Abstract
The ubiquitous mammalian chromatin-remodeling SWI/SNF-like BAF complexes play critical roles in tumorigenesis. It was suggested that the direct interaction of BRG1 with the retinoblastoma protein pRB is required for regulation of cell cycle progression by pRB. We present evidence that the BRG1-containing complexes regulate the expression of the cdk inhibitor p21CIP1/WAF1/SDI. Furthermore, we show that the physical interaction between BRG1 and pRB is not required for induction of cell growth arrest and transcriptional repression of E2F target genes by pRB. Instead, BRG1 activates pRB by inducing its hypophosphorylation through up-regulation of the cdk inhibitor p21. The hypophosphorylation of pRB is reinforced by down-regulation of critical components, including cdk2, cyclin E, and cyclin D, in the pRB regulatory network. We demonstrate that up-regulation of p21 by BRG1 is necessary to induce formation of flat cells, growth arrest, and finally, cell senescence. Our results suggest that the BRG1-containing complexes control cellular proliferation and senescence by modulating the pRB pathway via multiple mechanisms.
Increasing genetic evidence indicates that the mammalian chromatin-remodeling SWI/SNF-like BAF or hSWI/SNF complexes (28, 32, 66) play an important role in controlling cell proliferation and differentiation and in inhibiting cancer formation (reviewed in reference 35). Various homozygous mutations in the INI1/hSNF5/BAF47 subunit are linked to malignant rhabdoid tumors (MRTs), which are aggressive pediatric tumors in children under 5 years of age (14, 53, 65). Mouse models with targeted disruptions of the BAF complex have also provided evidence supporting a role for the BAF complexes in inhibiting tumorigenesis. While homozygous deletions of the INI1/BAF47 gene in mouse are embryonic lethal, the heterozygous mice develop tumors at a high frequency (23, 34, 49). Similarly, homozygous deletions of BRG1, the essential ATPase subunit of the BAF complex, are lethal, while the heterozygous mice are predisposed to cancer formation (8). Interestingly, reintroduction of BRG1 into SW-13 cells that do not express detectable levels of BRG1 is sufficient to reverse their transformed phenotype by inducing growth arrest and a flattened shape, which requires the activity of the retinoblastoma protein (pRB) (17).
pRB is a major tumor suppressor that is frequently disrupted in cancer cells (55). It is a nuclear phosphoprotein that arrests cells in G0/G1 phase by repressing genes required for the G1/S phase transition (68). The transcriptional repression by pRB is mediated by interaction with the E2F family of transcription factors, whose binding sites are found in the promoters of many genes involved in cell cycle progression (reviewed in references 18 and 47). The interaction of pRB with E2F is controlled by the phosphorylation status of several serine and threonine residues. Inactivation of pRB by phosphorylation releases E2F and therefore the repression of its target genes, which allows the cell cycle to progress through G1 and S phase (7, 9), (12, 13, 19, 25-27, 31, 39, 42). Cyclin-dependent kinases (cdk's), which are implicated in the phosphorylation of pRB, are positively regulated by association with cyclins and negatively regulated by association with cyclin-dependent kinase inhibitors (CKIs) (reviewed in references 37 and 45).
Chromatin structure can be modified by covalent bond formation by acetylation, phosphorylation, methylation, and ubiquitination of histone molecules and/or by noncovalent action by ATP-utilizing remodeling enzymes (1, 21, 24, 29, 46, 48, 60, 64, 69) (3). Histone acetylation by histone acetyltransferases is often required for transcriptional activation, while histone deacetylation by histone deacetylases (HDACs) is associated with transcriptional repression. It has been suggested that HDAC is required for pRB to inhibit E2F activity by forming an HDAC-pRB-E2F repressor complex (5, 43, 44). Furthermore, genetic studies also implicate the ATP-dependent SWI/SNF complex as having an important impact on the function of E2F in Drosophila (56). These observations suggest that modification of chromatin structure plays important roles in the Rb pathway.
pRB can bind to both BRG1 and hBRM (17, 59, 63). Deletion of the pRB-binding domain from BRG1 inhibited its ability to induce cell growth arrest and flat cell formation of SW-13 cells (17, 59), and overexpression of hBRM enhanced the ability of pRB to block the transcriptional activation by E2F-1 (63). Based on these critical observations, it is thought that the direct interaction of pRB with BRG1 and hBRM is required for regulating cell cycle progression by pRB (58, 71). However, in vitro studies have demonstrated that pRB can repress transcriptional activation mediated by the E2F transcription factor in the absence of the BAF complex (50). Additionally, E2F activity was effectively blocked by pRB in BRG1/hBRM-deficient C33a cells (74). Furthermore, the critical pRB-binding motif, LXCXE, is not conserved in the Drosophila BRM protein, suggesting that interacting with BRG1 may not be critical for pRB's function in Drosophila. In this report, we provide evidence that the activity of pRB does not require the direct interaction with BRG1. Rather, the BAF complex regulates the phosphorylation of pRB through the cyclin-dependent kinase inhibitor p21CIP1/WAF1/SDI. The activity of BRG1 to induce the formation of flat cells and cell growth arrest is mainly mediated by p21.
MATERIALS AND METHODS
Constructs and antibodies.
pBJ5, pBJ5-BRG1, pBJ5-BRG1 (K785R), pREP4-luc, and pREP7-Rluc were described previously (40, 41). pBJ5-drBRG1, pBJ5-snfBRG1, and pBJ5-sthBRG1 were constructed by replacing the amino acid region 1339 to 1449 of human BRG1 with the corresponding region of Drosophila BRM (1326 to 1417), yeast SNF2 (1315 to 1421), and yeast STH1 (1020 to 1110). pBJ5-mE7BRG1 was constructed by mutating the LXCXE sequence of BRG1 to FXYXY. The pREP7-BRG1, pREP7-BRG1(K785R), pREP7-mE7BRG1, pREP7-drBRG1, pREP7-snfBRG1, and pREP7-sthBRG1 plasmids were constructed by subcloning the corresponding hemagglutinin (HA)-tagged BRG1 cDNA from the pBJ5 vector into the pREP7 vector. For the yeast two-hybrid analysis, full-length pRB was fused to the GAL4 DNA-binding domain in the pGBKT7 vector, and the BRG1 fragment (amino acid region 1324 to 1536) was fused to the GAL4 activation domain in the pGADT7 vector. The same amino acid region of BRG1 was fused with the maltose-binding protein in a bacterial expression vector for the in vitro binding assay. pGL3-p21pr-luc and pREP4-p21pr-luc were constructed by inserting the PCR-amplified p21 promoter, using the −297 forward primer (5′-GAGCTAGCCAGATTTGTGGCTCACTTCGTG-3′) and +8 reverse primer(5′-CAAAGCTTGACTTCGGCAGCTGCTCACACC-3′), into the NheI-HindIII sites of pGL3-basic and pREP4-luc. The following sequences were used to construct small interference vectors in pBS/U6 (61): p21, 5′GGGTCGAAAACGGCGGCAGACC3′; and BAF47, 5′GGACATGTCAGAGAAGGAGAACA3′. The U6 promoter and the small interference RNA together were then subcloned into pREP4 episomal vector.
The following antibodies were used in this study: BRG1 (40), HA tag (Y-11; Santa Cruz), p21 (Upstate), p107 (SD-9; Santa Cruz), cdc2 (B-6; Santa Cruz), cyclin A (H-432; Santa Cruz), pRB (IF8 and IF8-HRP; Santa Cruz), cyclin D1 (A-12; Santa Cruz), cyclin E (C-19; Santa Cruz), cyclin E2 (N-20; Santa Cruz), cdk2 (D-12; Santa Cruz), cdk4 (H-22; Santa Cruz), and p27 (F-8; Santa Cruz). Phospho-Rb(ser780), phospho-Rb(ser795), and phospho-Rb(ser807/811) antibodies were purchased from Cell Signaling, and BrdU was from Pharmingen.
Cell culture, transient, stable transfection, and luciferase assay.
SW13 cells were cultured as described previously (40). Cells were transfected by using Superfect (Qiagen) transfection reagent according to the vendor's manual. Luciferase assays were performed as described previously (40).
Western blot and immunoprecipitation kinase assays.
Whole cell extracts were prepared by incubating SW-13 cells with the lysis buffer containing 1× phosphate-buffered saline (PBS), 1 mM EDTA, 10% glycerol, 0.5% NP-40, 5 mM NaF, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail for 30 min at 4°C. The extracts (50 μg) were resolved by sodium dodecyl sulfate (SDS)-4 to 12% polyacrylamide gel electrophoresis and then transferred onto Immobilon-P membranes (Millipore) for detection with different antibodies. Immunoprecipitation kinase assays were performed as described previously (57).
BrdU incorporation and immunostaining.
SW-13 cells transfected with pcDNA-p21HA were plated onto a coverslip. After 48 to 96 h of transfection, BrdU (final concentration, 10 μM) was added to the culture medium and incubated for 60 min. After being fixed with 3.7% formaldehyde in 1× PBS for 10 min at room temperature, the cells were permeabilized with 0.5% Triton X-100 in 1× PBS for 3 min, followed by digestion with 50 U of DNase I/ml for 45 min at 37°C in a buffer containing 0.15 M NaCl, 4.2 mM MgCl2, and 10 μM HCl. The cells were blocked with 5% bovine serum albumin in 1× PBS for 30 min at room temperature before binding of the first antibodies (rabbit anti-HA, 1:25; mouse anti-BrdU, 1:25 in 0.05% Tween 20-1× PBS [PBST]) for 30 min at room temperature. The cells were washed three times (5 min each wash) with 1 × PBST, followed by incubation with goat anti-rabbit immunogloblin G-biotin (1:1,000 in 1× PBST) for 15 min at room temperature. Following washes with PBST, the cells were incubated with strepavidin-tetramethyl rhodamine isocyanate (1:3,000) plus goat anti-mouse immunoglobulin G-fluorescein isothiocyanate plus DAPI (100 ng/ml) in PBST. After final washes with PBST, the coverslip was mounted and photographed with a Nikon 800 microscope.
ChIP.
The chromatin immunoprecipitation (ChIP) assays were carried out as described previously (40) using HeLa cell or SW13 cells that were transfected with pREP7 vector or pREP7-HA-BRG1 and selected with hygromycin for 5 days. The anti-HA polyclonal antibody (Y-11; Santa Cruz) and J1 anti-BRG1 polyclonal antibody were used. The following primers were used for detecting p21 promoter sequences: −297F, 5′CAGATTTGTGGCTCACTTCGTG3′; +8R, 5′GACTTCGGCAGCTGCTCACACC3′. The following were primers for the CSF1 upstream control sequence: −5234R, 5′CTCTTCCTCCTGATAGCTCCATGA3′; −5436F, 5′CACTATGTTAGCCAGGATGGTCTC3′. Twenty percent of the ChIP DNA was subjected to PCR amplification using Taq polymerase with the following conditions: 94°C, 30 s; 55°C, 30 s; 72°C, 30 s; repeat 32 times for the p21 primers and 35 times for the CSF1 control primers.
RESULTS
Small interference RNA of BAF47 inhibited both the BRG1- and hBRM-containing complexes.
In order to evaluate the function of the BAF complexes in cell proliferation and differentiation, we decided to inhibit human BAF47 by RNA interference, which is a shared subunit of BRG1- and hBRM-containing BAF complexes (66). Six small interference RNA target sequences were selected for BAF47 and cloned into the pREP4 episomal vector under the control of the U6 promoter. One target sequence was efficient in inhibiting BAF47 mRNA and protein in HeLa cells and MG63 cells (Fig. 1A and B and data not shown).
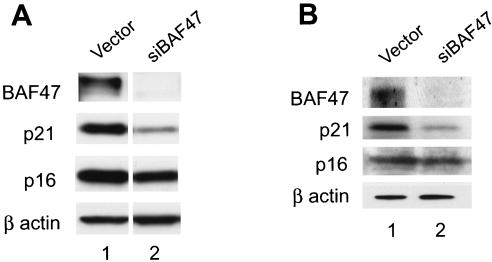
FIG. 1.
The BAF complex is required for the expression of the p21 gene. (A) The expression of p21 but not p16 requires the presence of the BAF complex in HeLa cells. HeLa cells were transfected with the small interference RNA construct targeting BAF47 (siBAF47) and selected for 3 days with puromycin. The surviving cells were harvested and analyzed by Western blotting for expression of the proteins indicated on the left side of the panel. (B) Expression of p21 but not of p16 requires the presence of the BAF complex in MG63 cells. MG63 cells were transfected with the control vector or the siBAF47 construct and analyzed as described for panel A.
The expression of the cdk inhibitor p21 is down-regulated by inhibition of BAF47.
Cell cycle progression is controlled by pRB family proteins that are regulated by cdk's. The cdk's are negatively regulated by association with cdk inhibitors, the Cip/Kip and Ink4 family of proteins that are prototyped by p21 and p16. Western blotting showed that the expression of p21 was significantly down-regulated by inhibition of the BAF47 protein, while the expression level of p16 was not significantly changed in HeLa cells (Fig. 1A). We used MG63, a human fibroblast cell line, to reconfirm that the endogenous BAF complex regulates the expression of p21 in different cell types (Fig. 1B).
The p21 promoter is a direct target of the BAF complex.
To determine if the p21 promoter is directly regulated by the BAF complexes, we cloned 300 bp of p21 promoter region into the episomal pREP4 reporter vector and cotransfected the construct with the small interference RNA constructs into HeLa cells. Consistent with the Western blotting results, inhibition of BAF47 significantly down-regulated the p21 promoter activity (Fig. 2A). The ChIP experiments using HeLa cells showed that the BRG1 antibody but not the preimmune serum precipitated the endogenous p21 promoter sequence, while the preimmune serum and the BRG1 antibody pulled down similar amounts of the unrelated control sequence (Fig. 2B, compare lanes 4 and 5). These results indicate that the BAF complexes are specifically associated with the p21 promoter in vivo.
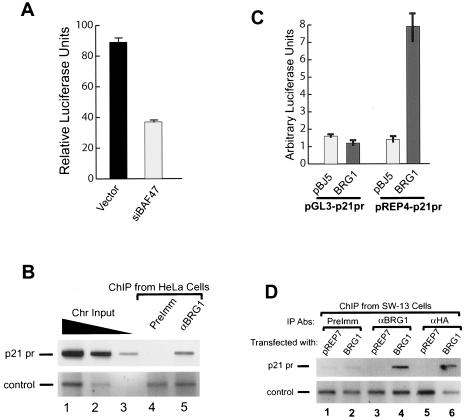
FIG. 2.
p21 is a direct target gene of the BAF complex. (A) The activity of the p21 promoter was inhibited by small interference RNA targeting BAF47. The p21 promoter construct in the pREP-luc vector was cotransfected with either the control vector or the RNA interference construct into HeLa cells and selected for 3 days with puromycin. The luciferase activity was analyzed using a dual luciferase assay kit as described previously (40). (B) BRG1 is bound to the endogenous p21 promoter (p21 pr) in HeLa cells. Chromatin was prepared by sonication from HeLa cells cross-linked with formaldehyde as described previously (41). DNA purified from immunoprecipitates with antibodies against BRG1 (αBRG1) or preimmune serum (PreImm) was analyzed by PCR with primers covering the p21 promoter (−1 to −300) and the negative control (−5436 to −5234 of the CSF1 upstream sequence). The p21 promoter sequence was amplified for 32 cycles (upper panel). The CSF1 control sequence was amplified for 35 cycles in order to detect the products (lower panel). The PCR products were resolved by agarose gel electrophoresis and detected by ethidium bromide staining. Images were inverted. The chromatin (Chr) input was diluted five times at each step. (C) Activation of the p21 promoter by BRG1 requires formation of proper chromatin structure. The p21 promoter (300 bp) was cloned into the pGL3 reporter vector or the episomal pREP4-luc reporter vector. The constructs were cotransfected with either pBJ5 or BRG1 into SW-13 cells. The luciferase activity was analyzed as in panel A. (D) The transiently expressed BRG1 is bound to the endogenous p21 promoter in SW-13 cells. Chromatin fractions were prepared by sonication following cross-linking with formaldehyde of SW-13 cells transfected with pREP7 or HA-tagged BRG1 expression constructs and selected with hygromycin B for 7 days. The chromatin immunoprecipitation assays were performed as described for panel B. The p21 promoter sequence was amplified for 32 cycles (upper panel). The PCR was run for 35 cycles in order to detect the control products in the lower panel. The pull-down of the p21 promoter sequence by anti-BRG1 and anti-HA antibodies (Abs) was dependent on the presence of the BRG1 protein in the cells.
BRG1 and hBRM are not expressed at any detectible levels in SW-13 cells. Reexpression of BRG1 or hBRM in the cells results in cell growth arrest and formation of flat cells (17). Since knocking down the BAF complex by inhibiting BAF47 via RNA interference down-regulated p21 gene expression and its promoter activity, reconstitution of the active BAF complex by transient expression of BRG1 should activate the p21 promoter in SW-13 cells. Indeed, cotransfection of the BRG1 expression vector stimulated the p21 promoter reporter activity about fivefold in the episomal vector but not in the pGL3 vector, as shown in Fig. 2C, suggesting that the formation of chromatin structure is required for the activation of the p21 promoter by the BRG1-containing complex. The ChIP experiments, using SW-13 cells transfected with the pREP7 control vector or an HA-tagged BRG1 expression construct, showed that the p21 promoter sequence was pulled down by both the BRG1 and HA antibodies but not by the preimmune serum. Moreover, the pull-down was dependent on the presence of the BRG1 protein in the cells (Fig. 2D, lanes 4 and 6). In contrast, the control sequence was nonspecifically pulled down by the preimmune, BRG1, and HA antibodies independently of the presence of the BRG1 protein in the cells. Furthermore, the control sequence was detected only after 35 cycles of PCR amplification. These results indicate that the expressed BRG1 is specifically associated with the p21 promoter in vivo. To determine if BAF47 regulates the expression of the p21 gene independently of BRG1 or hBRM, we transfected SW-13 cells with the small interference RNA construct targeting BAF47. We found that inhibition of BAF47 did not down-regulate the mRNA level of p21 and its promoter activity (data not shown), indicating that the regulation of the p21 promoter is mediated by the BRG1- or hBRM-containing BAF complex.
These data indicate that the BRG1- or BRM-containing complexes directly regulate the p21 promoter. The p21 promoter is regulated by both p53 and Sp1, both of which have been suggested to mediate targeting of the BRG1-containing complexes (30, 40). Therefore, the recognition of the p21 promoter by BRG1 could be mediated by the p53 and Sp1 proteins.
Expressing BRG1 inhibited the activities of cdk2 and cdk4.
p21 is a critical determinant of G1 arrest by inhibiting cdk2. We found that ectopic expression of BRG1 in SW-13 cells increased the mRNA and protein levels of p21 (Fig. 3A, upper panel and data not shown). Moreover, the level of cdk2 protein was reduced, while the level of cdk4 protein was not significantly changed (Fig. 3A, upper panel). Coimmunoprecipitation using cdk2 antibody showed that much more p21 was associated with cdk2 when BRG1 was expressed in SW-13 cells (Fig. 3A, middle panel, compare lanes 3 and 4). Unexpectedly, less p21 was bound to cdk4, even though the p21 level was higher in the presence of BRG1 in the cells (Fig. 3A, lower panel, compare lanes 5 and 6). This might have resulted from the reduced levels of cyclin D in the presence of BRG1 (Fig. 4A, panel k), since simultaneous interaction of p21 with cdk4 and cyclin D stabilizes the complex formation. These results suggest that the activities of cdk2 and cdk4 might be inhibited in the presence of BRG1. Therefore, we measured the kinase activities of the immunoprecipitated cdk2 and cdk4 proteins on a recombinant Rb protein. As expected, the kinase activity of the immunoprecipitated cdk2 in the whole-cell extracts was significantly lower in the presence of wild-type but not dominant-negative BRG1 (Fig. 3B, top panel, lanes 2 and 3 versus lane 1). Interestingly, the activity of cdk4 was also dramatically reduced (Fig. 3B, top panel, lanes 5 and 6 versus lane 4), even though expression of BRG1 did not noticeably increase the levels of p16 (Fig. 4A, panel c). The reduction of cdk4 activity might have resulted from the decrease of cyclin D levels in the presence of BRG1 (Fig. 4A, panel k).
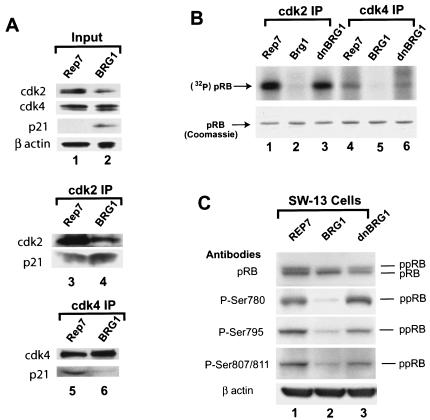
FIG. 3.
Reexpression of BRG1 in SW-13 cells induces hypophosphorylation of pRB. (A) Reexpression of BRG1 in SW-13 cells up-regulates p21. SW-13 cells transfected with the pREP7 vector or the BRG1 expression construct were selected with hygromycin B for 7 days. Whole-cell lysates were prepared for analysis of the various protein levels by Western blotting (lanes 1 and 2) or coimmunoprecipitation of the p21 protein using cdk2 (lanes 3 and 4) and cdk4 (lanes 5 and 6) antibodies. (B) Reexpression of BRG1 in SW-13 cells inhibits the activities of cdk2 and cdk4. Whole-cell extracts were prepared from cells transfected with the pREP7 vector, BRG1, or dominant-negative BRG1 (K785R) (dnBRG1) and selected with hygromycin B for 7 days. The immunoprecipitates using cdk2 and cdk4 antibodies were incubated with the bacterially expressed C terminus of pRB in the presence of [32P]ATP. The resulting mixture was resolved by SDS-polyacrylamide gel electrophoresis and visualized by Coomassie blue staining and exposure to X-ray film. (C) Reexpression of BRG1 in SW-13 cells induces hypophosphorylation of pRB. SW-13 cells transfected with pREP7 vector, BRG1, or dominant negative BRG1 (K785R) (dnBRG1) were selected with hygromycin B for 7 days. The cells were harvested for analysis of pRB by Western blotting with various phospho-specific antibodies indicated on the left side of the panel. ppRB, hyperphosphorylated retinoblastoma protein; pRB, hypophosphorylated retinoblastoma protein.
FIG. 4.
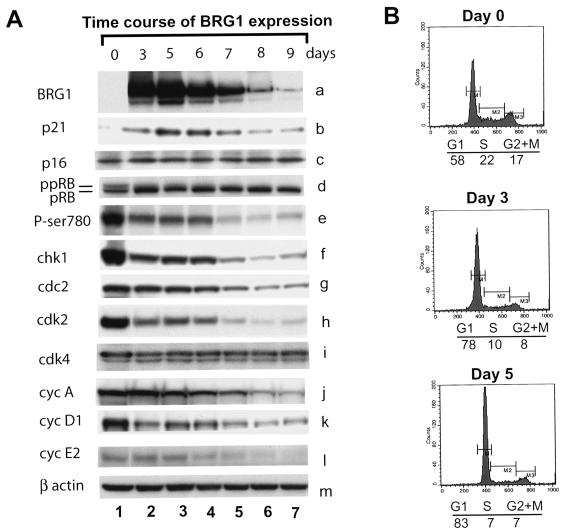
Reexpression of BRG1 in SW-13 cells induces hypophosphorylation of pRB and G1 arrest. (A) Reexpression of BRG1 in SW-13 cells induces expression of p21 but represses E2F target genes. SW-13 cells were transfected with the pREP7-BRG1 expression construct and selected with hygromycin B for the indicated period of time. The cells were harvested and analyzed for the proteins indicated on the left side of the panel. (B) SW-13 cells were transfected with the pREP7-BRG1 expression construct and selected with hygromycin for the time indicated above each panel. The cell cycle was analyzed using propidium iodide staining. The cell cycle distribution is indicated under each panel. ppRB, hyperphosphorylated retinoblastoma protein; pRB, hypophosphorylated retinoblastoma protein; cyc, cyclin.
The pRB protein was hypophosphorylated by expression of BRG1.
The major substrate of the cdk's is pRB, which regulates the cell cycle by its status of phosphorylation. Since the cdk activities were dramatically inhibited, we expect that pRB becomes hypophosphorylated in the presence of BRG1. As shown in Fig. 3C, expression of wild-type BRG1 increased the ratio of the faster-migrating hypophosphorylated pRB to the slower-migrating hyperphosphorylated pRB (first panel). Using phospho-specific antibodies, we found that the phosphorylation of serine 780 was significantly reduced (second panel), which appears to be necessary for pRB to bind with E2F (33). Similarly, Ser795, whose phosphorylation is critical for the inactivation of pRB-mediated growth suppression (11), was hypophosphorylated in the presence of BRG1 (third panel). The phosphorylation of 807/811, which disrupts binding with the ubiquitously expressed c-Abl tyrosine kinase (36), was also down-regulated (fourth panel, compare lanes 1 and 2). Expression of the ATP-binding site mutant (K785R), which inactivates BRG1 (32), had little effect on the phosphorylation of pRB (compare lanes 1 and 3). Thus, pRB is activated by ectopic expression of BRG1 in SW-13 cells.
E2F target genes are repressed by expression of BRG1.
Hypophosphorylated pRB forms complexes with the E2F family of transcription factors, which recognize the E2F-binding sites in the promoters of many genes required for cell cycle and proliferation control (18). To determine if the BRG1-induced hypophosphorylation of pRB results in repression of E2F target genes, we examined the expression levels of some known E2F target genes by Western blotting. Accompanying the hypophosphorylation of pRB, the known E2F target genes, such as chk1, cdk2, cyclin A, cyclin E2, and p107, were significantly repressed (Fig. 4A and data not shown). Though not a target gene of E2F, cyclin D1 was also repressed by expression of BRG1, possibly by a mechanism that requires HDAC and INI1/hSNF5/BAF47 (72). Even though the cdk4 protein was not down-regulated by expression of BRG1 (Figs. 3A and 4A), its activity was significantly repressed (Fig. 3B), which could be attributed to the repression of its positive regulator, cyclin D1 (Fig. 4A, panel k).
Hypophosphorylation of pRB induces G1 arrest. Therefore, we tested if BRG1-induced hypophosphorylation of pRB causes cell cycle arrest. As shown in Fig. 4A (panels d and e), expression of BRG1 for 3 days induced significant hypophosphorylation of pRB. The cell cycle analyses showed that the G1 cell population at day 3 increased to 78% from 58% at day 0 (Fig. 4B). The G1 population further increased to 83% at day 5. These results indicate that the hypophosphorylation of pRB induced by expression of BRG1 in SW-13 cells resulted in G1 arrest.
Inhibiting p21 by RNAi blocked the formation of flat cells induced by BRG1.
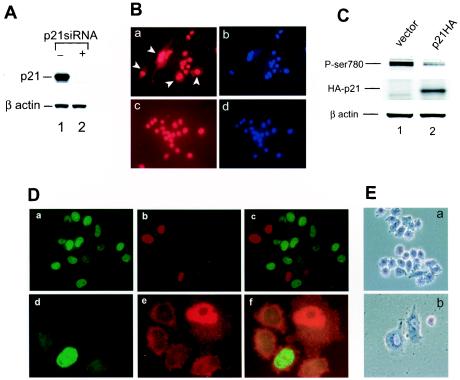
CKI family proteins inhibit the activity of cdk's to phosphorylate pRB and therefore arrest cells in the G0/G1 phase. p16 does not seem to be involved in cell growth control in SW-13 cells, since it was not significantly up-regulated by reexpression of BRG1 (Fig. 4A, panel c) and its overexpression failed to arrest the cells (71). In contrast, p21 was highly induced by expression of BRG1, suggesting that it may play important roles in BRG1-induced cell growth arrest and flat cell formation. To determine if p21 is required for BRG1-induced flat cell formation, we inhibited p21 by RNA interference. As shown in Fig. 5A, transfection of the p21 small interference RNA but not a control vector completely inhibited the expression of the p21 protein (compare lanes 1 and 2). We then cotransfected the pREP4-p21siRNA construct or the pREP4 control vector with a BRG1-expressing construct into SW-13 cells and selected with hygromycin B for 5 days. As shown in Fig. 5B, immunostaining with BRG1 antibody showed that all of the surviving cells expressed BRG1 protein (panels a and c), compared to the DNA staining by DAPI (panels b and d). About 30% of the cells in the absence of the p21 small interference RNA acquired a flattened shape (Fig. 5B, panel a). However, in the presence of the p21 small interference RNA, only 3% of the cells showed the flattened shape (Fig. 5B, panel c), indicating that p21 is required for BRG1-induced flat cell formation.
FIG. 5.
p21 is the main downstream mediator of BRG1 for inducing cell growth arrest and formation of flat cells. (A) Expression of the p21 small interference RNA inhibits expression of the p21 protein. SW-13 cells were transfected with pREP4-p21siRNA or the pREP4 control vector. After 3 days of selection with hygromycin B, the cells were harvested for analysis of p21 expression by Western blotting. β-actin was used as a control. (B) Inhibition of p21 blocks BRG1-induced flat cell formation. SW-13 cells were cotransfected with pBJ5-BRG1 plus control vector (panels a and b) or with pBJ5-BRG1 plus pREP4-p21siRNA (panels c and d). After 5 days of selection with hygromycin B, the cells were stained with DAPI (blue) and an antibody against BRG1 (red). The images were taken with high background to reveal the flattened shape, indicated by arrowheads in panel a. Magnification, ×20. (C) Overexpression of p21 in SW-13 cells induces hypophosphorylation of pRB. SW-13 cells were transfected with pcDNA-p21HA or a control vector. After 3 days of selection with 200 μg of zeocin/ml, the cells were harvested and analyzed for expression of p21 with an HA antibody and the phosphorylation of pRB at serine 780 with phospho-specific antibody by Western blotting. β-Actin was used as a control. (D) Overexpression of p21 in SW-13 inhibits BrdU incorporation. SW-13 cells were transfected with pcDNA-p21HA. After 48 (panels a, b, and c) or 96 h (panels d, e, and f) of transfection, the cells were labeled with BrdU for 60 min. Following fixation with formaldehyde, the cells were immunostained for HA-p21 expression (red) and BrdU incorporation (green). The high background signals in panels d to f were used to show the flattened shape of the p21-expressing cell. The images were taken with ×60 magnification. Panels d to f were further magnified to show the cell shape. (E) Overexpression of p21 in SW-13 cells induces formation of flat cells. SW-13 cells were transfected with pREP4-p21 expression construct and selected with hygromycin B for 6 days. The images were taken at magnification ×20.
Overexpression of p21 is sufficient to induce hypophosphorylation of pRB and cell growth arrest.
If p21 is the main downstream mediator of the BRG1 activity, overexpression of p21 alone may be sufficient to induce growth arrest of SW-13 cells. As shown in Fig. 5C, overexpression of p21 induced hypophosphorylation of pRB. Remarkably, BrdU incorporation was completely inhibited in cells overexpressing the p21 protein. As shown in Fig. 5D, after 48 h of p21 transfection, none of the p21-positive cells shown in red were labeled by BrdU (green), while about 50% of p21-negative cells were labeled by BrdU (Fig. 5D, panels a, b, and c), indicating that expression of the p21 protein efficiently arrested cells. Furthermore, after 4 days of p21 transfection, about 30% of p21-positive cells acquired a flatted shape (Fig. 5D, panels d, e, and f). All of the p21-positive cells became flat cells after 6 days of selection (Fig. 5E, panel b).
Expression of BRG1 induced cell senescence.
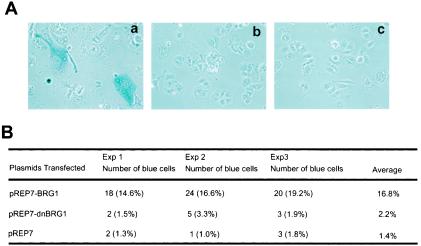
Up-regulation of p21 is often associated with cell senescence (reviewed in references 37 and 45). In contrast, disruption of the p21 gene in normal diploid human fibroblasts bypassed senescence (6). The mechanisms by which the reexpression of BRG1 in SW-13 cells induces flat cell formation and finally cell death have not been unambiguously elucidated. Our observation that p21 was up-regulated by expression of BRG1 in SW-13 cells implies that the cells undergo senescence, as suggested by a previous report (54). Therefore, we examined the activity of β-galactosidase, a marker of senescent cells, in the absence or presence of BRG1. As shown in Fig. 6A (c) and B, 16.8% of the cells transfected with BRG1 were β-galactosidase positive compared to only 1.4% with the control vector. Most of the β-galactosidase-positive cells were flat cells, consistent with the notion that flat cell shape is a sign of cell senescence (62). The dominant-negative BRG1 (K785R) did not significantly increase the population of β-galactosidase-positive cells (Fig. 6A, b, and 6B), consistent with its inability in up-regulating the p21 protein and inducing hypophosphorylation of pRB.
FIG. 6.
Ectopic expression of BRG1 in SW-13 cells induces cell senescence. (A) SW-13 cells were transfected with pREP7 (c) or BRG1 (a) or the dominant-negative BRG1 (b) expression constructs and selected with hygromycin B for 10 days. The cells were stained for β-galactosidase activity as described previously (16). The images were taken at magnification ×20. (B) Quantification of β-galactosidase-positive cells.
Drosophila BRM, yeast SNF2, and STH1 do not interact with pRB.
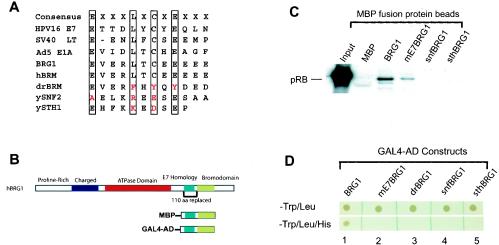
The above results suggest that BRG1 induces flat cell formation and cell growth arrest by inducing p21 protein, which activates pRB and leads to the observed phenotype. However, BRG1 interacts directly with pRB via its LXCXE motif, which, together with its downstream sequence, is required for binding to pRB and for the formation of flat cells (17). It was suggested that this interaction is required for BRG1 and pRB to cooperatively regulate cell cycle progression (71). Therefore, we decided to determine if the interaction is required for induction of p21 protein and cell growth arrest. Sequence alignment surrounding the BRG1 E7 homology domain revealed that in contrast to the well-characterized ATPase domain and bromodomain (32), the LXCXE motif is not conserved in yeast SNF2, STH1, and Drosophila BRM, which are the homologues of BRG1 (Fig. 7A). Therefore, we asked if the corresponding Drosophila and yeast sequences can replace the BRG1 sequence for binding to pRB. The strategy we took was to mutate the LXCXE motif of human BRG1 to the corresponding Drosophila BRM sequence, FXYXY (named mE7BRG1) or replace the entire E7 homology domain and its downstream sequence of 110 amino acids (1339 to 1449) of BRG1 with the corresponding Drosophila or yeast sequences. It was shown that deletion of the 110-amino-acid region from BRG1 abrogated the ability of BRG1 to bind pRB and to induce cell growth arrest and formation of flat cells (17, 71). To determine the pRB-binding activity of mE7BRG1 or the chimera BRG1 proteins, a fusion maltose-binding protein (MBP) covering the E7 homology domain and the Bromo domain was expressed in bacteria (Fig. 7B). The fusion protein purified onto amylose resin was incubated with nuclear extracts of SW-13 cells. The bound proteins were detected by Western blotting with pRB antibody. As shown in Fig. 7C, mutation of the LXCXE motif of BRG1 to the FXYXY sequence significantly reduced the binding of pRB. Replacement with the Drosophila and yeast E7 homology domain practically abolished pRB binding.
FIG. 7.
The pRB-binding activity of hBRG1 is not conserved in its Drosophila and yeast homologs. (A) Sequence comparison of the LXCXE motif in the E7 homology domain of several pRB-binding proteins with hBRG1 and hBRM and their Drosophila and yeast homologues. (B) Protein domains in hBRG1. The 110-amino-acid region surrounding the E7 homology domain and its downstream sequence replaced with the corresponding Drosophila and yeast sequences is indicated under the graph (110 aa replaced). The MBP fusion proteins for the in vitro pull-down assay and the GAL4-AD fusion proteins for the yeast two-hybrid assay are also indicated. (C) In vitro pull-down assay. Nuclear extracts of SW-13 cells were incubated with Sepharose beads bound with the MBP fusion proteins in 250 mM salt. The bound proteins were resolved by SDS-PAGE and detected by Western blotting with the pRB antibody. MBP, maltose-binding protein; BRG1, MBP fusion with wild-type BRG1 shown in panel B; mE7BRG1, BRG1 bearing the LXCXE-to-FXYXY mutation; snfBRG1, the E7 homology domain and its downstream sequence of BRG1 were replaced with the corresponding yeast SNF2 sequence; sthBRG1, the E7 homology domain and its downstream sequence of BRG1 were replaced with the corresponding yeast STH1 sequence; drBRG1, the E7 homology domain and its downstream sequence of BRG1 were replaced with the corresponding Drosophila BRM sequence. (D) Yeast two-hybrid assay showing that the mutation or replacement of the E7 homology domain of BRG1 inhibits its pRB-binding activity. The yeast cotransformed with the pRB and BRG1 constructs was plated onto a double dropout plate (−Trp/−Leu) (top panel) or triple dropout plate (−Trp/−Leu/−His) (lower panel). The BRG1 constructs are shown as in panel B and explained in the legend to panel C.
To confirm these in vitro binding data, we used the yeast two-hybrid system by constructing plasmids expressing the GAL4 activation domain fused to the wild-type and mutant BRG1 sequences as shown in Fig. 7B. These constructs were cotransformed into Saccharomyces cerevisiae with a plasmid expressing the GAL4 DNA-binding domain fused to pRB. Transformed yeast cells were plated onto medium lacking histidine so that growth required activation of the His3 gene driven by a GAL1 promoter. No growth occurred after transformation with single constructs (data not shown). Robust growth was observed when both the pRB and wild-type BRG1 constructs were present (Fig. 7D, lane 1, lower panel). In contrast, no growth was observed when the E7 homology domain and its downstream sequence of BRG1 were replaced with the corresponding sequences from yeast SNF2, SHT1, or Drosophila BRM (Fig. 7D, lower panel, lanes 3, 4, and 5). Interestingly, no growth occurred when the LXCXE motif of BRG1 was mutated to FXYXY, even though the same mutation showed some binding in the in vitro MBP pull-down assay (Fig. 7D, lane 2, lower panel). These results were confirmed by the β-galactosidase assay (data not shown).
The above results indicate that replacing the E7 homology region and its downstream sequence of BRG1 with the corresponding Drosophila or yeast sequence abolished the ability of BRG1 to bind to pRB, suggesting that the Drosophila pRB may not need to bind to BRM for its normal function. Since yeast does not have the pRB system, the pRB-binding activity of SWI2/SNF2 or STH1 may not be relevant.
The pRB-binding activity of BRG1 is not required for induction of p21 and activation of pRB.
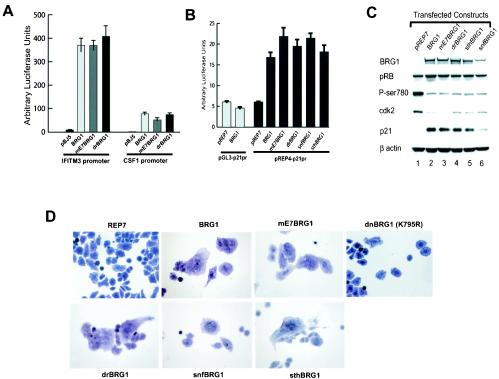
Even though a conserved E7 homology domain does not appear to be important for the function of the yeast SNF2, STH1, and Drosophila BRM, it might be required for the activity of human BRG1. Therefore, we tested these mutant BRG1 constructs on the known BRG1 target genes, including CSF1 and IFITM3 (40, 41). Surprisingly, our results indicate that neither the mutation of the LXCXE motif nor replacement of the whole E7 homology domain with its downstream sequence affected the activity of BRG1 in activation of its target promoters (Fig. 8A). Furthermore, all the BRG1 mutants that do not bind pRB activated the p21 promoter (Fig. 8B). Consistent with the reporter assay, the endogenous p21 protein was induced (Fig. 8C). As expected, accompanying p21 up-regulation, pRB became significantly hypophosphorylated (Fig. 8C). Expression of cdk2 and other E2F target genes was repressed (Fig. 8C and data not shown), indicating that pRB was fully activated.
FIG. 8.
The pRB-binding activity of BRG1 is not required for activation of its target genes and for inducing formation of flat cells. (A) The pRB-binding activity of BRG1 is not required for activation of the CSF1 and IFITM3 promoters. The CSF1 or IFITM3 promoter reporter construct was cotransfected into SW-13 cells with the control vector or the different BRG1 expression constructs. The luciferase activity was analyzed as described previously (40). (B) The pRB-binding activity of BRG1 is not required for activation of the p21 promoter. p21 promoter reporter constructs were cotransfected into SW-13 cells with wild-type BRG1 or mutant BRG1 that does not bind pRB. The luciferase activity was analyzed as described previously (40). (C) The pRB-binding activity of BRG1 is not required for inducing the endogenous p21 protein and hypophosphorylation of pRB. SW-13 cells were transfected with various BRG1 expression constructs and selected with hygromycin B for 7 days. The cells were harvested for Western blot analysis of proteins indicated on the left side of the panel. The constructs transfected were indicated on the top of the panel. (D) The mutant BRG1 proteins that do not bind pRB are capable of inducing formation of flat cells. SW-13 cells were transfected with pREP7 empty vector or the mutant BRG1 expression constructs and selected with hygromycin B for 7 days. The cells were photographed after staining with crystal violet. All of the images were taken at magnification ×20. See the legend to Fig. 7C for explanation of designations.
Disruption of BRG1/pRB interaction did not inhibit the function of the BAF complex in flat cell formation.
We reasoned that pRB-BRG1 interaction might be required for assays requiring the activity of pRB, such as cell growth arrest and formation of flat cells induced by reexpression of BRG1 in SW-13 cells. Therefore, we transfected SW-13 cells with pREP7 episomal constructs which express the wild-type and mutant BRG1 proteins. While the empty pREP7 vector did not induce a significant number of flat cells, the LXCXE mutant and the chimeric BRG1 constructs induced the flattened shape as efficiently as wild-type BRG1. Some of the results are shown in Fig. 8D. In contrast, expression of the dominant-negative BRG1 (K785R) did not induce a significant number of flat cells, as reported previously (17). BrdU incorporation and cell cycle analysis revealed that all of the pRB binding-deficient mutant BRG1 proteins arrested cells as well as the wild-type BRG1 (data not shown).
We conclude from these results that the pRB-BRG1 interaction is not required for activation of BRG1 target genes and induction of cell growth arrest and flat cell formation by the BAF complex.
DISCUSSION
BRG1 regulates the activity of pRB by up-regulating CKI p21.
The pRB system is disrupted in the majority of human tumors (55). Ectopic expression of pRB in tumor cell lines results in cell growth arrest and senescence manifested by formation of flat cells (70). The ability of pRB to induce cellular senescence has been suggested to require the function of BRG1 (17, 58, 59, 63, 71). It is thought that BRG1 regulates the activity by directly binding to pRB (17). However, our data show that even though the mutant BRG1 proteins did not bind pRB, they were fully active in activating the BRG1 target promoters, repressing E2F target genes, and inducing flat cell formation. Therefore, it appears that BRG1 does not regulate the activity of pRB by direct binding to it. Instead, it regulates the phosphorylation of pRB by controlling the transcription of the p21 gene, which in turn inhibits the activity of cdk2 and results in a hypophosphorylated state of pRB. The hypophosphorylated pRB then represses transcription of E2F target genes, including cyclin E1, E2, cdk2, etc., to reinforce the active status of pRB. Furthermore, cdk2 can be activated by cdc25A and cdc25B phosphatases that are transcriptionally activated by c-Myc (22). The expression of c-Myc was repressed by expression of BRG1 in SW-13 cells (data not shown), which might be mediated by the E2F binding site in its promoter. Therefore, the hypophosphorylated state of pRB is reinforced by transcriptional repression of several critical components in the pRB regulatory network by pRB family proteins mediated by E2F. Taken together, these results suggest that disruption of the chromatin-remodeling BAF complex will down-regulate CKI family proteins, which in turn will disrupt the balance of the pRB regulatory network and result in dysregulation of the cell cycle and cell proliferation. This is consistent with the observed phenomena that mice are predisposed to cancer formation by disruption of the BAF complexes. It will be interesting to examine the pRB pathway in the cancer cells derived from mice with mutations of the BAF subunits.
It appears that the BAF complex regulates the expression of different CKI proteins in different tissues. For example, reconstitution of the BAF complex by ectopic expression of the BAF47 subunit in MRT-derived cell lines up-regulated p16 (4). However, overexpression of dominant-negative BRG1 (K785R) did not affect the expression levels of p21 in NIH3T3 cells (15). The particular member of the CKI proteins regulated by the BAF complex could be cell type specific and determined by the developmental history. These results suggest that the chromatin structure of a promoter is programmed early in development for a specific response to a regulatory signal later.
Mechanisms of tumor formation when the BAF complex is disrupted.
Ample evidence implicates the SWI/SNF-like BAF complexes as tumor suppressors. However, the mechanism underlying tumor formation by disruption of the BAF complexes is not understood. Our data that the BAF complex regulates the expression of p21 and therefore the phosphorylation status of pRB shed new light on the mechanisms of tumorigenesis caused by mutations of the complexes. The cdk2 inhibitor p21 is a downstream mediator of p53 and contributes to p53-dependent arrest (10). Interestingly, oncogenic ras signaling induces premature cell senescence that is accompanied by up-regulation of the p53, p16, and p21 proteins (38, 52, 73). However, disruption of either p53 or p16 is sufficient to prevent ras-induced arrest and allows ras-induced immortalization of primary mouse fibroblasts (52). The up-regulation of p16 and p21 is also observed during the replicative senescence of normal human fibroblasts (2). Furthermore, disruption of p21 bypasses senescence in normal human diploid fibroblasts (6). Overexpression of p21 in human cancer cells induces senescence independently of p53 (20, 67). Interestingly, reexpression of pRB in pRB-deficient tumor cells results in senescence (62, 70). Taken together, these observations indicate that the p21/p16/pRB pathway is directly involved in senescence. Reconstitution of the BAF complexes by reexpression of the missing subunit up-regulated p21 in SW-13 cells (this study) and p16 (4) in MRT-derived cells, which result in the observed senescence (Fig. 5) (4, 54, 72).
Cellular senescence has been suggested as a mechanism of tumor suppression (51, 52). We propose that disruption of the chromatin-remodeling BAF complexes down-regulates expression levels of p16 and p21 and therefore inhibits senescence of aging cells and ultimately contributes to tumor formation.
Acknowledgments
We thank Tian Chi, Gerald R. Crabtree, Warren J. Leonard, David Levens, Mimi Wan, and Carl Wu for critical reading of the manuscript. K.Z. thanks Warren Leonard for continuous encouragement and support.
This work was supported by intramural grants to NHLBI, NIH.
REFERENCES
- 1.Aalfs, J. D., and R. E. Kingston. 2000. What does ‘chromatin remodeling’ mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Alcorta, D. A., Y. Xiong, D. Phelps, G. Hannon, D. Beach, and J. C. Barrett. 1996. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc. Natl. Acad. Sci. USA 93:13742-13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 4.Betz, B. L., M. W. Strobeck, D. N. Reisman, E. S. Knudsen, and B. E. Weissman. 2002. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G(1) arrest associated with induction of p16ink4a and activation of RB. Oncogene 21:5193-5203. [DOI] [PubMed] [Google Scholar]
- 5.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. P., W. Wei, and J. M. Sedivy. 1997. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science 277:831-834. [DOI] [PubMed] [Google Scholar]
- 7.Buchkovich, K., L. A. Duffy, and E. Harlow. 1989. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 58:1097-1105. [DOI] [PubMed] [Google Scholar]
- 8.Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287-1295. [DOI] [PubMed] [Google Scholar]
- 9.Chen, P. L., P. Scully, J. Y. Shew, J. Y. Wang, and W. H. Lee. 1989. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell 58:1193-1198. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X., J. Bargonetti, and C. Prives. 1995. p53, through p21 (WAF1/CIP1), induces cyclin D1 synthesis. Cancer Res. 55:4257-4263. [PubMed] [Google Scholar]
- 11.Connell-Crowley, L., J. W. Harper, and D. W. Goodrich. 1997. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol. Biol. Cell 8:287-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeCaprio, J. A., Y. Furukawa, F. Ajchenbaum, J. D. Griffin, and D. M. Livingston. 1992. The retinoblastoma-susceptibility gene product becomes phosphorylated in multiple stages during cell cycle entry and progression. Proc. Natl. Acad. Sci. USA 89:1795-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeCaprio, J. A., J. W. Ludlow, D. Lynch, Y. Furukawa, J. Griffin, H. Piwnica-Worms, C. M. Huang, and D. M. Livingston. 1989. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell 58:1085-1095. [DOI] [PubMed] [Google Scholar]
- 14.DeCristofaro, M. F., B. L. Betz, W. Wang, and B. E. Weissman. 1999. Alteration of hSNF5/INI1/BAF47 detected in rhabdoid cell lines and primary rhabdomyosarcomas but not Wilms' tumors. Oncogene 18:7559-7565. [DOI] [PubMed] [Google Scholar]
- 15.de la Serna, I. L., K. Roy, K. A. Carlson, and A. N. Imbalzano. 2001. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J. Biol. Chem. 276:41486-41491. [DOI] [PubMed] [Google Scholar]
- 16.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 18.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 19.Ewen, M. E., H. K. Sluss, C. J. Sherr, H. Matsushime, J. Kato, and D. M. Livingston. 1993. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell 73:487-497. [DOI] [PubMed] [Google Scholar]
- 20.Fang, L., M. Igarashi, J. Leung, M. M. Sugrue, S. W. Lee, and S. A. Aaronson. 1999. p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53. Oncogene 18:2789-2797. [DOI] [PubMed] [Google Scholar]
- 21.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 22.Galaktionov, K., X. Chen, and D. Beach. 1996. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature 382:511-517. [DOI] [PubMed] [Google Scholar]
- 23.Guidi, C. J., A. T. Sands, B. P. Zambrowicz, T. K. Turner, D. A. Demers, W. Webster, T. W. Smith, A. N. Imbalzano, and S. N. Jones. 2001. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21:3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 25.Hatakeyama, M., J. A. Brill, G. R. Fink, and R. A. Weinberg. 1994. Collaboration of G1 cyclins in the functional inactivation of the retinoblastoma protein. Genes Dev. 8:1759-1771. [DOI] [PubMed] [Google Scholar]
- 26.Hinds, P. W., S. Mittnacht, V. Dulic, A. Arnold, S. I. Reed, and R. A. Weinberg. 1992. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70:993-1006. [DOI] [PubMed] [Google Scholar]
- 27.Hu, Q. J., J. A. Lees, K. J. Buchkovich, and E. Harlow. 1992. The retinoblastoma protein physically associates with the human cdc2 kinase. Mol. Cell. Biol. 12:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imbalzano, A. N., H. Kwon, M. R. Green, and R. E. Kingston. 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370:481-485. [DOI] [PubMed] [Google Scholar]
- 29.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 30.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato, J., H. Matsushime, S. W. Hiebert, M. E. Ewen, and C. J. Sherr. 1993. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 7:331-342. [DOI] [PubMed] [Google Scholar]
- 32.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 33.Kitagawa, M., H. Higashi, H. K. Jung, I. Suzuki-Takahashi, M. Ikeda, K. Tamai, J. Kato, K. Segawa, E. Yoshida, S. Nishimura, and Y. Taya. 1996. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 15:7060-7069. [PMC free article] [PubMed] [Google Scholar]
- 34.Klochendler-Yeivin, A., L. Fiette, J. Barra, C. Muchardt, C. Babinet, and M. Yaniv. 2000. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klochendler-Yeivin, A., C. Muchardt, and M. Yaniv. 2002. SWI/SNF chromatin remodeling and cancer. Curr. Opin. Genet. Dev. 12:73-79. [DOI] [PubMed] [Google Scholar]
- 36.Knudsen, E. S., and J. Y. Wang. 1996. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J. Biol. Chem. 271:8313-8320. [DOI] [PubMed] [Google Scholar]
- 37.Lee, M. H., and H. Y. Yang. 2001. Negative regulators of cyclin-dependent kinases and their roles in cancers. Cell. Mol. Life Sci. 58:1907-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 12:3008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, B. T., S. Gruenwald, A. O. Morla, W. H. Lee, and J. Y. Wang. 1991. Retinoblastoma cancer suppressor gene product is a substrate of the cell cycle regulator cdc2 kinase. EMBO J. 10:857-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, H., H. Kang, R. Liu, X. Chen, and K. Zhao. 2002. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol. Cell. Biol. 22:6471-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, R., H. Liu, X. Chen, M. Kirby, P. O. Brown, and K. Zhao. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106:309-318. [DOI] [PubMed] [Google Scholar]
- 42.Ludlow, J. W., J. Shon, J. M. Pipas, D. M. Livingston, and J. A. DeCaprio. 1990. The retinoblastoma susceptibility gene product undergoes cell cycle-dependent dephosphorylation and binding to and release from SV40 large T. Cell 60:387-396. [DOI] [PubMed] [Google Scholar]
- 43.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 44.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 45.Mittnacht, S. 1998. Control of pRB phosphorylation. Curr. Opin. Genet. Dev. 8:21-27. [DOI] [PubMed] [Google Scholar]
- 46.Muchardt, C., and M. Yaniv. 1999. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J. Mol. Biol. 293:187-198. [DOI] [PubMed] [Google Scholar]
- 47.Nevins, J. R. 1998. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 9:585-593. [PubMed] [Google Scholar]
- 48.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 49.Roberts, C. W., S. A. Galusha, M. E. McMenamin, C. D. Fletcher, and S. H. Orkin. 2000. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. USA 97:13796-13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ross, J. F., X. Liu, and B. D. Dynlacht. 1999. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol. Cell 3:195-205. [DOI] [PubMed] [Google Scholar]
- 51.Sager, R. 1991. Senescence as a mode of tumor suppression. Environ. Health Perspect. 93:59-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 53.Sevenet, N., A. Lellouch-Tubiana, D. Schofield, K. Hoang-Xuan, M. Gessler, D. Birnbaum, C. Jeanpierre, A. Jouvet, and O. Delattre. 1999. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum. Mol. Genet. 8:2359-2368. [DOI] [PubMed] [Google Scholar]
- 54.Shanahan, F., W. Seghezzi, D. Parry, D. Mahony, and E. Lees. 1999. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol. Cell. Biol. 19:1460-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherr, C., and F. McCormick. 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103-112. [DOI] [PubMed] [Google Scholar]
- 56.Staehling-Hampton, K., P. J. Ciampa, A. Brook, and N. Dyson. 1999. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics 153:275-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strobeck, M. W., A. F. Fribourg, A. Puga, and E. S. Knudsen. 2000. Restoration of retinoblastoma mediated signaling to Cdk2 results in cell cycle arrest. Oncogene 19:1857-1867. [DOI] [PubMed] [Google Scholar]
- 58.Strobeck, M. W., K. E. Knudsen, A. F. Fribourg, M. F. DeCristofaro, B. E. Weissman, A. N. Imbalzano, and E. S. Knudsen. 2000. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 97:7748-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strober, B. E., J. L. Dunaief, Guha, and S. P. Goff. 1996. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol. 16:1576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sudarsanam, P., and F. Winston. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 61.Sui, G., C. Soohoo, B. Affar el, F. Gay, Y. Shi, and W. C. Forrester. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Templeton, D. J., S. H. Park, L. Lanier, and R. A. Weinberg. 1991. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc. Natl. Acad. Sci. USA 88:3033-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trouche, D., C. Le Chalony, C. Muchardt, M. Yaniv, and T. Kouzarides. 1997. RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl. Acad. Sci. USA 94:11268-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Varga-Weisz, P. 2001. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene 20:3076-3085. [DOI] [PubMed] [Google Scholar]
- 65.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203-206. [DOI] [PubMed] [Google Scholar]
- 66.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, Y., G. Blandino, and D. Givol. 1999. Induced p21waf expression in H1299 cell line promotes cell senescence and protects against cytotoxic effect of radiation and doxorubicin. Oncogene 18:2643-2649. [DOI] [PubMed] [Google Scholar]
- 68.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 69.Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25:619-623. [DOI] [PubMed] [Google Scholar]
- 70.Xu, H. J., Y. Zhou, W. Ji, G. S. Perng, R. Kruzelock, C. T. Kong, R. C. Bast, G. B. Mills, J. Li, and S. X. Hu. 1997. Reexpression of the retinoblastoma protein in tumor cells induces senescence and telomerase inhibition. Oncogene 15:2589-2596. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, Z. K., K. P. Davies, J. Allen, L. Zhu, R. G. Pestell, D. Zagzag, and G. V. Kalpana. 2002. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol. Cell. Biol. 22:5975-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu, J., D. Woods, M. McMahon, and J. M. Bishop. 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 12:2997-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu, L., G. Enders, J. A. Lees, R. L. Beijersbergen, R. Bernards, and E. Harlow. 1995. The pRB-related protein p107 contains two growth suppression domains: independent interactions with E2F and cyclin/cdk complexes. EMBO J. 14:1904-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]