown to be functionally interconnected with p53 (36, 48). Since NF-κB is regulated by the Rho family GTPases and could mediate cell growth regulation by the Rho proteins (14, 29, 52), we determined the effect of p19Arf or p53 deletion on the activity and expression of NF-κB and examined the contribution of Rac1, RhoA, and Cdc42 to its modulation in the Arf- or p53-null background. The p19Arf−/− and p53−/− cells were transiently transfected with the NF-κB-luciferase reporter plasmid that contains the promoter response elements of NF-κB, and the relative luciferase activities were compared with that of wild-type MEFs and that of Arf−/− or p53−/− cells reconstituted with the respective tumor suppressor genes. The NF-κB transcriptional activity was upregulated by p19Arf and p53 deletions by ∼3.5- and ∼5.5-fold, respectively, and the observed activity changes were completely reversed when the Arf or p53 gene was reintroduced into the knockout cells (Fig. 3A). These results demonstrate that p19Arf and p53 negatively regulate NF-κB activity. To address whether the Rho proteins contribute to NF-κB regulation in Arf−/− and p53−/− cells, we transduced the dominant negative mutants of Rac1, RhoA, and Cdc42 (Rac1N17, RhoAN19, and Cdc42N17, respectively) as well as the GFP marker into the mutant cells and assayed the NF-κB reporter activities of these cells. Rac1N17 and Cdc42N17 had no detectable effect on the elevated reporter activities of NF-κB in Arf−/− and p53−/− cells (Fig. 3B), nor did they show significant inhibition of the increased nuclear expression level of NF-κB in these cells (Fig. 3C). In comparison, RhoAN19 attenuated the NF-κB activity and its nuclear expression level in p53-deficient cells but not in p19Arf-deficient cells (Fig. 3B and C). The dominant negative Rac1N17, RhoAN19, and Cdc42 N17 mutants were expressed similarly in these MEFs (12) and displayed various degrees of inhibition on the respective Rho protein activity and cell growth (Fig. 2). Moreover, Rac1N17 reversed the migration phenotype of these cells (14), indicating that these dominant negative Rho mutants were functionally expressed. These results suggest that RhoA participates in NF-κB regulation by p53 but that Rac1 and Cdc42 do not contribute to the upregulation of its activity, due to p19Arf or p53 defects.
FIG. 3.
Involvement of Rac1, RhoA, and Cdc42 in NF-κB activation induced by p19Arf or p53 deletion. The NF-κB promoter-driven luciferase reporter was transiently expressed in the indicated MEF cells together with a vector expressing Arf, p53, or dominant negative mutants of Rho GTPases (A and B). After a 30-hour recovery followed by a 12-hour starvation, the luciferase activities in the cells were measured. The luciferase activities were expressed as the fold of activation relative to the activity induced by the empty vector alone in the wild-type (WT) cells and were normalized to an internal transfection control (β-galactosidase coexpressed with pCMV vector). To detect NF-κB protein levels (C), nuclear extracts containing 20 μg of proteins were separated by 10% SDS-polyacrylamide gel electrophoresis and the amount of NF-κB present was probed by anti-NF-κB p65 antibody. Anti-beta actin blotting was carried out in parallel as a loading control.
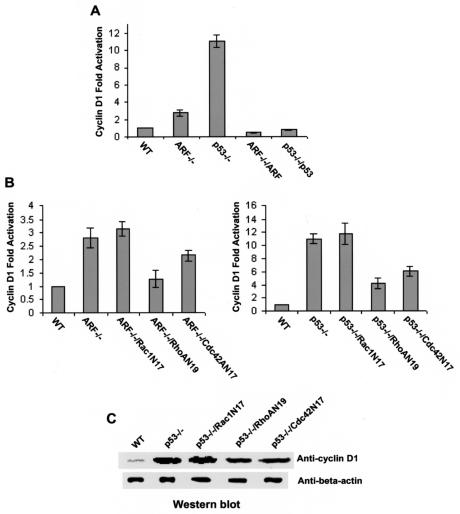
Cyclin D1 has been suggested as one of the key downstream effectors that mediate cell growth control by the Rho GTPases (49, 50, 51, 58). Deletion of p19Arf and p53 resulted in significant upregulation of cyclin D1 activity (∼3- and ∼11-fold, respectively) and nuclear expression (Fig. 4A and C). Reconstitution of p19Arf and p53 into the respective mutant cells readily reversed the cyclin D1 activity (Fig. 4A), indicating that both p19Arf and p53 negatively regulate its activity. Since Rac1, RhoA, and Cdc42 can each stimulate cyclin D1 activation and deletions of p19Arf and p53 result in activation of the Rho protein activities, we examined the possible involvement of the Rho GTPases in cyclin D1 regulation by Arf and p53. Transduction of dominant negative RhoA and Cdc42 in Arf−/− and p53−/− cells partially inhibited cyclin D1 at both the activity and the nuclear expression levels, whereas dominant negative Rac1 had no detectable effect (Fig. 4B and C). These results provide evidence that RhoA and Cdc42 but not Rac1 are involved in cyclin D1 regulation by p19Arf and p53. It is therefore possible that RhoA and Cdc42 work through NF-κB and/or cyclin D1 modulation to contribute to the growth phenotype of the p19Arf−/− and p53−/− cells, while Rac1 adopts a distinct, NF-κB- and cyclin D1-independent pathway to influence p19Arf−/− and p53−/− cell growth.
FIG. 4.
Contribution of Rac1, RhoA, and Cdc42 to cyclin D1 regulation in p19Arf- or p53-deficient MEF cells. One microgram of cyclin D1-luciferase reporter plasmid was cotransfected with a vector expressing Arf, p53, or the dominant negative mutants of the Rho proteins into the indicated cells. The luciferase activities in the cell lysates were measured to determine the relative cyclin D1 transcriptional activities (A and B) and were normalized to those of a β-galactosidase transfection control. To directly compare the protein levels of cyclin D1, nuclear extracts containing 20 μg of proteins were separated by 10% SDS-polyacrylamide gel electrophoresis and the amount of cyclin D1 was probed with an anti-cyclin D1 antibody (C). Anti-beta actin blotting was done in parallel. WT, wild-type.
Active Rho GTPases cooperate with p19Arf or p53 deletion to promote hyperproliferation and transformation.
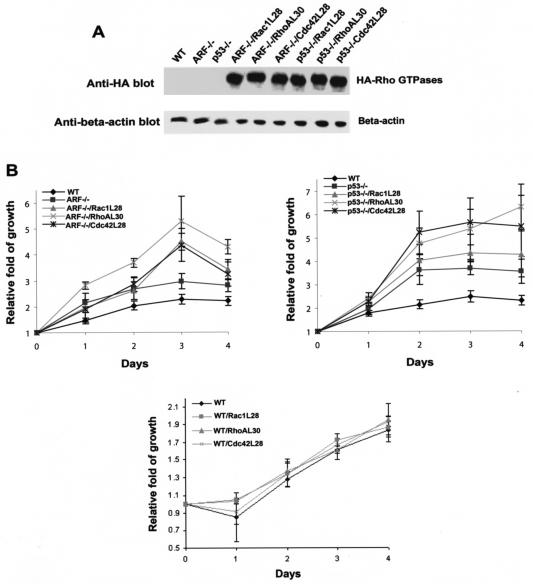
Since Rac1, RhoA, and Cdc42 appear to contribute to the growth regulation of p19Arf−/− and p53−/− cells, we next asked if hyperactive Rho proteins could further stimulate the proliferation of these cells. For this purpose, we introduced a set of fast-cycling mutants of Rac1, RhoA, and Cdc42 (Rac1L28, RhoAL30, and Cdc42L28, respectively), which possess an increased intrinsic rate of exchange of GDP by GTP-mimicking mitogenic stimulation (21, 53), into the p19Arf−/− or p53−/− cells. The expression of Rac1L28, RhoAL30, and Cdc42L28 were confirmed by Western blot analysis (Fig. 5A). As shown in Fig. 5B, these fast-cycling Rho GTPase mutants were able to further enhance the growth rates of p19Arf−/− and p53−/− cells to various extents under conditions in which they had only minor effects on wild-type MEF growth, suggesting that active Rho proteins can cooperate with p19Arf and p53 defects to promote hyperproliferation of the cells.
FIG. 5.
The active Rac1, RhoA, and Cdc42 mutants can stimulate hyperproliferation of p19Arf- or p53-null cells. (A) Expression of the fast-cycling mutants of Rac1, RhoA, and Cdc42 that carry an N-terminal HA tag in p19Arf- or p53-null cells was probed by anti-HA Western blotting. (B) The indicated cells were plated in a culture medium containing 5% fetal bovine serum. At the indicated time points, the amount of incorporated [3H]thymidine was quantified by scintillation counting. The data are representative of three independent experiments and are presented as the fold of growth relative to the respective cells at day 0. Error bars represent the standard deviations of four repeats. WT, wild type.
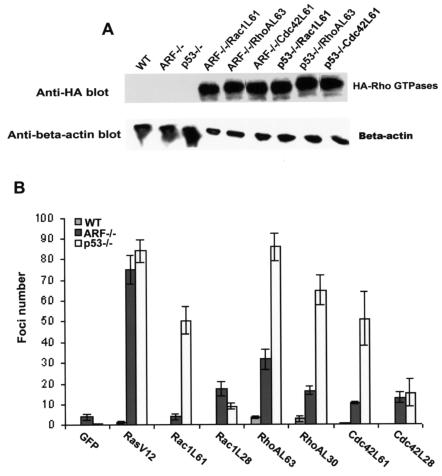
Oncogenic Ras was able to cooperate with p19Arf or p53 deletion to promote MEF transformation in vitro and in vivo (16). Because Rac1, RhoA, and Cdc42 are all important downstream mediators of Ras signaling (17, 31-33), we wondered if the hyperactive Rho proteins could be sufficient to promote Arf−/− and p53−/− cell transformation. For this purpose, in addition to the fast-cycling Rho protein mutants, we generated Arf−/− and p53−/− primary MEFs expressing the constitutively active forms of Rac1, RhoA, and Cdc42 (Rac1L61, RhoAL63, and Cdc42L61, respectively). These mutants expressed equally well in the p53−/− cells (Fig. 6A) and in the p19Arf−/− cells (data not shown). Both forms of the activating Rac1, RhoA, and Cdc42 mutants, i.e., the fast-cycling and the constitutively active forms, displayed various foci-forming activity in p53−/− MEFs as well as in p19Arf−/− MEFs (Fig. 6B). Some of these mutants, e.g., RhoAL63, displayed activities as potent as that of oncogenic Ras, while others, such as Rac1L61, was only weakly transforming (Fig. 6B). Thus, like oncogenic Ras, active Rac1, RhoA, or Cdc42 can cooperate with p53 or p19Arf deletion to promote primary cell transformation.
FIG. 6.
The active Rac1, RhoA, and Cdc42 mutants cooperate with p19Arf or p53 deletion to promote cell transformation. (A) Expression of the constitutively active mutants of Rac1, RhoA, and Cdc42 containing an N-terminal HA tag in p19Arf- or p53-null cells was probed by anti-HA Western blotting. (B) Five thousand MEF cells expressing the indicated proteins were mixed with 5 × 104 parental p19Arf−/− or p53−/− cells and cultured in 100-mm plates. The cell cultures were fed every 2 days with fresh culture medium. Fourteen days postplating, the foci were fixed, stained, and quantified under a microscope. The data are representative of two independent experiments. WT, wild type.
RhoA signaling modulates cell cycle progression and apoptotic response of p53-null cells.
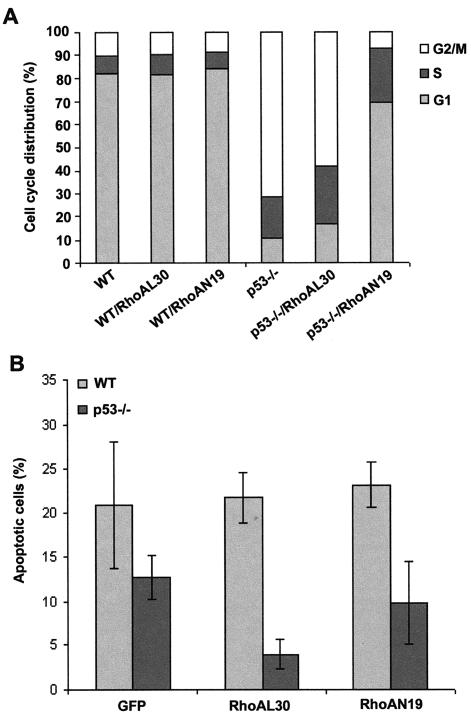
To further address the role of Rho in p53-mediated cell proliferation, we carried out a set of experiments comparing the cell cycle and apoptotic properties of p53−/− MEFs expressing the fast-cycling active mutant of RhoA or dominant negative mutant of RhoA. As shown in Fig. 7A, when the cell cycle progression of wild-type and p53−/− MEFs was analyzed by PI staining followed by FACS, the dominant negative RhoA mutant was found to effectively extend the G1 phase and suppress the G2/M phase of p53−/− MEFs that were altered due to a p53 defect, whereas the active RhoA mutant did not significantly alter the relative phases of the cell cycle. Moreover, p53 deficiency led to a decrease in the cellular apoptotic response to gamma irradiation compared with that of wild-type cells, which could be further suppressed by the expression of the active RhoA mutant (Fig. 7B). These results indicate that RhoA may contribute to p53-regulated cell proliferation by modulating cell cycle progression and that RhoA activation may cause hyperproliferation of p53−/− cells by further suppressing a p53-independent apoptotic signal.
FIG. 7.
Dominant negative RhoA affects cell cycle progression of p53−/− MEFs, while active RhoA suppresses gamma irradiation-induced apoptosis. (A) p53−/− cells transduced with GFP alone or with retrovirus expressing RhoAL30 or RhoAN19 were analyzed by FACS for cell cycle progression after PI staining in a culture medium containing 5% fetal bovine serum. The data are representative of the results of two independent experiments. (B) Various retroviral transduced MEF cells were analyzed by FACS for apoptotic cell population after gamma irradiation (20 Gy). Wild-type (WT) MEFs transduced with GFP alone were compared in parallel.
Multiple pathways regulated by RhoA are involved in promoting transformation of p53−/− MEFs.
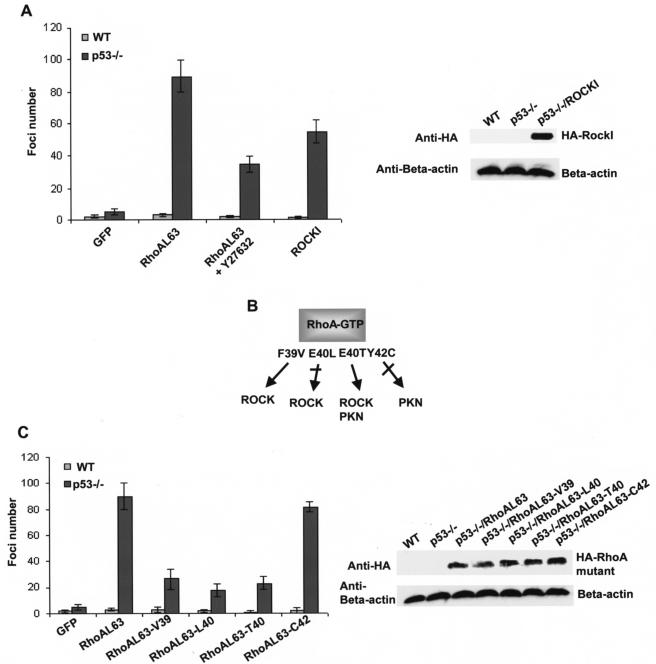
To begin to unveil the molecular pathways regulated by Rho GTPases that are important for promoting transformation of p53−/− cells, we next examined the involvement of a key effector of RhoA, ROCK, in RhoA-mediated transformation. As shown in Fig. 8A, treatment of the RhoAL63-expressing p53−/− MEFs with a ROCK inhibitor, Y27632, led to a partial inhibition of the foci-forming activity elicited by RhoAL63, and ectopic expression of ROCKI in p53−/− cells was able to only partially recapitulate the transforming activity. To confirm the contribution of ROCK to RhoA-mediated transformation and to assess the involvement of additional effector pathways downstream of RhoA, we further tested a set of effector-domain mutants of RhoA for their ability to induce transformation of p53−/− primary MEFs. As previously characterized in vitro (10, 38) and depicted in Fig. 8B, RhoA-F39V is defective in PKN binding but retains ROCK binding, RhoA-E40L is defective for ROCK recognition, RhoA-E40T retains ROCK and PKN binding but is defective in kinectin and mDia binding, and RhoA-Y42C is selectively defective in PKN binding. Consistent with the partial loss of transforming activity in the case of ROCK inhibitor-treated cells, RhoAL63-L40 that is defective in ROCK binding suffered partial loss of foci-forming activity (Fig. 8C). Since ROCK was found to be significantly upregulated in the mRNA level in the RhoA-transduced p53−/− cells in a gene array assay (unpublished data), these results strongly suggest that the RhoA-ROCK pathway is required for the RhoA-induced transformation of primary p53−/− MEFs, corroborating previous findings with NIH 3T3 cells (39). Interestingly, the RhoA mutant that retained ROCK and PKN binding but was defective for kinectin and mDia and possibly other effector pathways (RhoAL63-E40T) was partially active in transforming p53−/− cells, whereas the RhoA mutants that have lost PKN-binding ability (RhoA-F39V and RhoA-Y42C) remained partially or fully active (Fig. 8C). Therefore, additional effector pathways other than ROCK that emanated from active RhoA but not PKN appear to also be involved in the RhoA-mediated transformation of p53−/− cells. These results provide clues on the contribution of RhoA-regulated signaling cascades to the transformation phenotype of a primary cell system.
FIG. 8.
Multiple effector pathways regulated by RhoA contribute to the transformation phenotype of p53−/− MEF cells. (A) Involvement of ROCK in the RhoAL63-elicited transformation of p53−/− MEFs. Colony-forming activities of p53−/− MEFs transduced with the RhoAL63 mutant or ROCK1 in the presence or absence of Y27632 (20 μM) were determined 14 days postplating. (B) Effector domain mutants of RhoA allow selective uncoupling of RhoA with ROCK, PKN, or other effectors. Single-point mutants made in the switch I domain of RhoA impair or retain specific effector binding as depicted. (C) The effect of effector domain mutations on active RhoA-induced transformation of p53−/− cells. Expression of the respective RhoAL63 mutants in the p53−/− cells was detected by anti-HA Western blot. The data shown are representative of two independent experiments. WT, wild type.
DISCUSSION
In the present study, we demonstrate that Rho family GTPases Rac1, RhoA, and Cdc42 contribute to p19Arf- and p53-regulated gene transcription and cell growth and that activation of these Rho GTPases can cooperate with p19Arf or p53 defects to promote hyperproliferation and transformation. The contributions of RhoA and Cdc42 to the growth phenotype of p19Arf- and p53-deficient cells may come in part through modulation of the transcriptional activities of a few key cell growth regulators, including the transcription factor NF-κB and the cell division kinase regulator cyclin D1, whereas Rac1 appears to be involved in p19Arf- p53 mediated cell growth independently of NF-κB and cyclin D1. Significantly, we show that RhoA is involved in p53-regulated cell proliferation by modulating the cell cycle and a p53-independent apoptotic signal and that multiple RhoA-regulated pathways, including that of ROCK, appear to be important for promoting transformation of p53−/− cells. Although more detailed mechanisms of the connection and cooperativeness between the Rho proteins and the p19Arf-p53 tumor suppressor pathway remain to be explored, these results help establish an important functional relationship of Rho GTPases with the p19Arf-p53 pathway, defects of which occur in many cases of human cancer (13, 43). The findings may have important implications for strategies that target Rho proteins in anticancer therapy.
Previous studies have shown by genetic disruption of the tumor suppressor genes that p19ARF and p53 but not p27Kip1 or pRb have a profound negative effect on cell motility and migration (12). p19ARF likely depends on p53 for cell migration regulation, and a specific transcriptional activity or specific target genes controlled by p53 may serve as the link between p53 and actin cytoskeleton to promote the migration phenotype. In particular, we found that the endogenous PI 3-kinase and Rho GTPase activities were significantly elevated in Arf−/− and p53−/− cells and that both PI 3-kinase and Rac1 were required for the p19Arf- and p53-regulated migration (12). In the present study, we further determined the relationship between PI 3-kinase and Rac1/RhoA activities in p19Arf- and p53-deficient MEFs and revealed that activation of Rac1/RhoA by a p19Arf or p53 defect depended on the elevated PI 3-kinase activity and did not occur if PI 3-kinase activity was not elevated. Therefore, a pathway initiated from p19Arf or p53 deficiency could lead to PI 3-kinase activation, which in turn activates Rac1 and RhoA (Fig. 9). We are currently examining the candidate genes whose expression profiles are altered by p19ARF and p53 deficiencies in an effort to identify the molecule(s) responsible for PI 3-kinase and Rho GTPase activation. One possibility is that the gene(s) controlled by p53 (55) provides an autocrine mechanism that feeds back to stimulate the cells leading to PI 3-kinase activation and subsequently to Rho GTPase activation.
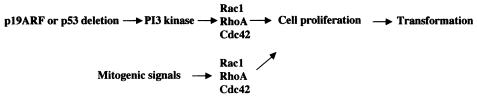
FIG. 9.
A model depicting the relationship between the p19Arf-p53 tumor suppressor pathway and the Rho GTPase signaling module in the regulation of cell proliferation and transformation. In addition to malfunction in checkpoint and apoptosis control, deficiency of p19Arfand/or p53 may alter transcriptional balance and promote cell growth by upregulating PI 3-kinase and Rho GTPase activities. Mitogenic signals that cause activation of RhoA, Rac1, or Cdc42 could further modulate cell cycle and apoptotic machineries and cooperate with p53 deficiency to promote cell hyperproliferation and transformation.
Aberrant activation of Rho GTPases can promote cell hyperproliferation and growth transformation (3, 5, 25, 58). The mechanism of Rho protein-stimulated cell growth leading to transformation appears to be at least twofold: activation of cell cycle promoting regulators such as cyclin D1 (49, 50) and inhibition of negative regulators of cell cycle progression such as p21Cip1 and p27Kip1 (28, 47). We found that Arf and p53 deletion caused a markedly increased cyclin D1 activity which is in part dependent on RhoA and Cdc42, suggesting that these Rho proteins contribute to the growth phenotype of the Arf−/− and p53−/− cells by transducing signals to stimulate cyclin D1 activity. Surprisingly, Rac1 does not appear to be involved in the cyclin D1 regulation by p19Arf or p53, but it must employ another mechanism to affect the proliferation of these cells, because Rac1 is required for the increased proliferation of the Arf−/− and p53−/− cells.
A number of mechanisms have been proposed for cyclin D1 activation and growth regulation by Rho GTPases. Rac1 and RhoA can activate NF-κB, which in turn activates cyclin D1 (14, 29). Rac1 and Cdc42 might promote extracellular signal-regulated kinase 1 and 2 activation by means of PAKs, which can directly phosphorylate and activate Raf and MEK (51). In addition, Rac1-mediated activation of c-Jun NH2-terminal kinase and p38 mitogen-activated protein kinase cascades can lead to increased phosphorylation and activity of the AP-1 components Jun and ATF (58), and the recently identified Cdc42-Par6 atypical protein kinase C pathway may also stimulate NF-κB, causing cyclin D1 activation (7). We have observed that RhoA but not Rac1 or Cdc42 plays a role in NF-κB upregulation by Arf or p53 deletion, raising the possibility that the contribution of RhoA to the growth phenotype of the cells may be due partly to activation of NF-κB, which in turn stimulates cyclin D1. The differential regulation of NF-κB and cyclin D1 activities by RhoA, Rac1, and Cdc42 in the p19Arf−/− and p53−/− cells suggests that each member of Rho family GTPases may utilize a distinct mechanism to contribute to the growth phenotype. The finding that Rac1 does not appear to contribute to the p19Arf- or p53-mediated NF-κB or cyclin D1 regulation is somewhat surprising, since previous work by a number of laboratories using cloned fibroblast cell lines have implicated Rac1 as one important regulator of NF-κB and cyclin D1 activity (29, 50, 52). It is likely that the involvement of Rac1 in these transcription processes is cell context and pathway specific. Furthermore, our observations that individual Rho proteins may be involved differentially in p19Arf- and p53-regulated gene induction and cell proliferation add further evidence that p19Arf and p53 have overlapping and interdependent as well as independent functions in the regulation of cell growth (24, 34, 43).
Whether the Rho proteins, Rac1 and Cdc42 in particular, can further regulate cell growth through suppression of cell cycle inhibitors such as p21Cip1 or p27Kip1 in the Arf−/− and p53−/− background remains to be examined further. It is clear, however, that RhoA could have an impact on cell proliferation in both a p53-dependent and a p53-independent manner through modulation of cell cycle and apoptotic machineries, given our observation that the dominant negative RhoA mutant could effectively extend the G1 phase and suppress the G2/M phase of p53−/− MEFs, while the active RhoA mutant could suppress the gamma irradiation-induced apoptosis independently of p53.
Constitutively active or fast-cycling Rho GTPase mutants, as well as many of their GEFs, can induce foci formation or anchorage-independent growth of NIH 3T3 cells (21, 35, 52, 56, 58). We found that the activating mutants of Rac1, RhoA, and Cdc42 can induce a hyperproliferative and transforming phenotype in both the Arf−/− and p53−/− primary MEFs to an extent similar to that caused by oncogenic Ras in certain cases (e.g., active RhoA) (Fig. 6). Since a number of downstream effectors of Ras other than the Rho proteins, including PI 3-kinase and Raf, combine to contribute to the transforming phenotype of oncogenic Ras (3), the potency of some of the Rho members suggests that they may turn on distinct signaling components from Ras or may be more efficient in turning on the same set of growth-promoting factors as Ras in inducing cell transformation. In this regard, we have tested the latter possibility, that further enhancement of NF-κB and cyclin D1 activities by the active Rho mutants could be responsible for the efficient induction of transformation in the Arf−/− and p53−/− cells. We found that both the luciferase reporter activities and the nuclear expression levels of NF-κB and cyclin D1 remain unchanged with or without the expression of the active Rho protein mutants (data not shown). It is therefore possible that the active Rho GTPases stimulate additional factors that cooperate with the p53 pathway defects to promote cell transformation. It is also worth noting that the two different forms of active Rho GTPases, i.e., the fast-cycling and constitutively GTP-bound forms, may have distinct transcription profiles, as we have observed in a gene array study (unpublished data), which could help explain the quantitative differences of these mutants in promoting p19Arf−/− or p53−/− MEF transformation.
In an attempt to further dissect the requirement of RhoA downstream effectors for the transforming phenotype of active RhoA, we utilized ROCKI and the ROCK inhibitor, Y27632, as well as a set of the effector domain mutants of RhoA that impair or retain coupling with specific effectors in the transformation assay. Our results confirm an important role of the ROCK pathway in RhoA-mediated transformation of p53−/− cells, while also implying that multiple effectors downstream of RhoA may collaborate to allow the optimal effect of RhoA.
Our results present a seemingly paradoxical relationship between Rho GTPases and the p53 tumor suppressor pathway. On the one hand, basal activities of RhoA, Rac1, and Cdc42 are all part of the required proliferative signals of the p19Arf- or p53-regulated networks. On the other hand, hyperactive Rho proteins can cooperate with p19Arf or p53 deletion or mutation to promote transformation, suggesting that events leading to the activation of Rho family GTPases may constitute second hits in tumor induction (Fig. 9). The later synergism between the Rho GTPases and the p53 pathway further indicates that signaling components of the Rho GTPases are capable of delivering quantitatively different and/or additional inputs to cell proliferation regulation by the p53 pathway. Given the intertwining, complex nature of cell growth regulatory mechanisms of the p53 pathway and Rho proteins, one remaining challenge would be to delineate which of the known or unknown pathways controlled by p19Arf-p53 and each Rho GTPase might cooperate to promote cell hyperproliferation and transformation. Rho family GTPases have been implicated in many aspects of tumor development. Overexpression, upregulation, or rearrangement of RhoA, RhoC, Rac1, Rac2, Rac3, Cdc42, and RhoH have been detected in human tumors ranging from colon, breast, lung, and myeloma to head and neck squamous-cell carcinoma (6, 9, 26, 30, 40, 45). Due to their recognized roles in Ras transformation (58), growth factor and integrin signaling (41), cell cycle control (27), apoptosis (1, 44), and invasion and metastasis (54), Rho GTPases have been proposed as potential anticancer therapeutic targets (37). Interestingly, to date no activating mutations like those found in oncogenic ras have been discovered in Rho family members, suggesting that Rho proteins might primarily serve as signaling links from upstream mitogenic signals to play a modifier role in tumor induction. Our studies demonstrating a role of Rho proteins in p19Arf- and p53-controlled cell growth and migration and a synergistic effect of active RhoA, Rac1, and Cdc42 with p19Arf or p53 deletion on cell transformation further strengthen the view that these Rho GTPases may contribute to and/or cooperate with p53 deficiency in tumorigenesis and tumor progression. It remains to be seen if upregulation of Rho GTPase activities in the p19Arf−/− or p53−/− genetic background could result in a shortened latency in tumorigenesis in an animal model. Should that be the case, targeting individual Rho GTPases would be a worthy endeavor among future anticancer strategies.
Acknowledgments
We thank members of the Zheng laboratory staff for help with retroviral production and cDNA constructs and Martine Roussel (St. Jude Children's Research Hospital) for the kind gifts of MEFs and Arf cDNA.
This work was supported in part by grants from the National Institutes of Health (grants GM60523 and GM53943) and the Department of Defense Breast Cancer Program (grant BC990290) (to Y.Z.).
REFERENCES
- 1.Aznar, S., and J. C. Lacal. 2001. Rho signals to cell growth and apoptosis. Cancer Lett. 165:1-10. [DOI] [PubMed] [Google Scholar]
- 2.Balint, E., and K. H. Vousden. 2001. Activation and activities of the p53 tumour suppressor protein. Br. J. Cancer 85:1813-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar-Sagi, B., and A. Hall. 2000. Ras and Rho GTPases: a family reunion. Cell 103:227-238. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, A. L., and A. Hall. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241-255. [PMC free article] [PubMed] [Google Scholar]
- 5.Boettner, B., and L. Van Aelst. 2002. The role of Rho GTPases in disease development. Gene 286:155-174. [DOI] [PubMed] [Google Scholar]
- 6.Clark, E. A., T. R. Golub, E. S. Lander, and R. O. Hynes. 2000. Genomic analysis of metastasis reveals an essential role for RhoC. Nature 406:532-535. [DOI] [PubMed] [Google Scholar]
- 7.Etienne-Manneville, S., and A. Hall. 2002. Rho GTPases in cell biology. Nature 420:629-635. [DOI] [PubMed] [Google Scholar]
- 8.Ferbeyre, G., E. de Stanchina, A. W. Lin, E. Querido, M. E. McCurrach, G. J. Hannon, and S. W. Lowe. 2002. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol. Cell. Biol. 22:3497-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fritz, G., I. Just, and B. Kaina. 1999. Rho GTPases are over-expressed in human tumors. Int. J. Cancer 81:682-687. [DOI] [PubMed] [Google Scholar]
- 10.Fujisawa, K., P. Madaule, T. Ishizaki, G. Watanabe, H. Bito, Y. Saito, A. Hall, and S. Narumiya. 1998. Different regions of Rho determine Rho-selective binding of different classes of Rho target molecules. J. Biol. Chem. 273:18943-18949. [DOI] [PubMed] [Google Scholar]
- 11.Gu, Y., M-D. Filippi, J. E. Siefring, E. P. Williams, A. Jasti, R, Prabhakar, D. J. Kwiatkowski, and D. A. Williams. The highly related Rho GTPases, Rac1 and Rac2, separately control hematopoietic cell survival and cycle progression, but together regulate adhesion and migration. Science, in press.
- 12.Guo, F. K., Y. Gao, L. Wang, and Y. Zheng. 2003. p19Arf-p53 tumor suppressor pathway regulates cell motility by suppression of phosphoinositide 3-kinase and Rac1 GTPase activities. J. Biol. Chem. 278:14414-14419. [DOI] [PubMed] [Google Scholar]
- 13.Hahn, W. C., and R. A. Weinberg. 2002. Modelling the molecular circuitry of cancer. Nat. Rev. Cancer 2:331-341. [DOI] [PubMed] [Google Scholar]
- 14.Joyce, D., B. Bouzahzah, M. Fu, C. Albanese, M. D'Amico, J. Steer, J. U. Klein, R. J. Lee, J. E. Segall, J. K. Westwick, C. J. Der, and R. G. Pestell. 1999. Integration of Rac-dependent regulation of cyclin D1 transcription through a nuclear factor-B-dependent pathway. J. Biol. Chem. 274:25245-25249. [DOI] [PubMed] [Google Scholar]
- 15.Kaibuchi, K., S. Kuroda, M. Fukata, and M. Nakagawa. 1999. Regulation of cadherin-mediated cell-cell adhesion by the Rho family GTPases. Curr. Opin. Cell Biol. 11:591-596. [DOI] [PubMed] [Google Scholar]
- 16.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 17.Khosravi-Far, R., P. A. Solski, G. J. Clark, M. S. Kinch, and C. J. Der. 1995. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol. Cell. Biol. 15:6443-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 19.Li, R., B. Debreceni, B. Jia, Y. Gao, G. Tigyi, and Y. Zheng. 1999. Localization of the PAK1-, WASP-, and IQGAP1-specifying regions of the small GTPase Cdc42. J. Biol. Chem. 274:29648-29654. [DOI] [PubMed] [Google Scholar]
- 20.Liliental, J., S. Y. Moon, R. Lesche, R. Mamillapalli, N. Gavrilova, Y. Zheng, H. Sun, and H. Wu. 2000. Genetic deletion of the PTEN tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr. Biol. 10:401-404. [DOI] [PubMed] [Google Scholar]
- 21.Lin, R., R. A. Cerione, and D. Manor. 1999. Specific contributions of the small GTPases Rho, Rac, and Cdc42 to Dbl transformation. J. Biol. Chem. 274:23633-23641. [DOI] [PubMed] [Google Scholar]
- 22.Ma, P., M. Magut, X. Chen, and C.-Y. Chen. 2002. p53 is necessary for the apoptotic response mediated by a transient increase of Ras activity. Mol. Cell. Biol. 22:2928-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malliri, A., R. A. van der Kammen, K. Clark, M. van der Valk, F. Michiels, and J. G. Collard. 2002. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature 417:867-871. [DOI] [PubMed] [Google Scholar]
- 24.McKeller, R. N., J. L. Fowler, J. J. Cunningham, N. Warner, R. J. Smeyne, F. Zindy, and S. X. Skapek. 2002. The Arf tumor suppressor gene promotes hyaloid vascular regression during mouse eye development. Proc. Natl. Acad. Sci. USA 99:3848-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mira, J. P., V. Benard, J. Groffen, L. C. Sanders, and U. G. Knaus. 2000. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by p21-activated kinase-dependent pathway. Proc. Natl. Acad. Sci. USA 97:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montaner, S., R. Perona, L. Saniger, and J. C. Lacal. 1998. Multiple signaling pathways lead to the activation of the nuclear factor κB by the Rho family of GTPases. J. Biol. Chem. 273:12779-12785. [DOI] [PubMed] [Google Scholar]
- 27.Olson, M. F., A. Ashworth, and A. Hall. 1995. An essential role for rho, rac, and cdc42 GTPases in cell cycle progression through G1. Science 269:1270-1272. [DOI] [PubMed] [Google Scholar]
- 28.Olson, M. J., H. F. Paterson, and C. J. Marshall. 1998. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature 394:295-299. [DOI] [PubMed] [Google Scholar]
- 29.Perona, R., S. Montaner, L. Saniger, P. I. Sanchez, R. Bravo, and J. C. Lacal. 1997. Activation of the nuclear factor-κB by Rho, Rac, and Cdc42 proteins. Genes Dev. 11:463-475. [DOI] [PubMed] [Google Scholar]
- 30.Preudhomme, C., C. Roumier, M. P. Hildebrand, E. Dallery-Prudhomme, D. Lantoine, J. L. Lai, A. Daudignon, C. Adenis, F. Bauters, P. Fenaux, J. P. Kerckaert, and S. Galiegue-Zouitina. 2000. Nonrandom 4p13 rearrangements of the RhoH/TTF gene, encoding a GTP-binding protein, in non-Hodgkin's lymphoma and multiple myeloma. Oncogene 19:2023-2032. [DOI] [PubMed] [Google Scholar]
- 31.Qiu, R., J. Chen, D. Kirn, F. McCormick, and M. Symons. 1995. An essential role for Rac in Ras transformation. Nature 374:457-459. [DOI] [PubMed] [Google Scholar]
- 32.Qiu, R., J. Chen, F. McCormick, and M. Symons. 1995. A role for Rho in Ras transformation. Proc. Natl. Acad. Sci. USA 92:11781-11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu, R., A. Abo, F. McCormick, and M. Symons. 1997. Cdc42 regulates anchorage-independent growth and is necessary for Ras transformation. Mol. Cell. Biol. 17:3449-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocha, S., K. J. Campbell, and N. D. Perkins. 2003. p53- and Mdm2-independent repression of NF-κB transactivation by the ARF tumor suppressor. Mol. Cell 12:15-25. [DOI] [PubMed] [Google Scholar]
- 35.Roux, P., C. Gauthier-Rouviere, S. Doucet-Brutin, and P. Fort. 1997. The small GTPases Cdc42Hs, Rac1 and RhoG delineate Raf-independent pathways that cooperate to transform NIH3T3 cells. Curr. Biol. 7:629-637. [DOI] [PubMed] [Google Scholar]
- 36.Ryan, K. M., M. K. Ernst, N. R. Rice, and K. H. Vousden. 2000. Role of NF-κB in p53-mediated programmed cell death. Nature 404:892-897. [DOI] [PubMed] [Google Scholar]
- 37.Sahai, E., and C. J. Marshall. 2002. Rho-GTPases and cancer. Nat. Rev. Cancer 2:133-142. [DOI] [PubMed] [Google Scholar]
- 38.Sahai, E., A. S. Alberts, and R. Treisman. 1998. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 17:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahia, E., T. Ishizaki, S. Narumiya, and R. Treisman. 1999. Transformation mediated by RhoA requires activity of ROCK kinases. Curr. Biol. 9:136-145. [DOI] [PubMed] [Google Scholar]
- 40.Schnelzer, A., D. Prechtel, U. Knaus, K. Dehne, M. Gerhard, H. Graeff, N. Harbeck, M. Schmitt, and E. Lengyel. 2000. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform. Oncogene 19:3013-3020. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz, M. A., and S. J. Shattil. 2000. Signaling networks linking integrins and rho family GTPases. Trends Biochem. Sci. 25:388-391. [DOI] [PubMed] [Google Scholar]
- 42.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 43.Sherr, C. J. 2001. The Ink4a/ARF network in tumor suppression. Nat. Rev. Mol. Cell Biol. 2:731-737. [DOI] [PubMed] [Google Scholar]
- 44.Subauste, M. C., M. Von Herrath, V. Benard, C. E. Chamberlain, T. S. Chuang, K. Chu, G. M. Bokoch, and K. M. Hahn. 2000. Rho family proteins modulate rapid apoptosis induced by cytotoxic T lymphocytes and Fas. J. Biol. Chem. 275:9725-9733. [DOI] [PubMed] [Google Scholar]
- 45.Suwa, H., G. Ohshio, T. Imamura, G. Watanabe, S. Arii, M. Imamura, S. Narumiya, H. Hiai, and M. Fukumoto. 1998. Overexpression of RhoC gene correlates with progression of ductal adenocarcinoma of the pancreas. Br. J. Cancer 77:147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295-2322. [DOI] [PubMed] [Google Scholar]
- 47.Vidal, A., S. S. Millard, J. P. Miller, and A. Koff. 2002. Rho activity can alter the translation of p27 mRNA and is important for RasV12-induced transformation in a manner dependent on p27 status. J. Biol. Chem. 277:16433-16440. [DOI] [PubMed] [Google Scholar]
- 48.Webster, G. A., and N. D. Perkins. 1999. Transcriptional cross talk between NF-κB and p53. Mol. Cell. Biol. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welsh, C. F., et. al. 2001. Timing of cyclin D1 expression within G1 phase is controlled by Rho. Nat. Cell Biol. 3:950-957. [DOI] [PubMed] [Google Scholar]
- 50.Westwick, J. K., R. J. Lee, Q. T. Lambert, M. Symons, R. G. Pestell, C. J. Der, and I. P. Whitehead. 1998. Transforming potential of Dbl family proteins correlates with transcription from the cyclin D1 promoter but not with activation of Jun NH2-terminal kinase, p38/Mpk2, serum response factor, or c-Jun. J. Biol. Chem. 273:16739-16747. [DOI] [PubMed] [Google Scholar]
- 51.Westwick, J. K., Q. T. Lambert, G. J. Clark, M. Symons, L. Van Aelst, R. G. Pestell, and C. J. Der. 1997. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol. Cell. Biol. 17:1324-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitehead, I. P., Q. T. Lambert, J. A. Glaven, K. Abe, K. L. Rossman, G. M. Mahon, J. M. Trzaskos, R. Kay, S. L. Campbell, and C. J. Der. 1999. Dependence of Dbl and Dbs transformation on MEK and NF-κB activation. Mol. Cell. Biol. 19:7759-7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, W. J., S. Tu, and R. A. Cerione. 2003. Activated Cdc42 sequesters c-Cbl and prevents EGF receptor degradation. Cell 114: 715-725. [DOI] [PubMed] [Google Scholar]
- 54.Yoshioka, K., F. Matsumura, H. Akedo, and K. Itoh. 1998. Small GTP-binding protein Rho stimulates the actomyosin system, leading to invasion of tumor cells. J. Biol. Chem. 273:5146-5154. [DOI] [PubMed] [Google Scholar]
- 55.Zhao, R., K. Gish, M. Murphy, Y. Yin, D. Notterman, W. H. Hoffman, E. Tom, D. H. Mack, and A. J. Levine. 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14:981-993. [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, Y. 2001. Dbl family guanine nucleotide exchange factors. Trends Biochem. Sci. 26:724-732. [DOI] [PubMed] [Google Scholar]
- 57.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zohn, I. M., S. L. Campbell, R. Khosravi-Far, K. L. Rossman, and C. J. Der. 1998. Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene 17:1415-1438. [DOI] [PubMed] [Google Scholar]