Enteropathogenic Escherichia coli bacterial load in stool samples measured by quantitative real-time polymerase chain reaction (qRT-PCR) is higher in children with diarrhea than in healthy control subjects. qRT-PCR may be useful for study of the relationship between disease and colonization in settings where the infection is endemic.
Abstract
Background. Enteropathogenic Escherichia coli (EPEC) strains are pediatric pathogens commonly isolated from both healthy and sick children with diarrhea in areas of endemicity. The aim of this study was to compare the bacterial load of EPEC isolated from stool samples from children with and without diarrhea to determine whether bacterial load might be a useful tool for further study of this phenomenon.
Methods. EPEC was detected by polymerase chain reaction (PCR) of colonies isolated on MacConkey plates from 53 diarrheal and 90 healthy children aged <2 years. DNA was isolated from stool samples by cetyltrimethylammonium bromide extraction. To standardize quantification by quantitative real-time PCR (qRT-PCR), the correlation between fluorescence threshold cycle and copy number of the intimin gene of EPEC E2348/69 was determined.
Results. The detection limit of qRT-PCR was 5 bacteria/mg stool. The geometric mean load in diarrhea was 299 bacteria/mg (95% confidence interval [CI], 77–1164 bacteria/mg), compared with 29 bacteria/mg (95% CI, 10–87 bacteria/mg) in control subjects (P = .016). Bacterial load was significantly higher in children with diarrhea than in control subjects among children <12 months of age (178 vs 5 bacteria/mg; P = .006) and among children with EPEC as the sole pathogen (463 vs 24 bacteria/mg; P = .006).
Conclusions. EPEC load measured by qRT-PCR is higher in diarrheal than in healthy children. qRT-PCR may be useful to study the relationship between disease and colonization in settings of endemicity.
Enteropathogenic Escherichia coli (EPEC) strains are among the most important pathogens infecting children worldwide, because they are common and are often associated with prolonged illness with its attendant risk of malnutrition [1]. These pathogens induce a distinctive histopathology known as attaching effacing (A/E) lesion, which is characterized by the intimate contact between bacteria and the epithelial surface of the enterocyte [2]. The protein intimin (eaeA), necessary for the A/E lesion, has been used for the molecular identification of EPEC. The current average prevalence of EPEC in pediatric diarrheal episodes in developing countries, using molecular methods, is 5%–10% [1] There is a trend, although often not statistically significant, for more frequent isolation of this pathogen among children with diarrhea than among healthy control subjects [3–6]. Recently, in a passive surveillance diarrhea cohort study involving 1034 Peruvian children, we isolated EPEC with a similar frequency from children with diarrhea (7.6%) and from asymptomatic controls (9.9%) [7]. However, multiple studies have established the virulence of EPEC, and remarkable progress has been made to identify the virulence determinants required to mediate the pathogenesis of these infections [8]. Our hypothesis was that presence of symptoms in EPEC infection relates to the bacterial load. The aims of this study were to develop, standardize, and validate a quantitative real-time polymerase chain reaction (qRT-PCR) assay for EPEC and to determine whether bacterial load relates to symptom status.
METHODS
Patients and Samples
Patients and samples included in this study were selected from a passive surveillance diarrhea cohort study conducted with 1034 children <2 year of age from Lima, Perú [7]. A total of 143 stool specimens previously found to be positive for EPEC by PCR of colonies isolated on MacConkey plates of stool culture were analyzed in this study. All stool samples had been tested for specific genes of all 6 pathotypes of diarrheagenic E. coli, including stx1 and sxt2 of Shigatoxin-producing E. coli [9]. We excluded for this study eaeA-positive samples positive for Shigatoxin (stx1 and stx2). Fifty-three stool specimens were from patients with diarrhea (26 aged <12 months and 27 aged ≥12 months), and 90 stool samples were collected from randomly selected healthy children without diarrhea 1 week before or after the stool sample collection (31 aged <12 months and 59 aged ≥12 months). Twenty-six samples were from children with a coinfection or cocolonization that included both an EPEC and another common enteric pathogen (Campylobacter, enteroaggregative E. coli, enterotoxigenic E. coli, or diffusely adherent E. coli). In 117 children, EPEC strains were the only pathogen isolated from the stool sample. Fresh stool samples were collected in capped containers and were stored at −20°C until evaluated by PCR. The prototypical strain EPEC E2348/69 was used as a positive control for standardization of the quantitative method.
Clinical information on the diarrheal episodes was obtained from the medical records filled by study personnel at the time of illness. We used a modified Vesikari score to determine the severity of an EPEC-associated diarrhea episode [7, 10]. The maximum possible score was 18. We defined illness as mild (score, 0–8 points), moderate (9–14 points), or severe (15–18 points).
DNA Isolation From Culture and Stool Samples
Genomic DNA was isolated from a pure culture of EPEC strain E2348/69, grown for 12–18 hours at 37°C, and extracted with the phenol-chloroform method. Ten-fold serial dilutions (equivalent to 101 to 106 bacilli) were prepared to determine the detection limit. According to GenBank (NC_011601), EPEC genome has 4 965 553 basepairs (bp), which is equivalent to 5.44 fg and represents 2 copies of eaeA gen. The DNA was isolated from stool specimens using a cetyltrimethylammoniumbromide extraction method modified from a method described elsewhere [11]. In brief, 50–300 mg of each stool specimen was dispersed in 250 μL of lysis buffer (0.25% sodium dodecyl sulfate in 0.1 M ethylenediaminetetraacetic acid [EDTA]; pH, 8.0), and 100 μg/mL of proteinase K was added. The lysate was incubated at 55°C for 2 hours. Then, 75 μL of 3.5 M sodium chloride (NaCl), followed by 42 μL of 10% CTAB/0.7 M NaCl (heated to 55°C), was added. After the components were mixed, the sample was incubated at 65°C for 30 minutes. This was followed by extractions with equal volumes of chloroform and then phenol-chloroform isoamyl alcohol (25:24:1), and the DNA was precipitated with ice-cold ethanol. The dried DNA pellet was dissolved in Tris-EDTA buffer and passed over a DNA clean-up spin column. The DNA was finally eluted from the spin column in 100 μL of Tris-EDTA 1× buffer; 2 μL of this DNA solution was used in the PCR.
RT-PCR Assay for Quantification of EPEC E2348/69
PCR was performed using a iCycler IQ™ Multicolor Real-Time PCR Detection System (Bio-Rad, California) monitoring by an Optical System Software Version 3.1. Each PCR assay contained 0.5 U of Phusion polymerase (Finnzyme OY) in high-fidelity Phusion buffer, with a final concentration of 350 μM of each deoxyribonucleotide triphosphates and 4 mM MgCl2. The primers used for the detection of the eaeA gene were forward 5′-ATGCTTAGTGCTGGTTTAGG-3′ and reverse 5′-GCCTTCATCATTTCGCTTTC-3′ [9], and they were used at final concentrations of 0.5 μM. SYBR Green I (Cambrex Bio Science) was diluted as recommended by the manufacturer and mixed with 10 nM fluorescein. Hot-start of 98°C for 30 seconds was used to prevent nonspecific amplification. The amplification protocol consisted of incubation at 98°C for 20 seconds, 61°C for 20 seconds, 72°C for 30 seconds, and 75°C for10 seconds. After 40 cycles, a melting curve using SYBR Green was determined with a ramp speed of 2.5°C per second at 73°C–95°C, with a reading every 0.2°C. Melting peaks were automatically calculated by the Opticon Monitor software 115, which plotted the negative derivative of fluorescence with respect to temperature (–d(F)/dT vs T). To make sure that we did not have inhibition of amplification because of the presence of contaminants in stool samples, we randomly selected some stool samples from children, mixed them with human DNA, and determined the presence of the ERV3 (endogenous retrovirus) gene as an internal control [12]. We obtained positive ERV3 results for all samples, including those that were negative for the eaeA gene.
Amplification Efficiencies and Quantitation of Bacterial Load
We constructed standard curves by using known quantities of genomic DNA (1.04 × 100 to 1.04 × 108 fg) extracted from EPEC E2348/69 samples. For standard curve analyses, the threshold cycles (CT) were plotted against the corresponding log of input quantity DNA determining the detection limit of the assay. For a comparison of PCR amplification efficiencies and detection sensitivities among experiments, slopes of the standard curves were calculated by a linear regression analysis with iCycler 3.1 software (Bio-Rad). Amplification efficiency (E) was estimated by using the slope of the standard curve and the formula E = (10 −1/slope) − 1. A reaction with 100% efficiency will generate a slope of −3.32. The amount of DNA was calculated as copies of eaeA gene per milligram of stool. From the quantification of the template DNA, an estimation of the relative bacterial load in the different samples was performed. To facilitate the interpretation, we have expressed the bacterial load as bacteria per milligram of stool instead of genome copies per milligram.
Statistical Analysis
To estimate bacterial load, an undetectable bacterial load in a previously positive isolated colony PCR was considered to be 0.01 bacteria/mg. To determine the differences in the bacterial load between diarrhea and control samples, the groups were compared using 2-sample Mann–Whitney U test. We fitted logistic regression models with random intercept to assess whether bacterial load is associated with diarrhea for children with EPEC as part of a coinfection or as a single infection. We tested in the models for effects of potential confounders, such as age, sex, breastfeeding status, and complementary feeding. Only those variables that remained significant to the .05 level were retained in the final model. We used Stata software, version 10 (StataCorp), for analysis.
Ethical Aspects
The study was approved by the institutional review boards of Universidad Peruana Cayetano Heredia and Instituto de Investigación Nutricional, Lima, Perú.
RESULTS
Standard Curves, Detection Limits, and Amplification Efficiencies
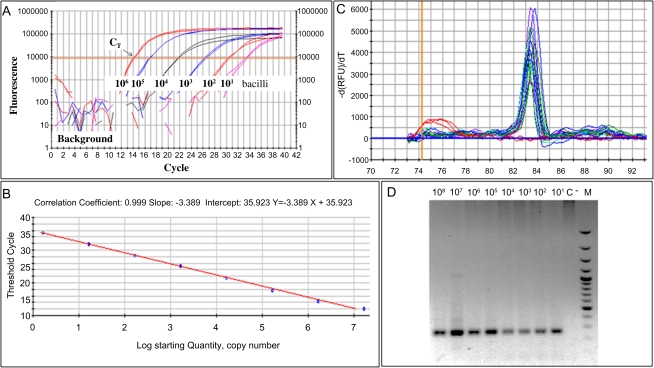
To demonstrate the detection range of the qRT-PCR, 10-fold serial dilutions containing 1.04 × 100 to 1.04 × 108 fg of genomic DNA mL−1 were assayed in triplicates for the eaeA gene of EPEC E2348/69 strain (Figure 1A). The results were reported as threshold cycle numbers versus log starting quantities of DNA. The correlation coefficient was 0.999, and the PCR efficiency was 97.3% (Figure 1B). The detection limit of this PCR assay (standard curve method) was 10 copies of eaeA gene/mg stool (equivalent to 5 bacteria/mg stool). The melting curve analysis of the qRT-PCR eaeA product is shown in Figure 1C. The mean melting temperature (SD)for the eaeA gene was 84.3°C (0.3°C). The electrophoresis ran for the PCR products showed a unique specific band of 248 pb corresponding to the eaeA gene (Figure 1D).
Figure 1.
Quantitative real-time polymerase chain reaction (RT-PCR) standardization for enteropathogenic Escherichia coli (EPEC). A, RT-PCR results from representative experiments using DNA from a pure culture of EPEC E2348/69. Fluorescence from the PCR products is plotted against the corresponding number of copies of intimin (eaeA) gene, corresponding to 101 to 106 bacilli, to obtain the threshold cycle (CT). B, Standard curve for the RT-PCR analysis was done from the same stock of DNA diluted 10-fold. We plotted CT against the log of the number of eaeA copies; the reaction efficiency was >97.3%. C, The melting temperature for the eaeA gene was 83.8 ± 0.23°C; curves are superimposed for the different DNA concentrations used in the analysis. D, Agarose gel (2%): (1–8) PCR products (248 pb) corresponding to the 10-fold dilutions (108 to 101 bacilli); C, No template control; (M) 100-bp molecular weight ladder.
Quantification of EPEC in Stool Samples
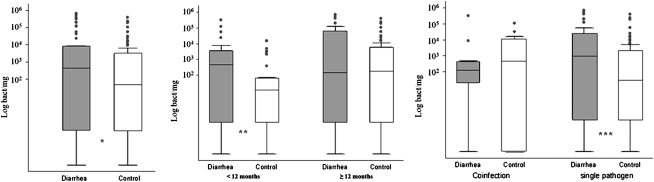
EPEC load in stool samples from infected or colonized children with a previously isolated colony positive by PCR ranged from ≤10 bacteria/mg to 4.03 × 106 bacteria/mg. The quantitative data showing EPEC load in diarrheal and healthy children are shown in Figure 2 and Table 1. EPEC load was significantly higher (P = .016) in the diarrhea group than in the control group (Figure 2A). We found 46 samples with a fecal bacteria load ≤10 bacteria/mg, from which 71% were from healthy infants (33 samples). Because most reports on diarrheagenic E. coli show age-dependent differences in incidence rates, we divided the samples according to age groups (Figure 2B). Among children <12 months of age, the bacterial load in the diarrhea group was significantly higher than that in the control group (P = .006); there were no statistically significant differences among older children. Coinfections were common, accounting for 13% of diarrheal samples and 6% of control samples; there was not a statistically significant difference in the EPEC load between diarrhea and control groups when present as part of a mixed infection (Figure 2C). There were no differences in the age distribution among sole pathogen and coinfection groups. The most common coinfections were EPEC with Campylobacter (n = 3) and EPEC with enterotoxigenic E. coli (n = 3). However, among stool samples with EPEC as a single pathogen, the bacterial load was significantly higher in diarrhea than in control samples (463 vs 24 bacteria/mg; P = .006; Figure 2C). Of interest, we have not found differences in the mean number of positive EPEC colonies per patient (5 colonies were collected per patient from the MacConkey plate) from the initial PCR between diarrhea and control samples. Similarly, there was no correlation between the number of positive colonies per patient from the initial PCR from the MacConkey plate and the bacterial load determined directly from the stool samples.
Figure 2.
Comparison of enteropathogenic Escherichia coli (EPEC) load among diarrhea and control samples. A, Diarrhea samples (gray bars) and samples from healthy control subjects (white bars). EPEC load in stool samples from children with (n = 53) and without (n = 90) diarrhea; *P = .016. B, EPEC load in children <12 months of age (diarrhea, n = 26; control, n = 31) and children 12–24 months of age (diarrhea, n = 27; control, n = 59); **P = .006. C, EPEC load in coinfections (diarrhea, n = 9; control, n = 13) and single-pathogen infection (diarrhea, n = 44; control, n = 77); ***P = .006.
Table 1.
Enteropathogenic Escherichia coli Load in Stool Samples From Children With Diarrhea and Without Diarrhea (Control), Measured by Quantitative Real-time Polymerase Chain Reaction
| Diarrhea geometric mean (95% CI) bacteria/mg | Control geometric mean (95% CI) bacteria/mg | P value | |
| All stool samples | 299 (77–1164) | 29 (9–87) | .016 |
| (n = 53) | (n = 90) | ||
| By age group | |||
| <12 months | 178 (27–1147) | 5 (1–26) | .006 |
| (n = 26) | (n = 31) | ||
| ≥12 months | 220 (18–2685) | 29 (5–156) | .188 |
| (n = 27) | (n = 59) | ||
| By type of EPEC infection | |||
| Sole pathogen isolated | 463 (97–2221) | 24 (7–81) | .006 |
| (n = 44) | (n = 77) | ||
| Coinfection | 44 (1–1289) | 35 (1–1657) | .732 |
| (n = 9) | (n = 13) |
Abbreviations: CI, confidence interval; EPEC, enteropathogenic Escherichia coli.
EPEC Load and Clinical Data
Among diarrhea cases, stool samples were collected on a variable number of days into illness. Overall, there was a tendency for a higher bacterial load during the first days of illness (Table 2). Complete clinical information was available in 39 episodes; 29 (74%) corresponded to a mild episode, 9 (23%) to a moderate episode, and 1 (3%) to a severe episode. The bacterial load was similar between mild and moderate cases (144 vs 95 bacteria/mg; P = .722). The duration of the diarrhea was known in 45 episodes; the mean duration (SD) was 6.8 (5.9) days (median, 5 days; range, 1–24 days). Twenty-eight episodes lasted <7 days, 10 lasted 7–14 days, and 7 lasted >14 days. We did not find any difference in the bacterial load as related to the duration of the episode (444 vs 184 vs 146 bacteria/mg for <7, 7–14, and >14 days, respectively). We also explored bacterial load as related to breastfeeding of the child at the time of the sample collection. Ninety-six children were breastfeeding (37 diarrheal and 59 controls), whereas 46 were not (16 diarrheal and 30 controls). Among children breastfeeding, there were no statistically significant differences in the bacterial load of diarrheal and control samples (299 vs 54 bacteria/mg; P = .133). Among children who were not breastfeeding at the time of sampling, a significantly higher bacterial load was found in diarrhea samples than in healthy control samples (300 vs 6 bacteria/mg, respectively; P = .038).
Table 2.
Enteropathogenic Escherichia coli Load in Relation to the Day of Illness
| Day of illness | No. (%) of samplesa | Geometric mean (95% CI) bacteria/mg |
| 1 | 4 (9) | 855 (.2–4 028 568) |
| 2 | 10 (22) | 154 (.7–34 157) |
| 3 | 9 (20) | 384 (4–34 476) |
| 4 | 6 (13) | 283 (11–7482) |
| 5 | 7 (15) | 149 (2–12 685) |
| 6 | 1 (2) | 3520 |
| ≥7 | 9 (20) | 188 (15–2318) |
Abbreviation: CI, confidence interval.
Forty-six samples were included; 7 samples were excluded because there was no recorded date of the beginning of the episode.
Association Between Bacterial Load and Diarrhea
The coefficients for the logistic regression with random effects for children with EPEC as part of a coinfection or as a single infection were determined after adjusting for age, sex, and an interaction of both. For a given child, the odds of diarrhea increased by 29% (odds ratio, 1.29; 95% CI, 1.08–1.53) for each log10 unit increase in bacterial load in the stool sample.
DISCUSSION
The interpretation of the isolation rate of a pathogen in stool samples from children with diarrhea, compared with asymptomatic colonization, is complex. Factors that should be considered in interpretation include bacterial factors (bacteria with more virulent genes), host factors (age, genetic susceptibility, previous exposure, development of immunity, and protective factors from breastfeeding), and environmental factors (poor hygiene and high and frequent fecal contamination). With the development of new diagnostic methods, such as RT-PCR, the detection limit is now very low; therefore, detection of pathogens can occur in samples from healthy control children. Our results show that EPEC load was significantly higher in children with diarrhea than in asymptomatic control subjects, especially in single pathogen. Therefore, we propose that, in addition to the aforementioned factors, the bacterial load should be considered in the interpretation of illness and colonization of pathogens in the gut.
Several epidemiological studies from developing countries have been conducted in children to determine age-related differences in diarrheagenic E. coli infections [7, 13–19]. Similar to these studies, we found a relationship between age and symptoms. Our analysis showed that, in children <12 months of age with diarrhea, the bacterial load was higher than that in healthy control subjects infected with the same pathogen, consistent with the generally accepted view that EPEC is a true pathogen in young children. There appears to be a reduced threshold of bacterial load required for the initiation of EPEC infection in younger infants; however, the bacterial load in patients with diarrhea appears to remain unchanged in younger and older infants, probably related to presence or absence of protective factors.
In developing countries, mixed infections are common, especially in diarrheal samples [7], making it difficult to determine which pathogen is responsible for symptoms. In this study, among mixed infections, we found a similar EPEC load in children with and without diarrhea, suggesting that EPEC may not have had a role in these episodes of diarrhea. However, among stool samples with EPEC as a single-pathogen infection, we found a strong association between EPEC and diarrhea, shown by the higher bacterial load in children with diarrhea than in healthy controls. This suggests that EPEC is a true pathogen when present as the sole pathogen isolated; however, in mixed infection, we hypothesize that higher EPEC loads would be required to overcome a mixed infection. Similarly, in children breastfeeding at the time of the sample collection, there was no difference in the bacterial load in diarrhea and control samples. However, in children not breastfeeding, we found a higher bacterial load in children with diarrhea. This finding supports the notion that breast milk protects these children from symptomatic infection [20–23]. Conversely, breast milk may have decreased the load but not totally prevented EPEC-related symptoms in these children. Further studies will be needed to confirm each of these important conclusions.
With the implementation of molecular tools, the detection limit of pathogens has improved, and quantification is now possible [24]. In this study, we developed, standardized, and validated a qRT-PCR for EPEC in stool samples. The approach reported here is not entirely new. RT-PCR assays have been developed in the past for the detection and quantification of enteropathogens in food, animal, and clinical samples [25], including assays for Campylobacter, Cryptosporidium, Salmonella, E. coli O157:H7, and other Shigatoxin-producing E. coli [26–32]. Overall, the most common DNA isolation method has been the use of commercial kits, which are expensive and are not easily available in developing countries. The current study proposes the application of a bromide-extraction method as an alternative DNA isolation method, which is cheaper than commercial kits. It should be noted that this study is, to our knowledge, the first study that applies a qRT-PCR analysis for EPEC in human samples and compares stool samples from children with and without diarrhea.
This study has some limitations. First, we did not search for all possible enteric pathogens (norovirus, astrovirus, enteric adenovirus, Yersinia, and parasites) to define the single-pathogen infection with EPEC. This could account for some overlap in groups of patients with only EPEC infection found, because in some cases, unrecognized mixed infections might have been present. However, we evaluated stool samples for the most common pathogens in children (rotavirus, all diarrheagenic E. coli, Shigella, Salmonella, and Campylobacter). Second, we did not evaluate factors that may explain why EPEC was excreted in asymptomatic individuals (eg, protective antibody levels, lower ingestion dose, transplacental maternal immunity, and zinc levels). Third, the number of diarrheal samples was not sufficient to optimally determine the relationship between bacterial load and duration of illness and diarrhea severity or the effect of breastfeeding and age on bacterial load. Future studies are needed with larger numbers of EPEC diarrheal samples to clarify these important issues.
In summary, we developed, standardized, and validated a qRT-PCR for EPEC in stool samples with use of an inexpensive bromide DNA extraction method that can be used as a diagnostic tool to clarify the differences between colonization and illness. We found that the bacterial load of EPEC, measured by qRT-PCR of stool samples, is higher in children with diarrhea than in healthy control subjects. We have identified areas, outlined above, for further investigation using this approach.
Notes
Financial support.
This work was supported by a Public Health Service award (grants 1K01TW007405 to T. J. O. and R01-HD051716 to T. G. C.) from the National Institutes of Health; by Agencia Española de Cooperación Internacional para el Desarrollo, Spain (D/019499/08 and D/024648/09 to J. R. and T. J. O.); and by Dr Lanata’s Institutional Research Funds.
Potential conflicts of interest.
All authors:No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Ochoa TJ, Barletta F, Contreras C, Mercado E. New insights into the epidemiology of enteropathogenic Escherichia coli infection. Trans R Soc Trop Med Hyg. 2008;102:852–6. doi: 10.1016/j.trstmh.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen HD. Frankel G. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol Rev. 2005;29:83–98. doi: 10.1016/j.femsre.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Alikhani MY, Mirsalehian A, Aslani MM. Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J Med Microbiol. 2006;55:1159–63. doi: 10.1099/jmm.0.46539-0. [DOI] [PubMed] [Google Scholar]
- 4.Araujo JM, Tabarelli GF, Aranda KR, et al. Typical enteroaggregative and atypical enteropathogenic types of Escherichia coli are the most prevalent diarrhea-associated pathotypes among Brazilian children. J Clin Microbiol. 2007;45:3396–9. doi: 10.1128/JCM.00084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol. 2005;43:755–60. doi: 10.1128/JCM.43.2.755-760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rappelli P, Folgosa E, Solinas ML, et al. Pathogenic enteric Escherichia coli in children with and without diarrhea in Maputo, Mozambique. FEMS Immunol Med Microbiol. 2005;43:67–72. doi: 10.1016/j.femsim.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Ochoa TJ, Ecker L, Barletta F, et al. Age-related susceptibility to infection with diarrheagenic Escherichia coli among infants from periurban areas in Lima, Peru. Clin Infect Dis. 2009;49:1694–702. doi: 10.1086/648069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contreras CA, Ochoa TJ, Lacher DW, et al. Allelic variability of critical virulence genes (eaeA, bfpA and perA) in typical and atypical enteropathogenic Escherichia coli in Peruvian children. J Med Microbiol. 2010;59:25–31. doi: 10.1099/jmm.0.013706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guion CE, Ochoa TJ, Walker CM, Barletta F, Cleary TG. Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. J Clin Microbiol. 2008;46:1752–7. doi: 10.1128/JCM.02341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruuska T, Vesikari T. Rotavirus disease in Finnish children: use of numerical scores for clinical severity of diarrhoeal episodes. Scand J Infect Dis. 1990;22:259–67. doi: 10.3109/00365549009027046. [DOI] [PubMed] [Google Scholar]
- 11.Sharma AK, Chibbar S, Bansal G, Kaur U, Vohra H. Evaluation of newer diagnostic methods for the detection and differentiation of Entamoeba histolytica in an endemic area. Trans R Soc Trop Med Hyg. 2003;97:396–7. doi: 10.1016/s0035-9203(03)90068-x. [DOI] [PubMed] [Google Scholar]
- 12.Yuan CC, Miley W, Waters D. A quantification of human cells using an ERV-3 real time PCR assay. J Virol Methods. 2001;91:109–17. doi: 10.1016/s0166-0934(00)00244-5. [DOI] [PubMed] [Google Scholar]
- 13.Albert MJ, Faruque AS, Faruque SM, Sack RB, Mahalanabis D. Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. J Clin Microbiol. 1999;37:3458–64. doi: 10.1128/jcm.37.11.3458-3464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine MM, Ferreccio C, Prado V, et al. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am J Epidemiol. 1993;138:849–69. doi: 10.1093/oxfordjournals.aje.a116788. [DOI] [PubMed] [Google Scholar]
- 15.Orlandi PP, Magalhães GF, Matos NB, et al. Etiology of diarrheal infections in children of Porto Velho (Rondonia, western Amazon region, Brazil) Braz J Med Biol Res. 2006;39:507–17. doi: 10.1590/s0100-879x2006000400011. [DOI] [PubMed] [Google Scholar]
- 16.Porat N, Levy A, Fraser D, Deckelbaum RJ, Dagan R. Prevalence of intestinal infections caused by diarrheagenic Escherichia coli in Bedouin infants and young children in southern Israel. Infect Dis J. 1998;17:482–8. doi: 10.1097/00006454-199806000-00010. [DOI] [PubMed] [Google Scholar]
- 17.Quiroga M, Oviedo P, Chinen I, et al. Asymptomatic infections by diarrheagenic Escherichia coli in children from Misiones, Argentina, during the first twenty months of their lives. Rev Inst Med Trop Sao Paulo. 2000;42:9–15. doi: 10.1590/s0036-46652000000100002. [DOI] [PubMed] [Google Scholar]
- 18.Ratchtrachenchai OA, Subpasu S, Hayashi H, Ba-Thein W. Prevalence of childhood diarrhoea-associated Escherichia coli in Thailand. J Med Microbiol. 2004;53:237–43. doi: 10.1099/jmm.0.05413-0. [DOI] [PubMed] [Google Scholar]
- 19.Spano LC, Sadovsky AD, Segui PN, et al. Age-specific prevalence of diffusely adherent Escherichia coli in Brazilian children with acute diarrhoea. J Med Microbiol. 2008;57:359–63. doi: 10.1099/jmm.0.47660-0. [DOI] [PubMed] [Google Scholar]
- 20.Duijts L, Jaddoe VW, Hofman A, Moll HA. Prolonged and exclusive breastfeeding reduces the risk of infectious diseases in infancy. Pediatrics. 2010;126:e18–25. doi: 10.1542/peds.2008-3256. [DOI] [PubMed] [Google Scholar]
- 21.Mihrshahi S, Oddy WH, Peat JK, Kabir I. Association between infant feeding patterns and diarrhoeal and respiratory illness: a cohort study in Chittagong, Bangladesh. Int Breastfeed J. 2008;24:28–37. doi: 10.1186/1746-4358-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plenge-Bönig A, Soto-Ramírez N, Karmaus W, Petersen G, Davis S, Forster J. Breastfeeding protects against acute gastroenteritis due to rotavirus in infants. Eur J Pediatr. 2010;169:1471–6. doi: 10.1007/s00431-010-1245-0. [DOI] [PubMed] [Google Scholar]
- 23.Morrow AL, Rangel JM. Human milk protection against infectious diarrhea: implications for prevention and clinical care. Semin Pediatr Infect Dis. 2004;15:221–8. doi: 10.1053/j.spid.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–94. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 25.Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166–77. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- 26.Persson S, Olsen KE. Multiplex PCR for identification of Campylobacter coli and Campylobacter jejuni from pure cultures and directly on stool samples. J Med Microbiol. 2005;54:1043–7. doi: 10.1099/jmm.0.46203-0. [DOI] [PubMed] [Google Scholar]
- 27.Parr JB, Sevilleja JE, Samie A, et al. Detection and quantification of Cryptosporidium in HCT-8 cells and human fecal specimens using real-time polymerase chain reaction. Am J Trop Med Hyg. 2007;76:938–42. [PMC free article] [PubMed] [Google Scholar]
- 28.Pusterla N, Byrne BA, Hodzic E, Mapes S, Jang SS, Magdesian KG. Use of quantitative real-time PCR for the detection of Salmonella spp. in fecal samples from horses at a veterinary teaching hospital. Vet J. 2010;186:252–5. doi: 10.1016/j.tvjl.2009.08.022. Epub 17 September 2009. [DOI] [PubMed] [Google Scholar]
- 29.Oberst RD, Hays MP, Bohra LK, et al. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with internal fluorogenic probe and the 5 nuclease (TaqMan) assay. Appl Environ Microbiol. 1998;64:3389–96. doi: 10.1128/aem.64.9.3389-3396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma VK, Carlson SA. Simultaneous detection of Salmonella strains and Escherichia coli O157:H7 with fluorogenic PCR and single-enrichment-broth culture. Appl Environ Microbiol. 2000;66:5472–6. doi: 10.1128/aem.66.12.5472-5476.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibekwe AM, Watt PM, Grieve CM, Sharma VK, Lyons SR. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands. Appl Environ Microbiol. 2002;68:4853–62. doi: 10.1128/AEM.68.10.4853-4862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bellin T, Pulz M, Matusset A, Hempen HG, Gunzer F. Rapid detection of enterohemorrhagic Escherichia coli by real-time PCR with fluorescent hybridization probes. J Clin Microbiol. 2001;36:370–4. doi: 10.1128/JCM.39.1.370-374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]