Abstract
Novel low-power adiabatic sequences are demonstrated for in-vivo localized two-dimensional (2D) correlated MR spectroscopy, such as COSY (Correlated Spectroscopy) and TOCSY (Total Correlated Spectroscopy). The design is based on three new elements for in-vivo 2D MRS: the use of gradient modulated constant adiabaticity GOIA-W(16,4) pulses for i) localization (COSY and TOCSY) and ii) mixing (TOCSY), and iii) the use of longitudinal mixing (z-filter) for magnetization transfer during TOCSY. GOIA-W(16,4) provides accurate signal localization, and more importantly, lowers the SAR for both TOCSY mixing and localization. Longitudinal mixing improves considerably (five-folds) the efficiency of TOCSY transfer. These are markedly different from previous 1D editing TOCSY sequences using spatially non-selective pulses and transverse mixing. Fully adiabatic (adiabatic mixing with adiabatic localization) and semi-adiabatic (adiabatic mixing with non-adiabatic localization) methods for 2D TOCSY are compared. Results are presented for simulations, phantoms, and in-vivo 2D spectra from healthy volunteers and patients with brain tumors obtained on 3T clinical platforms equipped with standard hardware. To the best of our knowledge this is the first demonstration of in-vivo adiabatic 2D TOCSY and fully adiabatic 2D COSY. It is expected that these methodological developments will advance the in-vivo applicability of multi(spectrally)dimensional MRS to reliably identify metabolic biomarkers.
Keywords: Total Correlated Spectroscopy (TOCSY), Correlated Spectroscopy (COSY), Gradient Offset Independent Adiabatic (GOIA), in-vivo 2D MRS
1. INTRODUCTION
While two-dimensional (2D) or even higher dimensional (3D, 4D) NMR spectroscopy comprise a large panoply of well established methods in chemistry (1) and structural biology (2), where they provide a wealth of structural information, the development of in-vivo multidimensional (2D) MR spectroscopy lags behind (by multidimensional MRS we mean MRS with multiple spectral dimensions, as opposed to MRS with multiple spatial dimensions, known as MR spectroscopic imaging (MRSI)). For the in-vitro applications, especially in the case of crowded biomolecular systems such as proteins (3–5) or brain biopsies (6), these methods are crucial for disentangling overlapped signals and obtaining unambiguous assignments and structural constraints. A similar situation exists in-vivo, with many metabolites overlapping in a reduced chemical shift range. Although there are considerably less visible in-vivo metabolites (approximately twenty are described for brain (7,8)) than aminoacids in a protein (several hundreds or more), the overlapping may be as severe in-vivo due to reduced chemical shift dispersion (lower B0 fields) and lower spectral resolution. Hence, complex fitting routines (9,10) have been developed to extract the metabolic information from one dimensional in-vivo spectra. 2D in-vivo MR spectroscopy could help in reliable assignment, quantification, and perhaps, identification of new metabolites (more than 60 brain metabolites are reported (11–13) by multidimensional NMR spectroscopy from tissues and cell cultures, which can be used for advanced molecular typing of brain tumors (14) or other diseases). To date, despite early interest, mainly two of the 2D NMR experiments, 2D COSY (15,16) and 2D J-resolved (17), have been adapted and mostly used for in-vivo 2D MR spectroscopy (exchange NOESY spectroscopy (18) has been shown in-vivo (19) but less applied). Each of the in-vivo 2D sequences comes in several different versions such as 2D J-PRESS (20,21), S-PRESS (22), CT-PRESS (23), and L-COSY (20,24,25), including the possibility of multiple-quantum (20) filtering (for recent reviews see Refs. (26–28)). This is largely due to the challenges of in-vivo MRS, such as lower RF power deposition (SAR), reduced SNR, shorter acquisition time, and the hardware of the clinical scanners. However, continuous improvements in pulse design and clinical hardware allow currently more advanced approaches.
In this work we demonstrate, for the first time in-vivo, the localized 2D TOCSY (Total Correlation Spectroscopy) experiment. Although, 1D editing versions of in-vivo TOCSY with adiabatic (29,30) and non-adiabatic (31) mixing have been reported before, a 2D version has not been communicated yet. TOCSY performed in a 2D mode retains all the metabolites compared to the 1D editing which filters out most of the metabolites in order to detect a specific metabolite. Besides extending the number of dimensions, in our adiabatic 2D approach we make considerable changes from previous adiabatic 1D editing sequences (29,30), that enable us to improve the efficiency of magnetization transfer and allow us to run the 2D experiment in a similar amount of time as the 1D edited. Specifically, we introduce three new elements in the design of the in-vivo fully adiabatic TOCSY pulse sequence: i) gradient modulated constant adiabaticity GOIA-W(16,4) pulses (32) for LASER (Localized Adiabatic Selective Refocusing (33)) localization, ii) GOIA-W(16,4) pulses for TOCSY mixing, and iii) the use of a z-filter (34,35) to enable longitudinal TOCSY mixing and magnetization transfer between coupled spins. GOIA-W(16,4) pulses require lower B1 field amplitude which reduces the power deposition (specific absorption rate, SAR) during LASER localization, but more importantly during the TOCSY mixing, hence allowing shorter TR times. TOCSY mixing on longitudinal magnetization is five times more efficient than mixing on transverse magnetization. Adiabatic pulses are particularly useful for in-vivo MRS because they have been shown (33,36,37) to provide sharp and uniform excitation slices with virtually no chemical shift displacement error (CSDE) or flip angle dependence on the B1 field. In addition, adiabatic pulses are able to enhance the efficiency of TOCSY mixing by better averaging-out the chemical shift interaction (6,38,39).
Compared to other methods such as 2D COSY or 2D J-resolved, the 2D TOCSY (40–42) is one of the most powerful experiments because it reveals the full connectivity of a spin network. For example, 2D COSY contains information only about the direct scalar coupled spins, while 2D TOCSY provides correlations for both direct and indirect coupled spins (via relayed transfer). This is useful, as in some cases metabolites might have pairs of neighboring spins with similar chemical shifts. In the case of 2D J-resolved spectroscopy the indirect (F1) dimension measuring the scalar couplings has very limited range (up to 20 Hz for protons) compared to the range of chemical shift (several hundreds Hz up to kHz) available in the F1 dimension of 2D TOCSY or 2D COSY experiments. Moreover, the scalar couplings do not change with increasing magnetic B0 field, while the chemical shifts increase linearly with the B0 field, so the benefit of high field can not be fully realized in 2D J-resolved spectroscopy. The potential of TOCSY has not been fully exploited in-vivo due to the more intense SAR associated with the requirement of a sustained train of RF pulses during TOCSY mixing.
Several sequence design strategies are proposed and compared in our paper. A fully adiabatic 2D TOCSY which uses LASER localization, and two semi-adiabatic 2D TOCSY sequences that use either PRESS (Point Resolved Spectroscopy (43)) or STEAM (Stimulated Echo (44)) localization are presented. In all sequences the TOCSY is performed using an adiabatic MLEV-16 scheme (45) and a gradient-enhanced z-filter (46,47) which enables mixing of longitudinal magnetization, hence we name our sequences Z-TOCSY-LASER, Z-TOCSY-PRESS and Z-TOCSY-STEAM, respectively. In addition, a sequence that performs simultaneously localization and transverse TOCSY mixing (named LT-TOCSY) is proposed. In all our 2D sequences we implemented the possibility to obtain phase-sensitive spectra with an echo-antiecho acquisition. This enables the full potential of 2D spectra, by obtaining the maximum possible sensitivity and spectral resolution.
Our sequences can be run also in the 1D editing mode, and this was used in the tune-up process. The efficiency of magnetization transfer was compared in the 1D edited mode for Z-TOCSY-LASER and the TOCSY-LASER that uses transverse mixing (as originally proposed in Ref. (29)).
A fully adiabatic 2D COSY that uses the same LASER localization like the Z-TOCSY-LASER sequence is also shown. A fully adiabatic in-vivo 2D COSY has not been demonstrated yet, although a semi-adiabatic approach has been recently communicated (48). The 2D COSY-LASER was compared also with the 2D L-COSY method (24) that uses non-adiabatic pulses.
Theoretical analysis and simulations that form the basis of our TOCSY sequence design are included. We present results from phantoms, healthy volunteers and patients with brain tumors (glioblastoma, GBM) obtained on 3T clinical scanners equipped with standard hardware.
2. THEORY
Editing pulses for the purpose of in-vivo MRS (28) are generally spatially non-selective and they are applied on the entire imaged object. Similarly, in previous examples (29,30) of the 1D edited TOCSY experiments the mixing is performed spatially non-selective. In our approach of 2D TOCSY we take advantage of the ability of gradient modulation to reduce the B1 amplitude and shorten the duration of adiabatic pulses for increased bandwidths (32,36). Although spatial localization is not necessary for editing or mixing, there is not any reason why mixing and localization can not be done simultaneously, using gradients and RF pulses together, when this can be turned in advantage. This can be the concept of a more general principle and utility in sequence design (49). The theoretical framework relevant to the simulations and sequence development is shortly presented.
The efficiency of a particular TOCSY mixing scheme can be understood through the buildup curves of magnetization transfer between scalar coupled spins which for a simple two spin system are governed by the Hamiltonian
| [1] |
assuming a mixing train of Np gradient modulated adiabatic pulses with the B1,k (t) amplitude modulation, φk (t) phase modulation, Gk (t) gradient modulation, and Ωn (n = 1,2) and J represent the chemical shifts and the scalar coupling, respectively (in frequency units), Îα,n (α = x,y,z; n = 1,2) denote the individual spin operators, and F̂α (α = x,y,z) the total spin system operators (F̂α = Îα,1 + Îα,2). In the case of most mobile metabolites we neglect the influence of anisotropic interactions such as chemical shielding anisotropy and dipolar couplings on the coherent evolution of the spin system (however, these interactions are important for relaxation mechanisms as discussed later).
RF pulses with a constant adiabatic factor Q can be obtained if the modulation functions fulfill the condition (32,36)
| [2] |
, where ωc is the center sweep frequency and φk (0) represents the initial phase of the pulse, which in our case follows the MLEV-16 scheme (i.e. φk (0) = 0 or π). In particular, we employ the GOIA-W(16,4) pulses defined in (32), that use the WURST-16 function (50) for B1(t) modulation and inverse WURST-4 function for G(t) modulation.
The buildup curves for magnetization transfer can be calculated through density matrix formalism, assuming that initially only one of the spins has magnetization (i.e. initial density matrix σ̂(0) = Îz,1 for longitudinal mixing, or σ̂(0) = Îx,1 for transverse mixing) and detecting the magnetization transferred on the second spin (Îz,2 or Îx,2) for increasing mixing times. In practice numerical integration offers an easier approach than analytical tools, such as Average Hamiltonian Theory (51) or Floquet Theory (52) for time dependent Hamiltonians.
The incoherent spin evolution (relaxation) during GOIA-W(16,4) pulses occurs in the rotating frame of the effective field , where B1(t) and G(t) are modulated according to WURST-16 and (inverse) WURST-4 functions (50), respectively, and frequency modulation is obtained from . In the case of protons the homonuclear dipolar interaction is the dominant relaxation mechanism, and considering the main pool of the free low-molecular-weight metabolites the dynamics is in the fast regime with regard to the effective field (ωeffτc << 1, τc is the correlation time in the range of 1 ps - 1 ns, and ωeff is in the range of 1–20 kHz for all RF pulses that are practically attainable in-vivo). Under these conditions, the time- and position-dependent (instantaneous) rotating frame relaxation rates (53) for longitudinal mixing (R1ρ,dd) and transverse mixing (R2ρ,dd) can be approximated as:
| [3] |
, and
| [4] |
where α (t, z) = arctan [γB1 (t)/(Δ ω(t)−γ zG(t))] is the polar angle of the effective field in the laboratory frame, and D = ℏμ0 γ2/4πr3 is the dipolar coupling constant (ℏ is Planck’s constant, μ0 is the magnetic permeability, γ is the gyromagnetic ratio, r is the intra-molecular distance between the two spins, z is the molecule position along the slice director, and ω0 = γ B0 is the Larmor frequency,).
From Eq.[3] and Eq.[4] it can be easily checked that at any time R2ρ,dd (t, z) > R1ρ,dd (t, z), hence during the entire MLEV-16 train of GOIA-W(16,4) pulses the rotating frame relaxation for longitudinal mixing (assuming GOIA-W(16,4) pulses of duration Tp and slice thickness Δz)
| [5] |
is more favorable (smaller relaxation rate) than rotating frame relaxation for transverse mixing
| [6] |
3. METHODS
3.1. Numerical Simulations
Quantum mechanical simulations were performed in GAMMA (54) for the buildup curves of magnetization transfer under TOCSY mixing. We assumed a simple two spin system at 3T having the chemical shifts and scalar coupling of lactate (Ω 1 = 1.3 ppm, Ω2 = 4.1 ppm, J = 6.93 Hz from Ref. (55)) which is subjected to the MLEV-16 scheme using GOIA-W(16,4) pulses. We investigated several conditions to find low-power GOIA-W(16,4) pulses that provide magnetization transfer. For all pulses we assumed the same (minimum) adiabatic factor or time-bandwidth product (R = 20): i) duration Tp = 2 ms, bandwidth BW = 10 kHz, maximum RF amplitude γB1,max = 0.76 kHz, ii) Tp = 1.5 ms, BW = 13.34 kHz, γB1,max = 1.02 kHz, and iii) Tp = 1.25 ms, BW = 16 kHz, γB1,max = 1.22 kHz. A summary of the relevant parameters for the adiabatic pulses used in simulations and experiments are given in Table 1.
Table 1.
Parameters for adiabatic RF pulses used in simulations and experiments
| Sequence block | RF pulse | B1 modulation | G modulation(a) | Tp (ms) |
BW (kHz) |
γB1,max (kHz) |
|---|---|---|---|---|---|---|
| TOCSY (high-power) | GOIA-W(16,4) | WURST-16 | WURST-4 | 1.25 | 16 | 1.22 |
| TOCSY (medium-power) | GOIA-W(16,4) | WURST-16 | WURST-4 | 1.5 | 13.34 | 1.02 |
| TOCSY (low-power) (b) | GOIA-W(16,4) | WURST-16 | WURST-4 | 2.0 | 10 | 0.76 |
| LASER | GOIA-W(16,4) | WURST-16 | WURST-4 | 3.5 | 20 | 0.82 |
for all pulses an inverse WURST-4 function was used with a gradient reduction factor f = 0.9 in the middle of the pulse;
the condition used for in-vivo MRS.
In all simulations the spin evolution was calculated using a piece-wise constant Hamiltonian. A minimum time step of 20 μs was found to be sufficient in simulations that produce the same results for shorter time steps (i.e. the number of points in the pulse shapes were, 100 for 2 ms, 75 for 1.5 ms, and 62 for 1.25 ms, which in all cases exceeded several times the minimum number of points to accurately reproduce the adiabatic pulse (36)). The amplitude, gradient, and phase modulation for GOIA-W(16,4) pulses was obtained as previously described (32). Due to the use of gradient modulation which implies slice-selective properties of the GOIA-W(16,4) pulses, we assumed a one-dimensional object divided into a large number of infinitesimal sections. The offset (γzG) induced by the gradient was considered to be constant across an infinitesimal section and the spin evolution was calculated independently for each section. The final buildup signal was obtained by averaging across all sections. We considered both longitudinal and transverse mixing. In the case of transverse mixing we calculated also the slice profile of an MLEV-16 train of GOIA-W(16,4) pulses that acts also as a refocusing element. Symmetry of the pulse shapes and periodicity in the spin evolution were exploited to speed-up calculations.
3.2. Experiments
3.2.1. Pulse sequences
All the measurements were done on whole-body 3T Magnetom Tim Trio systems (Siemens, Erlangen). The RF body coil was used for transmit and a 32-channel phased array head coil (Siemens, Erlangen) was used for receive. The maximum amplitude of B1 field delivered by the transmit body coil is limited to 1 kHz (23.4 μT) for in-vivo applications. The whole-body gradient system (TQ-engine) was used, having the specifications of a maximum nominal amplitude of 26 mT/m and a maximum slew rate of 170 mT/(m*ms).
Pulse and gradient modulations were implemented in the IDEA environment (VB17A) using arbitrary classes and calculated inside the sequence preparation block. Pulse shapes had the same number of points as used in simulations. The gradient hardware needs a raster time of 10 μs which is faster than the RF raster time of 20 μs. To match the time steps an initial gradient shape is calculated with the same number of points as the RF shape (20 μs raster) and than expanded to a double number of points (10 μs raster) in the final shape by duplicating each point with itself. For selecting off-isocenter slices the shift in carrier frequency was implemented by calculating the corresponding phase modulation as detailed in (32).
For TOCSY mixing the individual GOIA-W(16,4) pulses were concatenated into larger shapes containing all 16 pulses according to the MLEV-16 scheme (45) in order to eliminate the gaps imposed by hardware for setting the frequency and phase of the RF, and the ramp-up/down of the gradients. The number of MLEV-16 supercycles (Nmix) can be repeated for the desired mixing time. Based on the results obtained from simulations and in order to reduce the SAR we used GOIA-W(16,4) pulses of 2 ms duration and 10 kHz bandwidth which require 0.76 kHz B1 maximum amplitude. The preferred slice orientation during TOCSY mixing is transverse due to slightly better performance of the Z gradient coil. At the beginning and the end of the MLEV-16 gradient shape the ramp-up and ramp-down were 200 μs long.
The voxel of interest can be localized using LASER for fully adiabatic sequences, or PRESS and STEAM in the case of semi-adiabatic sequences. For the LASER localization we used the method detailed in Ref. (32) (i.e. GOIA-W(16,4) pulses of duration Tp = 3.5 ms, bandwidth BW = 20 kHz, maximum B1 amplitude γB1,max = 0.82 kHz, echo time TE = 45 ms), while for PRESS (TE = 30 ms) and STEAM (TE = 20 ms) localization we used the standard sequences of Siemens. In particular, STEAM localization lends itself naturally for longitudinal TOCY mixing by inserting MLEV-16 between the last two 90° pulses. In the case of LASER or PRESS localization, a gradient enhanced z-filter (46,47) was introduced for longitudinal mixing by bracketing the TOCSY block with two non-selective 90° BIR-4 (56) adiabatic pulses (4 ms duration, 10 kHz bandwidth) and spoiler gradients (trapezoidal shape, 3 ms total duration, 500 μs ramp-up/down times, and 11.5 mT/m amplitude) for removing unwanted transverse magnetization. For STEAM the last two 90° pulses can act also as a z-filter, and only the spoiler gradients surrounding the MLEV-16 block need to be added. Selection of the coherence transfer pathway (CTP) is performed with sine-bell shaped gradients of 1 ms duration and 11.5 mT/m amplitude placed before and after the z-filter. For transverse mixing the z-filter is removed. Importantly, for longitudinal mixing the TOCSY block does not contribute to the echo time such as in the case of transverse mixing. Virtually, in the former case the total echo time is given by the echo time of the localization block (LASER, STEAM, PRESS).
For 2D acquisition, the time between the first 90° excitation pulse and the TOCSY block is incremented in repeated experiments to introduce the t1 evolution. An increment time of 0.8 ms, corresponding to 10 ppm spectral width at 3T for the F1 (indirect) dimension, was used.
While all the sequences are single shot and can be run as such in 1D mode, for the 2D experiment a phase cycle with a minimum two steps for the first 90° excitation pulse improves the spectra quality by removing axial peaks and t1 noise coming from the water signal that starts to recover during the t1 evolution (due to the need of averaging for SNR a minimum phase cycle is not a problem). Enhanced sensitivity (47) can be obtained if both coherence pathways during t1 evolution are retained through echo-antiecho acquisition of phase-sensitive spectra. For this, the phase (eae) of the first 90° BIR-4 pulse of the z-filter bracketing the longitudinal TOCSY mixing is alternated between x and −x simultaneously with alternating the polarity of the CTP gradient following the TOCSY from positive to negative (47). Each t1 increment is stored once with the echo data (x phase and positive polarity) and once with anti-echo data (−x phase and negative polarity) and data are processed accordingly. The 2D spectrum is obtained upon Fourier transformation in both time dimensions.
If needed, a larger slice thickness can be used for the GOIA-W(16,4) pulse employed during TOCSY compared to the size of the voxel selected by the localization schemes (LASER, PRESS, STEAM), in order to decrease eddy currents during the long mixing time.
The fully adiabatic 2D COSY is a simple extension of the LASER sequence, and we name it 2D COSY-LASER. A second non-selective 90° adiabatic BIR-4 pulse is inserted in the LASER sequence before the three pairs of refocusing GOIA-W(16,4) pulses. We note that the 2D COSY-LASER sequence represents the adiabatic version of the 2D COSY using the VSR (volume selective refocusing) localization block proposed in Ref. (25). The last BIR-4 pulse of the 2D COSY-LASER is surrounded by CTP gradients having the same characteristics mentioned above. The duration between the two 90° BIR-4 pulses is incremented in a similar fashion as described for 2D TOCSY. While the technique is essentially single shot, the same principles for phase cycle and echo-antiecho can be applied. The 2D L-COSY method using non adiabatic pulses and the minimum echo time of 30 ms, as previously described in (24), was used for comparison purposes.
Both Z-TOCSY-LASER and COSY-LASER sequences can be run also in a 1D editing mode by replacing the first non-selective 90° BIR-4 adiabatic pulse with a Gaussian selective pulse. We used a selective Gaussian pulse with 20 Hz bandwidth in the tune-up phase of the TOCSY sequence and we compared the efficiency of different TOCSY and COSY transfers in phantoms.
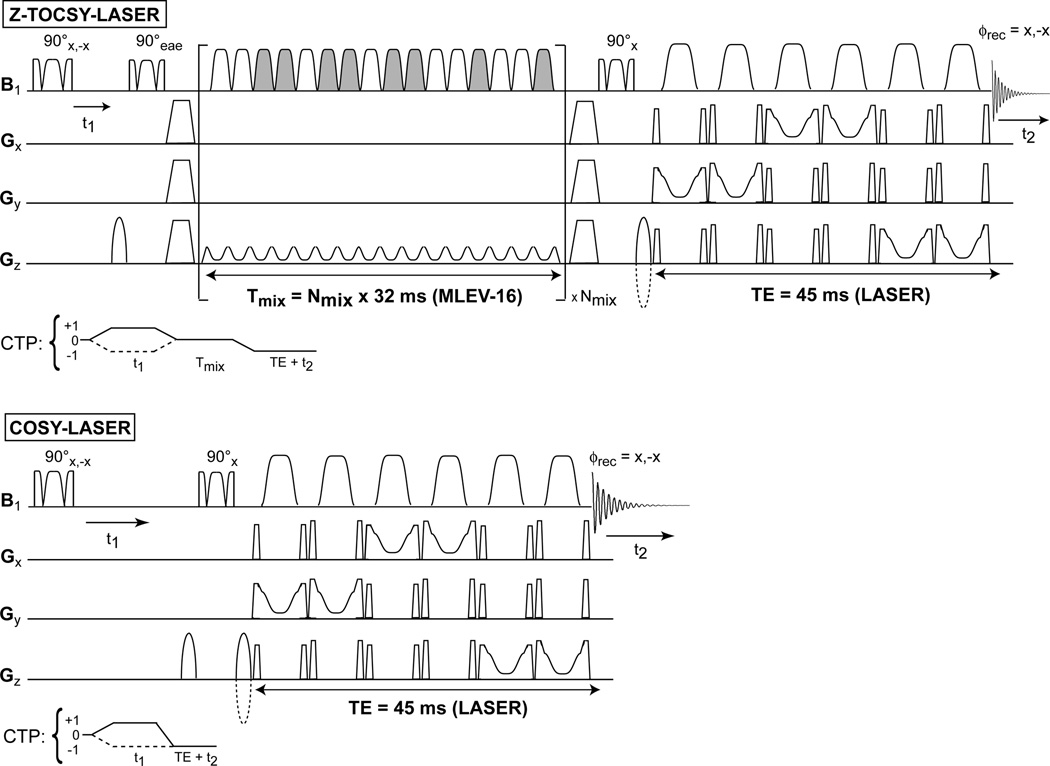
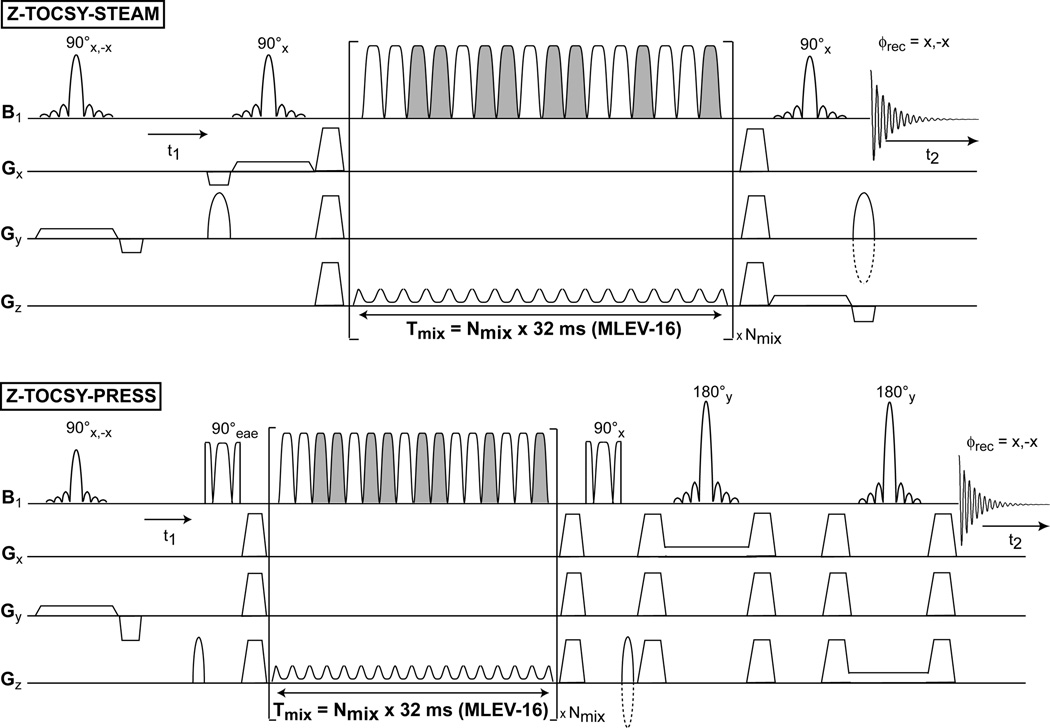
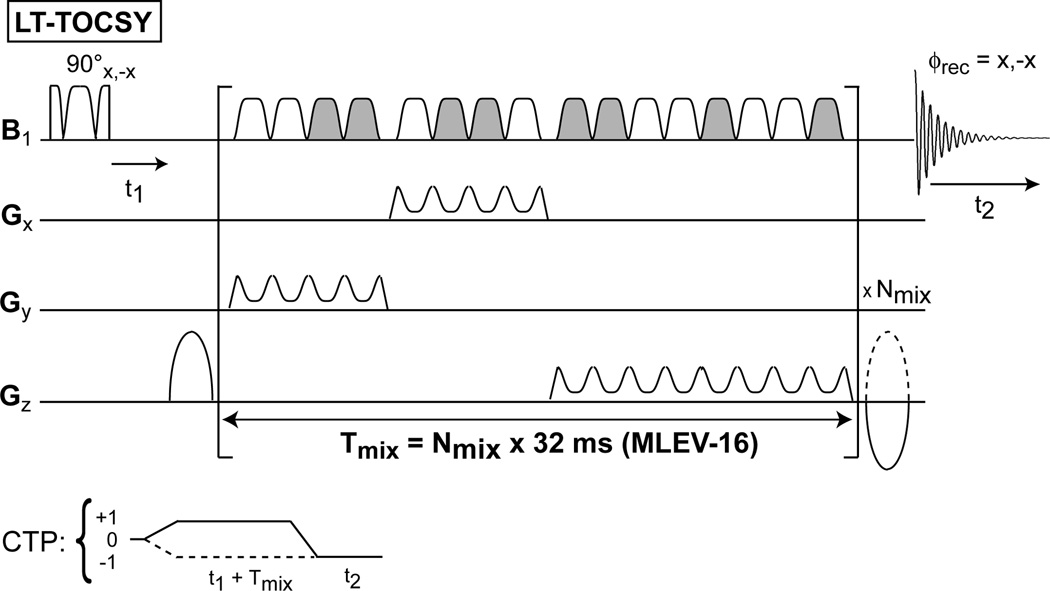
The diagrams of the 2D pulse sequences are presented in Fig. 1 for the fully adiabatic 2D Z-TOCSY-LASER and 2D COSY-LASER, in Fig. 2 for the semi-adiabatic 2D Z-TOCSY-STEAM and 2D Z-TOCSY-PRESS, and in Fig. 3 for the fully adiabatic 2D LT-TOCSY. In the 2D LT-TOCSY sequence that performs simultaneously localization and transverse mixing, the 16 pulses of the MLEV-16 can be used also for 3D volume localization by distributing them in three groups: 1) the first group of 4 pulses has the gradient applied on the y axis, 2) the second group of 4 pulses has the gradient applied on the x axis, and 3) the third group of 8 pulses applies the gradient on the z axis. No following LASER block is necessary for 3D localization. This design has the merit that can reduce considerably the SAR compared to the other TOCSY versions. In all sequences the water suppression is performed with a WET (57) scheme, preceding the first excitation pulse (the water suppression block is omitted from all figures).
Figure 1.
Fully adiabatic sequences for low-power in-vivo 2D TOCSY and 2D COSY experiments. The 2D Z-TOCSY-LASER experiment combines longitudinal TOCSY mixing and LASER localization, employing both the GOIA-W(16,4) pulses. A gradient enhanced z-filter, composed of 90° adiabatic BIR-4 pulses and spoilers (trapeze shape) that removes unwanted transverse magnetization, brackets the MLEV-16 block to perform transfer of longitudinal magnetization. The MLEV-16 block concatenates the individual GOIA-W(16,4) pulse shapes to eliminate gaps and can be repeated a number of times (Nmix) for the desired mixing time. The relative phase of the GOIA-W(16,4) pulses changes according to the MLEV-16 scheme (pulses which are grayed-out have an overall 180° phase shift compared to non-grayed pulses). The preferred gradient direction during MLEV-16 is selected for the gradient coil with the best performance. Sine-bell shaped gradients before and after the z-filtered MLEV-16 block select the coherence transfer pathway (CTP) depicted underneath the sequence. Echo-antiecho of 2D spectra can be performed if the phase (eae) of the first 90° pulse of the z-filter is alternated z to −x simultaneously with the change in polarity of the CTP gradient following the spoiler (the echo and antiecho FIDs are stored interleaved). The 2D COSY-LASER sequence is a simple extension of the LASER sequence with an additional 90° BIR-4 pulse, and sine-bell gradients to select the CTP depicted underneath the sequence (the polarity of CTP gradients can be alternated for interleaved echo-antiecho acquisitions). The t1 evolution time is incremented in successive experiments and a minimum two step phase cycle involving the first excitation pulse and the receiver phase can be used to remove t1 noise and axial peaks in both 2D spectra. 1D edited sequences can be simply obtained by replacing the first 90° BIR-4 pulse with a selective Gaussian pulse.
Figure 2.
Semi-adiabatic 2D TOCSY sequences. The same adiabatic longitudinal mixing TOCSY block from Fig 1 is used for magnetization transfer (grayed-out pulses have an overall 180° phase shift compared to non grayed pulses). The localization is performed using STEAM or PRESS methods. The same principles for phase cycle and echo-antiecho acquisition apply.
Figure 3.
A fully adiabatic sequence named 2D LT-TOCSY is proposed to perform simultaneously 3D volume localization and transverse mixing. The 16 GOIA-W(16,4) pulses of the MLEV-16 train are distributed in three groups: 1) the first group of four pulses for the y (coronal) gradient direction, 2) the second group of four pulses for x (sagital) gradient direction, and 3) the third group of eight pulses for z (axial) gradient direction. Grayed-out pulses have an overall 180° phase shift compared to non grayed pulses. Two short gaps (200 μs) are necessary for gradient ramp-up/down between the groups 1–2 and 2–3. Phase cycle and CTP gradients are similar with the previous sequences.
3.2.2. Phantom experiments
Magnetization transfer efficiency in the 1D editing mode and the quality of 2D spectra were tested in a uniform phantom containing a mixture of Lactate and GABA (50 mM each). Signal localization was further tested in a double layer phantom that contains an outer shell of oil and an inner core with a mixture of brain metabolites at physiological concentrations. The 2D acquisition parameters used for phantoms were: a voxel size of 3×3×3 cm3, a repetition time TR = 1.8 s, echo times specific to each localization (45 ms LASER, 30 ms PRESS, 20 ms STEAM, and 30 ms L-COSY), TOCSY mixing times of 64 ms (Nmix = 2), 64 t1 increments with 0.8 ms (10 ppm spectral width), each t1 increment with 8 averages (two steps phase cycle), 4 dummies scans for the first t1 increment, total acquisition time of 15:20 (min:s). Spectral width in the direct (t2) detected dimension was 10 ppm, and 1k points were acquired. Localizer images were used to position the MRS voxel.
3.2.3. Experiments on human subjects
2D Z-TOCSY-LASER spectra with longitudinal mixing were collected on healthy volunteers (4 subjects) and patients with brain tumor (GBM, 2 subjects). 2D L-COSY spectra were also acquired on the healthy volunteers for comparison purposes. The studies with human subjects were approved by the IRB of our institution.
In the case of volunteers a voxel size of 4×4×3 cm3 (AP-RL-FH) was placed in the visual cortex. The repetition time TR varied between a minimum of 2 s and a maximum of 2.65 s due to SAR limitations (subjects with bigger heads and more subcutaneous fat required higher voltage on the transmit body coil). The echo time was 45 ms for LASER, and a mixing time of 64 ms (Nmix = 2) was used for TOCSY. The 2D L-COSY used a minimum echo time of 30 ms, and the same TR as for TOCSY. For all experiments: 64 t1 increments with a 0.8 ms time step (10 ppm F1), 8 averages per t1 increment (two steps phase cycle), 4 dummies scans for the first t1 increment, total acquisition times between 17:12 (min:s) and 22:47 (min:s) according to the TR used.
In the case of the two patients slightly bigger voxels of 5×3×4 cm3 (AP-RL-FH) and 4×4×4 cm3 were used, respectively, in order to include most of the tumor visible on the FLAIR images. The other parameters were a repetition time TR = 2.65 s, echo time TE = 45 ms for LASER, 64 ms (Nmix = 2) TOCSY mixing time, 50 t1 increments with 0.8 ms (10 ppm F1), 8 averages per t1 increment (two steps phase cycle), 4 dummies scans for the first t1 increment, total acquisition times of 17:50 (min:s). Spectral width in the direct (t2) detected dimension was 10 ppm, and 1k points were acquired. FAST(EST)MAP (58) was used to shim the B0 field over the MRS voxel to a water linewidth of 10–12 Hz.
Images used to position the MRS voxel were acquired with: i) MEMPRAGE for volunteers (TR = 2.53 s, TE1/TE2/TE2/TE3/TE4 = 1.64/3.5/5.36/7.22 ms, TI = 1.2 s, FA=7°, 1 mm isotropic resolution, Ref. (59)), and ii) FLAIR for tumor patients (TR = 10000 ms, TE = 70 ms, 23 slices, 5 mm slice thickness (1 mm gap), and 0.6×0.45 mm2 in-plane resolution).
3.2.4. Data analysis
Raw 2D data were transferred to Matlab (The MathWorks, Inc., Natick, MA, USA) for signal combination from multiple elements followed by row concatenation into a 2D matrix. Commercial 2D spectral processing software (Felix-2007, Accelrys, San Diego, CA, USA) was used for spectral processing and analysis. The processing parameters used were: for F2 dimension – skewed sine-squared window, 2048 points, magnitude, and for F1 dimension – sine-squared window, linear prediction to 96 points, zero-filling to 512 points, magnitude. The 2D spectra were further analyzed and assigned in SPARKY 3.0 software (T. D. Goddard and D. G. Kneller, SPARKY 3, UCSF).
4. RESULTS
GAMMA (54) simulations were performed prior to experiments in order to check the efficiency of magnetization transfer of the TOCY mixing using GOIA-W(16,4) pulses and search for the low power conditions. The minimum time-bandwidth product of R = 20 for which pulses still behave adiabatically was used. In particular, three conditions were investigated: i) low power – Tp = 2 ms, BW = 10 kHz, γB1,max = 0.76 kHz, ii) medium power – Tp = 1.5 ms, BW = 13.34 kHz, γB1,max = 1.02 kHz, and iii) high power – Tp = 1.25 ms, BW = 16 kHz, γB1,max = 1.22 kHz. Typically a maximum γB1,max = 1.0 kHz is imposed on clinical scanners, hence we searched for values that are within 25% below and above this value (lower values render the transfer too inefficient, higher values would produce an SAR that is not feasible in-vivo).
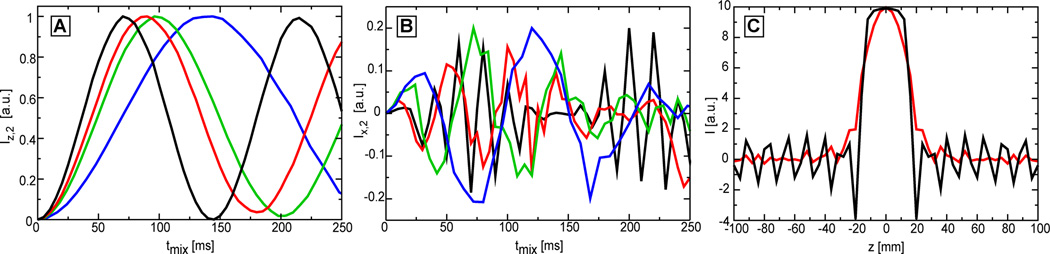
In Fig. 4A the buildup curves for longitudinal mixing are shown. Compared to the ideal magnetization transfer in the absence of chemical shifts (black curve, only the J-coupling in the case of high power RF condition), the buildup curves in the presence of the chemical shifts become progressively slower for decreasing B1,max fields. The first maximum of the buildup that corresponds to 72 ms (1/2J) in the ideal curve, shifts to 87 ms for γB1,max = 1.22 kHz (red curve), 96 ms for γB1,max = 1.02 kHz (green curve), and 139 ms for γB1,max = 0.76 kHz (blue curve). In practice limitations in SAR impose an upper limit of the mixing time. On our hardware 64 ms mixing is still possible within the limits of brain’s SAR for reasonable values of the TR in the range of 2–3 s (for other organs which have a lower SAR threshold longer mixing is possible). For 64 ms mixing the predicted transfer (in the absence of relaxation) amounts to 82% for γB1,max = 1.22 kHz, 73% for γB1,max = 1.02 kHz, and 44% for γB1,max = 0.76 kHz from the ideal case (black curve). Thus, under the most favorable conditions (γB1,max = 1.0 kHz) a transfer efficiency of 73% is possible on clinical scanners. Compared to the γB1,max = 1.0 kHz, the low power condition of γB1,max = 0.76 kHz achieves 60% transfer efficiency.
Figure 4.
Simulations of magnetization transfer buildups curves for longitudinal TOCSY mixing (panel A) and transverse TOCSY mixing (panel B). The lactate spin system is considered at 3T and the transfer is performed with MLEV-16 scheme employing GOIA-W(16,4) pulses. Three GOIA-W(16,4) pulses are investigated: 1) Tp = 1.25 ms, BW = 16 kHz, B1,max = 1.22 kHz (red curve); 2) Tp = 1.5 ms, BW = 13.34 kHz, B1,max = 1.02 kHz (green curve); 3) Tp = 2 ms, BW = 10 kHz, B1,max = 0.76 kHz (blue curve). The ideal buildup curve in the absence of chemical shifts is represented by the black curve (this was simulated considering only the J-coupling and the B1 field (B1,max = 1.22 kHz) in Eq.[1]). In panel C slice profiles are shown for the refocusing MLEV-16 train (red curve) and the doubly adiabatic spin echo (black curve), both employing the GOIA-W(16,4) pulses of Tp = 2 ms, BW = 10 kHz, B1,max = 0.76 kHz.
In Fig. 4B the case of transverse mixing is considered for the same situations: the ideal case with no chemical shifts (black curve, only the J-coupling in the case of high power RF condition), and including the chemical shifts for high- (γB1,max = 1.22 kHz, red curve), medium- (γB1,max = 1.02 kHz, green curve), and low-power (γB1,max = 0.76 kHz, blue curve) mixing. The transverse mixing buildup curves show two important differences compared to longitudinal mixing: i) faster oscillations that superimpose on the magnetization transfer curve driven by the slower scalar coupling, and ii) the maximum possible transfer is five times lower than in the case of longitudinal mixing. The faster oscillations are due to the fact that transverse magnetization is not aligned with the direction of the effective field (along z) at the beginning of the adiabatic pulse, hence the magnetization starts to precess around the effective field during mixing. The oscillations becomes faster as γB1,max and the effective field increase. By comparison the longitudinal magnetization is aligned with the effective field at the beginning of the adiabatic pulse and becomes spin locked during mixing. In addition to this, the transverse magnetization is also subjected to the faster R2ρ relaxation. Measurements of R2ρ and R1ρ relaxation rates at 4T in the case of HS4 and HS1 pulses (53) indicate that R2ρ can be more than four times faster than R1ρ. Simulations or measurements have not been attempted for R2ρ and R1ρ relaxation under GOIA-W(16,4) pulses, however similar results are expected (the difference seems even to increase for pulses with larger BW as indicated in Ref. (53)). The combination of faster oscillations around the effective field and damping of the transfer due R2ρ relaxation may result experimentally in an earlier maximum of an apparent faster buildup curve for transverse magnetization than would be expected from the scalar coupling (maximum at 1/2J) as observed previously in Ref. (29). The net result of all these factors is a reduced efficiency of the transverse mixing as shown also experimentally in Fig. 5B.
Figure 5.
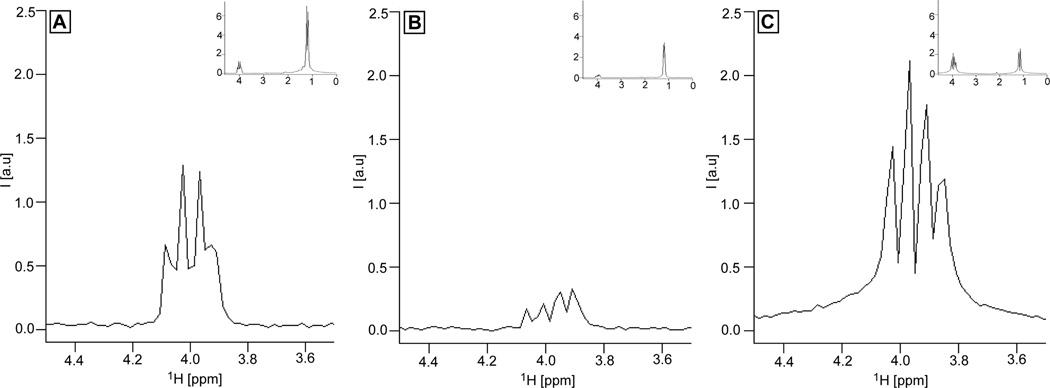
Selective magnetization transfer from the CH3 group to the CH group in the lactate spin system measured experimentally at 3T on a phantom containing equimolar (50 mM) mixture of Lactate and GABA using the 1D edited sequences: A) Z-TOCSY-LASER with longitudinal mixing, B) TOCSY-LASER with transverse mixing, C) COSY-LASER. In all three experiments the magnetization of the methyl group is excited selectively with Gaussian pulse (BW = 20 Hz). For TOCSY transfers (A and B) the same mixing time of 64 ms is used, employing MLEV-16 with GOIA-W(16,4) pulses of Tp = 2 ms, BW = 10 kHz, B1,max = 0.76 kHz. The same transfer time of 64 ms (t1 = 64 ms) is used in COSY-LASER. In all three experiments the same LASER localization is used: GOIA-W(16,4) pulses of Tp = 3.5 ms, BW = 20 kHz, B1,max = 0.82 kHz, and echo time of 45 ms. A voxel of 3×3×3 cm3, 1.8 s repetition time and 8 averages (4 dummy scans) are used. The signal transferred to the CH group is shown zoomed, and in the insets the full spectrum is included. Note that the GABA signal is not seen due to selective Lactate transfer, however, the GABA signal is seen in the 2D spectra of Fig. 7 using 2D non-selective sequences and the same phantom.
Simultaneous localization and transverse mixing can be performed when using GOIA-W(16,4) pulses according to the LT-TOCSY pulse sequence of Fig. 3. Simulations are performed for the slice profile of a refocusing MLEV-16 train using GOIA-W(16,4) pulses of 2 ms duration, 10 kHz bandwidth, and 0.76 kHz maximum RF amplitude. Figure 4C shows the comparison between the MLEV-16 slice profile (red curve) and the slice profile obtained with a double adiabatic spin echo sequence (60) (black curve) using the same pulses. Accurate localization is obtained in both cases. The advantage of the 2D LT-TOCSY sequence is the considerable reduction of SAR by eliminating the LASER block. However, the behavior of transverse mixing shown in Fig 4B, the faster R2ρ relaxation, and the need to introduce small gaps (200–400 μs) in the MLEV-16 train after each group of 4 or 8 pulses for the ramp-up/down of the gradients make this sequence less efficient for magnetization transfer.
Before attempting the 2D experiments, efficiency of our TOCSY mixing was verified by 1D edited selective magnetization transfer in a phantom containing an equimolar (50 mM) mixture of Lactate and GABA. Figures 5A and 5B show the results of the 1D editing selective TOCSY with longitudinal mixing and transverse mixing, respectively. Magnetization is transferred from the methyl group of Lactate, initially excited by a selective Gaussian pulse, to the methine group using a 64 ms mixing time for TOCSY with GOIA-W(16,4) pulses of 2 ms duration, 10 kHz bandwidth, and 0.76 kHz maximum amplitude. The results show that approximately five times more transfer is obtained with longitudinal mixing (1.45 maximum signal, Fig 5A) compared to transverse mixing (0.3 maximum signal, Fig 5B). Similar results are obtained for shorter mixing times of 32 ms. Indicative of the transfer is also the relative ratio of the CH to CH3 signal that can be seen in the insets shown in the upper right corners of Fig. 5A and 5B (0.21 ratio for longitudinal mixing and 0.06 for transverse mixing). The sequences employed are based on the diagram shown in Fig. 1 (Z-TOCSY-LASER), for which we replaced the first 90° BIR-4 pulse with a 90° selective Gaussian pulse (BW = 20 Hz), and in the case of transverse mixing the z-filter was removed according to the design proposed in Ref. (29). The same LASER localization (GOIA-W(16,4) pulses of 3.5 ms duration, 20 kHz bandwidth, and 0.82 kHz maximum amplitude, other acquisition parameters were identical TR = 1.8 s, NA = 8, 0 ms t1 evolution) was used for both transverse and longitudinal mixing. Comparison with COSY transfer is made employing the COSY-LASER sequence from Fig 1 which has the first 90° BIR-4 pulse replaced with 90° selective Gaussian pulse (BW = 20 Hz) for selective excitation of the Lactate methyl group (the same LASER localization and acquisition parameters as in the case of TOCSY are employed). In Fig. 5C the COSY transfer to the methine group obtained for 64 ms (t1 was set to 64 ms) is shown, which indicates that 50% more signal can be obtained compared to the longitudinal TOCSY mixing. However, an important aspect can be noticed for COSY: a much wider multiplet structure with a wider baseline is obtained due to the transfer of both in-phase and anti-phase magnetization components. TOCSY transfers in-phase magnetization, giving rise to narrower peaks (this is true for the longitudinal mixing from Fig. 1, the transverse mixing needs special purge pulses (61) to remove the anti-phase magnetization). Phase distorted multiplets can result in lower resolution of the 2D COSY compared to 2D TOCSY spectra, and this is important given the low resolution of in-vivo spectra. The increased resolution and the fact that long range (relayed) correlations can be obtained in TOCSY might represent an attractive counterbalance for less sensitivity compared to COSY. Examples that support this will be shown from both phantom and in-vivo data.
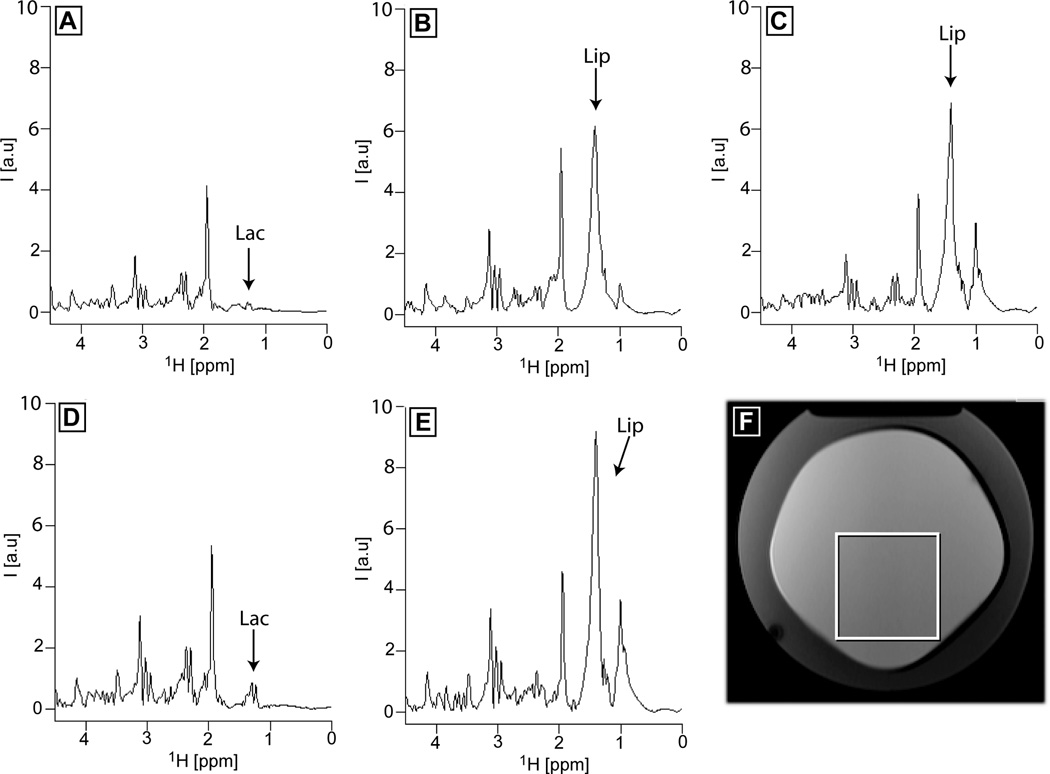
Accuracy of localization has been checked for all the sequences in a double layer phantom that contains and outer shell of oil and an inner core of brain metabolites. A voxel was placed inside the inner core close to the boundary with the oil shell, and only the first t1 increment (0 ms t1 evolution, 1D spectra) was acquired. In Fig. 6 results are shown for 1D Z-TOCSY-LASER (Fig. 6A), 1D Z-TOCSY-STEAM (Fig. 6B), 1D Z-TOCSY-PRESS (Fig. 6C), 1D COSY-LASER (Fig 6D), and 1D L-COSY (Fig 6E), for the voxel positioned according to Fig 6F. The fully adiabatic sequences Z-TOCSY-LASER and COSY-LASER show excellent localization and no signs of lipid contamination (the lactate peak from the inner core can be easily seen), while both semi-adiabatic sequences Z-TOCSY-STEAM and Z-TOCSY-PRESS show a large lipid signal (obscuring lactate), which is larger for the PRESS localization. The largest lipid contamination is shown by the L-COSY experiment. The lipid contamination in the non-adiabatic sequences is due to CSDE and wider slice profiles for localization pulses. In reality, a bigger voxel than the one prescribed is selected by these sequences. For the 3×3×3 cm3 voxel shown in Fig 6F, the selected voxel is approximately 5 mm larger in each direction, yielding a real voxel size of 4×4×4 cm3 (double volume of the voxel). For the LASER localization a much sharper excitation selects a voxel of the same size as the one prescribed. The difference in the size of the real voxel selected by the localization schemes explains also the SNR difference observed (vide infra) between the 2D spectra obtained with adiabatic and non-adiabatic localization. In principle, saturation bands can be placed around the voxel to reduce the lipid contamination for non-adiabatic localization. However, the typical saturation bands existing on our clinical scanner are not optimized to have very sharp edges, hence they do not suppress completely the lipids outside the voxel and in addition they might suppress slightly some of the metabolite signal inside the voxel (this can be checked in a homogeneous phantom with and without saturation bands). Optimized saturation bands (62) and their automatic placement (63) can eliminate errors and make this approach more robust.
Figure 6.
Accuracy of voxel localization at 3T for Z-TOCSY-LASER (A), Z-TOCSY-STEAM (B), Z-TOCSY-PRESS (C), COSY-LASER (D), and L-COSY (E) sequences in a double layer phantom containing an outer shell of oil and an inner core of brain metabolites at physiologic concentrations (the voxel position is shown in F). A large lipid contamination signal (Lip) originating from outside the voxel is observed for semi-adiabatic (B,C) and non-adiabatic (E) sequences. Instead, in the case of fully adiabatic sequences (A,D) the lactate signal (Lac) originating from inside the voxel can be detected. All TOCSY experiments used the same mixing as detailed in Fig. 5. The LASER block used in A and D was the same as in Fig. 5. STEAM and PRESS used echo times of 20 ms and 30 ms, respectively. The L-COSY sequence used an echo time of 30 ms. The same repetition time (TR = 1.8 s) and number of averages (NA = 8) were used in all experiments.
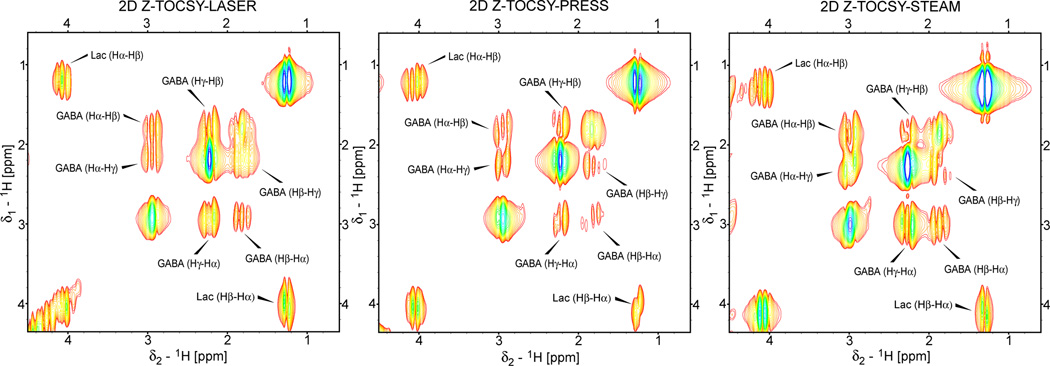
2D spectra have been acquired with all the sequences on the phantom containing lactate and GABA. GABA is in particular an important metabolite which is hard to observe in 1D spectra, and represents a good test case to show that long range cross-peaks can be obtained in 2D TOCSY. In Fig. 7 results are compared for the localized 2D TOCSY obtained with the fully adiabatic 2D Z-TOCSY-LASER (Fig. 7A), and the semi-adiabatic 2D Z-TOCSY-PRESS (Fig. 7B) and 2D Z-TOCSY-STEAM (Fig. 7C) sequences. As can be seen in all spectra the entire correlation network is obtained for each spin system. The intensity of the crosspeaks is greatest in 2D Z-TOCSY-STEAM, less in 2D Z-TOCSY-LASER and the lowest in 2D Z-TOCSY-PRESS (the same contour levels are chosen, the minimum contour level is 10 times the noise level, the crosspeaks for each metabolite are labeled according to the protons involved). However, the line-width in both dimensions is larger for the semi-adiabatic sequences compared to the fully adiabatic sequence. These can be explained by the larger selected voxel experiencing a greater B0 inhomogeneity, and by the fact that line shape modulation of coupled spins during the echo time is more pronounced for non-adiabatic localization (PRESS and STEAM).
Figure 7.
Example of localized 2D TOCSY spectra obtained at 3T with fully adiabatic and semi-adiabatic sequences in a phantom containing an equimolar (50 mM) mixture of lactate and GABA. The same TOCSY mixing as described in Fig 5 was used in all three experiments. The parameters for localization blocks are the same as described in Fig 5 and Fig 6. A voxel size of 3×3×3 cm3, 64 t1 increments, 0.8 ms (10 ppm) increment time, 8 averages for each t1 experiment, 1k points acquired for t2 dimension, and repetition time of 1.8 s yielding a total acquisition time of 15:20 (min:s). The data are zero-filled to 512×2048 (F1×F2) before FT, linear prediction in F1 dimension, sine-squared appodization in both dimensions, and displayed in magnitude mode with the same contour levels (the minimum contour level is 10 times above the noise level). The crosspeaks for both Lactate and GABA are labeled according to the protons involved.
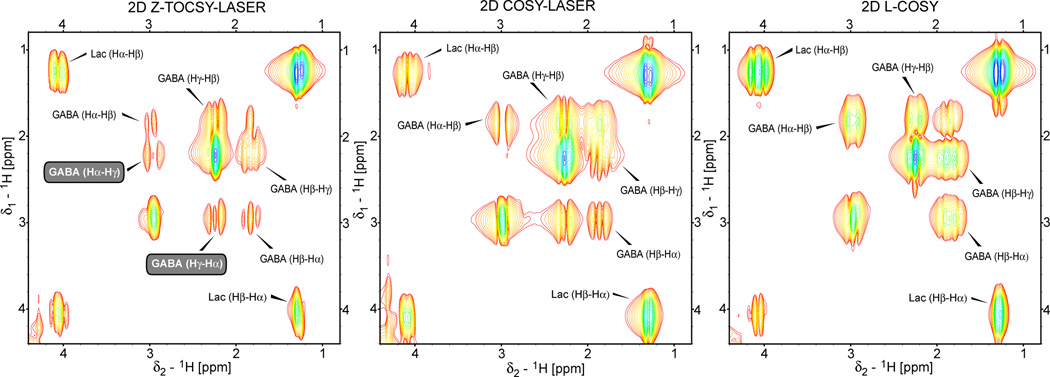
Localized 2D TOCSY (Z-TOCSY-LASER) and 2D COSY (COSY-LASER and L-COSY) spectra are compared on the lactate and GABA phantom in Fig. 8. It can be easily noticed that the pair of crosspeaks corresponding to the long range correlations between Hα and Hγ protons of GABA are not present in the 2D L-COSY spectra. However, one of the crosspeaks is present in the 2D COSY-LASER. The possible mechanism, which is verifiable by simulations, can be given by the fact that the train of LASER pulses may act, to a limited degree, also as mixing for magnetization transfer between strongly coupled spins with small chemical shift offsets such as GABA (the carrier was placed in the middle of the GABA spectrum, yielding offsets in the range of ±50 Hz, this effect of LASER is not seen for lactate protons that have more chemical shift dispersion). The difference in relaxation for the spins originating the magnetization could explain why only one of the crosspeaks is obtained (water suppression is unlikely to play a role since lactate crosspeaks are symmetric and the GABA crosspeaks are further away from water). Similar to Fig. 7, several observations can be made: i) the crosspeaks have the highest intensity for L-COSY, lower for COSY-LASER and the lowest for Z-TOCSY-LASER, and ii) the lines are sharper in the case of Z-TOCSY-LASER. These can be explained by larger voxel selected by L-COSY and transfer of both in-phase and anti-phase magnetization for L-COSY and COSY-LASER.
Figure 8.
Example of localized 2D TOCSY and COSY spectra obtained at 3T in a phantom containing an equimolar (50 mM) mixture of lactate and GABA using Z-TOCSY-LASER, COSY-LASER, and L-COSY sequences. The same TOCSY, COSY and LASER blocks as described in Figs 6 and 7 are used. The other acquisition and processing parameters are according to Fig 7. Metabolite crosspeaks are labeled according to the protons involved. Long range correlations between Hα and Hγ protons of GABA (labels highlighted by the gray boxes) can be obtained in 2D Z-TOCSY-LASER but not in 2D L-COSY. Only one of the Hα-Hγ crosspeaks is observed in 2D COSY-LASER. The crosspeaks’ intensity is stronger in 2D COSY but the resolution is higher (sharper crosspeaks) in 2D TOCSY. The same contour levels are displayed (the minimum contour level is 10 times above the noise level).
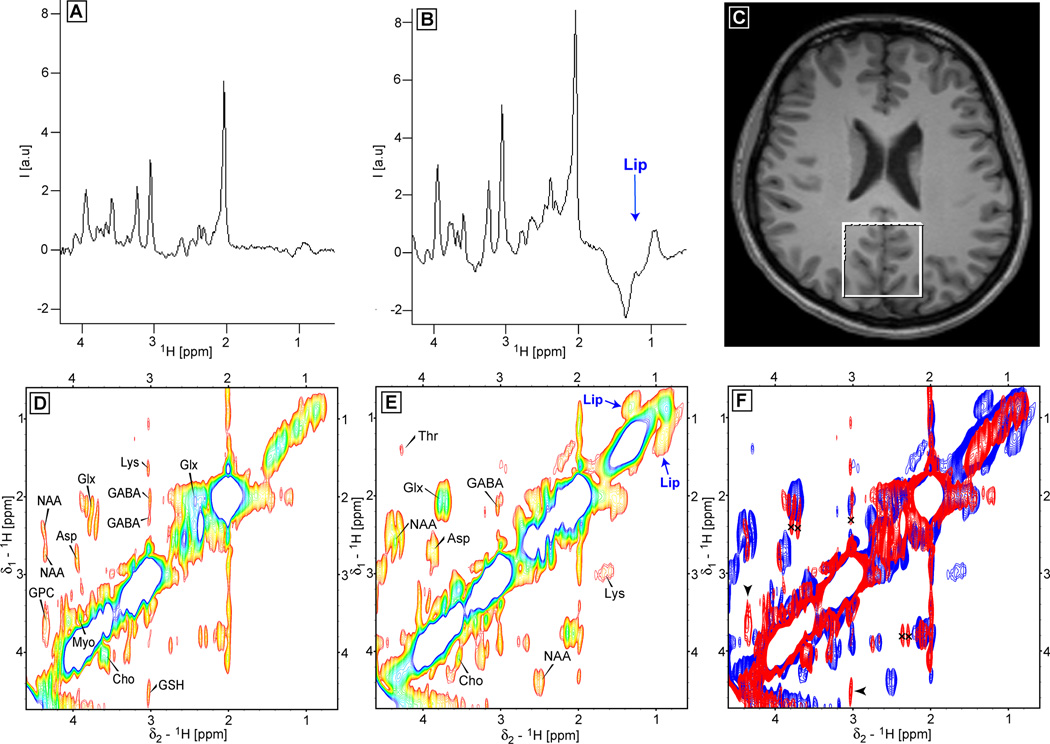
Representative in-vivo localized 2D TOCSY and COSY brain spectra are presented next. In Fig. 9 the comparison is made between 2D Z-TOCSY-LASER (Fig. 9D) and 2D L-COSY (Fig. 9E) obtained from a healthy volunteer (voxel position shown in Fig. 9F). The 1D spectra corresponding to the first t1 increment (0 ms t1 evolution) are shown in Fig. 9A for 2D Z-TOCSY-LASER and in Fig. 9B for 2D L-COSY. It is immediately apparent that large lipid contamination from subcutaneous fat is present in the L-COSY spectra due to a larger real voxel size that explains also the intensity difference. The crosspeaks of several metabolites can be easily identified in both 2D spectra (NAA, ASP-aspartate, Glx-glutamate/glutamine, Cho-choline, Myo-myoinositol, and tentatively for GABA and Lys-lysine). Some crosspeaks that seem to be present only in 2D Z-TOCSY-LASER have chemical shifts suggestive of glycerophospocholine (GPC) and gluthatione (GSH) which were found previously also in L-COSY (64) (albeit double the number of averages was used). On the other hand, threonine (Thr) appears to be present only in the 2D L-COSY (Thr was observed in 1D edited TOCSY (30), however in our low power implementation slower buildup might reduce the crosspeak intensity, this could be recovered potentially for the stronger mixing conditions). In addition, in the 2D L-COSY the crosspeaks corresponding to the contaminating lipids (Lip) are also present. Overall the crosspeaks in the 2D L-COSY spectrum are more intense, while the crosspeaks in the 2D Z-TOCSY-LASER are sharper. As explained and shown in phantoms sharper crosspeaks are not simply a scaling effect of the contour levels due lower intensity but the contribution of in-phase and anti-phase magnetization transfer in 2D L-COSY, while in 2D Z-TOCSY-LASER only in-phase magnetization transfer is ensured. In Fig. 9F an overlay between the 2D Z-TOCSY-LASER spectrum (red) and the 2D L-COSY (blue) is shown. Long range crosspeaks (marked by the black crosses) are present in the 2D Z-TOCSY-LASER for Glx and GABA. Crosspeaks that seem to be observed only in the 2D Z-TOCSY-LASER are indicated by black arrowheads.
Figure 9.
In-vivo brain spectra obtained at 3T with the 2D Z-TOCSY-LASER (D) and 2D L-COSY (E) sequences from a healthy volunteer (the voxel of 4×4×3 AP-RL-FH cm3 positioned in the occipital lobe is shown in C). 1D spectra corresponding to the first t1 experiment (0 ms t1 evolution) are shown in A for 2D Z-TOCSY-LASER, and in B for 2D L-COSY. A large lipid contamination is visible in L-COSY data. The 2D spectra of D and E allow the unambiguous identification of important metabolites: glutamate/glutamine (Glx), GABA, NAA, aspartate (ASP), lysine (Lys), choline (Cho), myoinositol (Myo). Few metabolites are seen only in 2D TOCSY such as glycerophosphocholine (GPC) and gluthatione (GSH), or only on the COSY spectra such as threonine (Thr). The crosspeaks corresponding to the contaminating lipid signal is present in the 2D COSY. Overlay between the 2D Z-TOCSY-LASER (red) and 2D L-COSY (blue) spectra is shown in panel F: i) the long range range TOCSY crosspeaks of Glx and GABA are marked by black crosses in the red spectrum, ii) and the metabolites (GSH, GPC) present only in TOCSY are indicated by black arrowheads. The crosspeaks are more intense in 2D L-COSY (explained also by a bigger size of the real excited voxel), but sharper (higher resolution) in 2D Z-TOCSY-LASER. Mixing time of 64 ms was used for TOCSY with MLEV-16 employing GOIA-W(16,4) pulses of Tp = 2 ms, BW = 10 kHz, B1,max = 0.76 kHz. LASER localization used GOIA-W(16,4) pulses of Tp = 3.5 ms, BW = 20 kHz, B1,max = 0.82 kHz, and echo time of 45 ms. The 2D L-COSY from Ref. (24) with and echo time of 30 ms was used. 64 t1 increments, 8 ms (10 ppm) time increment, 8 averages, TR = 2 s, total acquisition time of 17:04 (min:s). The same processing as described in Fig 7 was used, and the same contour levels were chosen (the minimum contour level is 5 times above the noise level).
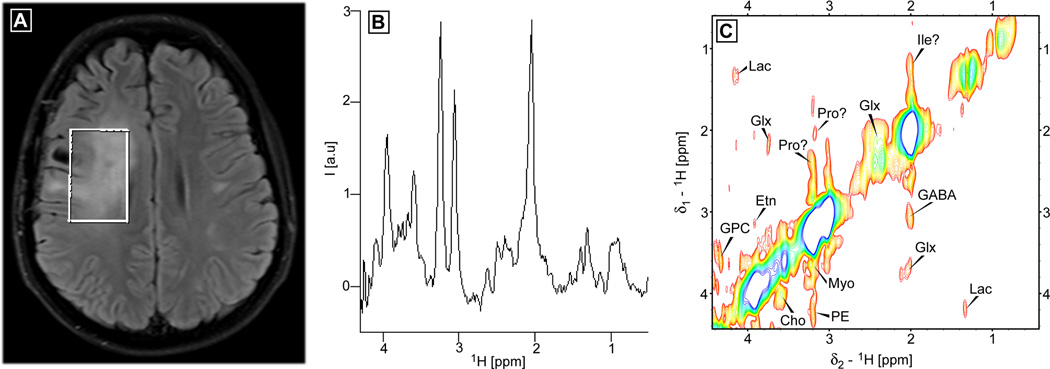
In Fig. 10 the 2D Z-TOCSY-LASER data obtained from a patient with glioblastoma is shown. The voxel position and size is chosen to include most of the FLAIR abnormality seen in Fig. 10A. In Fig. 10B the 1D spectrum corresponding to the first t1 increment (0 ms t1 evolution) is shown. Reduced NAA, increased Choline and the presence of Lactate can be noticed. Fig. 10C contains the 2D spectrum which shows crosspeaks for lactate (Lac), glutamate/glutamine (Glx), choline (Cho), glycerophospocholine (GPC), ethanolamine (Etn) and phosphoethanolamine (PE), and myoinositol (Myo). Notice that glutathione is absent, while important additional metabolites (Etn, PE) involved in the phospholipids turnover are present. Tentatively, some crosspeaks are assigned to the aminoacids proline (Pro) and Isoleucine (Ile). Note, that more robust methods (65) for automatic assignment and fitting of the 2D spectra could be employed. An added benefit (two for the price of one) of the 2D MRS methods is that a conventional 1D spectrum corresponding to the first t1 increment (0 ms t1 evolution) is also obtained, which can be analyzed and fitted with advanced routines (9,10) developed so far for in-vivo 1D MRS. A summary of the main experimental results obtained in-vivo and phantoms are given in Table 1.
Figure 10.
In-vivo 2D Z-TOCSY-LASER brain spectrum obtained at 3T from a patient with glioblastoma. A voxel of 5×3×4 AP-RL-FH cm3 is positioned to include most of the FLAIR abnormality (shown in A). The 1D spectrum corresponding to the first t1 experiment (0 ms t1 evolution) is shown in panel B, decreased NAA, high choline, and the presence of lactate can be observed. The 2D Z-TOCSY-LASER spectrum is shown in panel C. A number of metabolites can be unambiguously identified: glutamate/glutamine (Glx), GABA, lactate (Lac), myoinositol (Myo), choline (Cho), glycerophosphocholine (GPC), ethanolamine (Etn), and phosphoethanolamine (PE). Several crosspeaks are tentatively assigned to aminoacids proline (Pro) and isoluecine (Ile). The same acquisition and processing parameters are used as in Fig. 9, except for TR = 2.65 s and 50 t1 increments, yielding a total acquisition time of 17:40 (min:s). The minimum contour level is 5 times above the noise level.
5. DISCUSSIONS
In-vivo multidimensional MRS is a field that needs further development and validation. Optimized pulse sequences represent a major part in this effort together with technical developments that can improve SNR (such as 32-channel phased array head coil (66)) and shorten the acquisition times. The benefits of multidimensional MRS in resolving overlapped spectra and offering detailed metabolic information can motivate this effort. Perhaps, a close analogy with the situation of the multidimensional MRS is the diffusion tensor imaging (DTI), a field that is more known in the MR imaging community and faces a similar situation. The minimum number of six gradient directions that are necessary to calculate the diffusion tensor is not enough to resolve ambiguities in the direction of white matter fiber tracts that cross each other in the same voxel, hence a larger number of directions (40 or more in the case of High Angular Resolution Diffusion Imaging (HARDI)) is taken for this purpose, albeit to the cost of a much increased acquisition time (15 min or more for HARDI).
In this work we presented several novel approaches for a low power adiabatic in-vivo 2D TOCSY sequence, which to the best of our knowledge has not been demonstrated yet. The fully adiabatic sequence combines longitudinal TOCSY mixing and LASER localization, while the semi-adiabatic sequences use the same TOCSY mixing block and non-adiabatic (STEAM or PRESS) localization. The use of longitudinal mixing provides much improved (five times) transfer efficiency compared to previously proposed (29) transverse mixing of 1D editing sequences. The use of GOIA-W(16,4) pulses (32) reduces the SAR for both TOCSY and LASER blocks, allowing repetition times in the range of 2–2.7 s (for brain at 3T using the body transmit coil) and total acquisition times of 15–20 min for 2D TOCSY spectra. This acquisition times compare favorably with previously communicated times of in-vivo 1D editing TOCSY sequences (29–31), with the advantage that the full information is retained in the 2D spectra. For other organs (breast, liver, muscle, prostate), that have lower SAR thresholds, faster repetition and acquisition times are possible, or conversely, mixing with higher B1 fields that provide increased transfer. Also as an alternative to reducing SAR for brain, semi-adiabatic 2D TOCSY sequences can be performed with repetition times less than 2s. The 2D Z-TOCSY-STEAM provides less lipid contamination and more SNR compared to 2D Z-TOCSY-PRESS, and fits naturally the purpose of longitudinal mixing. Although compared to COSY the TOCSY seems to provide less SNR (60% for the low power condition, however, the medium and high power conditions could provide close to 100% transfer for mixing times in the 90 ms range), the higher resolution and relay crosspeaks represent certain advantages of TOCSY. Increased sensitivity by a factor of and improved resolution of TOCSY can be further obtained in phase sensitive spectra by acquisition of both coherence transfer pathways (47) with an echo-antiecho method. Other more efficient mixing schemes such as DIPSI-2 (67) could be investigated, although the need for arbitrary flip angles would make difficult the use of GOIA pulses (BIR-4 pulses could be used at the expense of increased SAR). On the other hand, all three TOCSY sequences can be easily run as exchange spectroscopy experiments (NOESY, Ref. (18)) if the B1 field is set to zero during mixing time. If desired, our sequences can be run also in the 1D edited mode as explained and demonstrated in our work.
Potential limitations and consequences that result due to the longer acquisition times of 2D MRS can be addressed with existing methods. First, the likelihood of motion that may degrade resolution and SNR is considerable, however, real-time motion correction schemes (68,69) can be incorporated in spectroscopy pulse sequences. Second, the brain coverage can be extended from a single voxel to multiple voxels within affordable acquisition time by using fast acquisition methods such as spiral readout gradients (70) that acquire simultaneously the k-space and the time-domain. Third, the acquisition time could be reduced by sparse sampling methods (71) of the t1 evolution time (the ultrafast approach of 2D NMR spectroscopy in a single scan (72) is less likely to be directly applicable due to the requirement of a homogenous sample, but it could be adopted with suitable modifications). Further technical developments of RF coils such as highly parallel phased arrays (73), cryo-probes (74), or dynamic nuclear polarization (75) could increase the SNR and allow to harvest the full metabolic information obtainable from multidimensional MRS.
6. CONCLUSIONS
In this work we presented novel strategies for localized in-vivo low power adiabatic 2D TOCSY and 2D COSY experiments. Our design shows improved magnetization transfer efficiency, precise localization, and reduced SAR. Importantly, these sequences can be run in a standard clinical environment with a feasible acquisition time. To the best of our knowledge a 2D TOCSY has not been yet realized in-vivo. It is expected that the proposed methods will contribute to the advancement of in-vivo multidimensional MRS and could promote its applicability to clinical questions.
Table 2.
Summary of experimental performance.
| Sequence | Transfer efficiency (b) | Localization accuracy | SNR | SAR (%) (d) |
|---|---|---|---|---|
| Z-TOCSY-LASER | 60% | ++++ | +++ | 100 |
| Z-TOCSY-STEAM | 60% | − | +++ (c) | 72 |
| Z-TOCSY-PRESS | 60% | − | +++ (c) | 72 |
| TOCSY-LASER (a) | 12% | ++++ | + | 94 |
| COSY-LASER | 100% | ++++ | ++++ | 28 |
| L-COSY | 100% | − | ++++ (c) | 7 |
The sequence design from Ref. (29) was used;
The efficiency calculated as percentage for the same mixing time of 64 ms (close to 1/2J = 72 ms for lactate) and the low-power condition γB1,max = 0.76 kHz in the case of TOCSY;
when the signal is adjusted to the size of the real voxel selected by the localization;
SAR calculated and measured at 3T (transmit body RF coil) by the watchdog monitor in percentage from the maximum of 3W/kg allowed for the brain in a volunteer case when using the same TR = 2 s and 64 ms mixing time at the low power condition for TOCSY (for other organs with lower SAR limits shorter TR < 2s is possible with the most intense Z-TOCSY-LASER sequence, the semi-adiabatic sequences Z-TOCSY-STEAM and Z-TOCSY-PRESS can be run on brain with TR = 1.5 s). Plus (+) and minus (−) symbols are given as a qualitative measure for the performance (for localization minus indicates poor localization as proven by the presence of contaminating signal from lipids).
Acknowledgements
Discussions with Dr. Thomas Benner from the Martinos Center, MGH are gratefully acknowledged and funding from the NIH grant R01 1200-206456.
References
- 1.Ernst RR, Bodenhausen G, Wokaun A. In: Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Halpern J, Green MLH, T M, editors. Oxford: Clarendon Press; 1987. [Google Scholar]
- 2.Cavanagh J, Fairbrother WG, Palmer AG, Skelton NJ. Protein NMR Spectroscopy, Principles and Practice. San Diego: Academic Press; 1996. [Google Scholar]
- 3.Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. Journal of the American Chemical Society. 2005;127(37):12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 4.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(44):15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andronesi OC, von Bergen M, Biernat J, Seidel K, Griesinger C, Mandelkow E, Baldus M. Characterization of Alzheimer's-like paired helical filaments from the core domain of tau protein using solid-state NMR spectroscopy. Journal of the American Chemical Society. 2008;130(18):5922–5928. doi: 10.1021/ja7100517. [DOI] [PubMed] [Google Scholar]
- 6.Andronesi OC, Mintzopoulos D, Struppe J, Black PM, Tzika AA. Solid-state NMR adiabatic TOBSY sequences provide enhanced sensitivity for multidimensional high-resolution magic-angle-spinning H-1 MR spectroscopy. Journal of Magnetic Resonance. 2008;193(2):251–258. doi: 10.1016/j.jmr.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 7.Pfeuffer J, Tkac I, Provencher SW, Gruetter R. Toward an in vivo neurochemical profile: Quantification of 18 metabolites in short-echo-time H-1 NMR spectra of the rat brain. Journal of Magnetic Resonance. 1999;141(1):104–120. doi: 10.1006/jmre.1999.1895. [DOI] [PubMed] [Google Scholar]
- 8.Mekle R, Mlynarik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR Spectroscopy of the Human Brain With Enhanced Singla Intensity at Ultrashort Echo Times on a Clinical Platform at 3T and 7T. Magnetic Resonance in Medicine. 2009;61(6):1279–1285. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- 9.Provencher SW. Estimation of Metabolite Concentrations from Localized in-Vivo Proton Nmr-Spectra. Magnetic Resonance in Medicine. 1993;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 10.Naressi A, Couturier C, Devos JM, Janssen M, Mangeat C, de Beer R, Graveron-Demilly D. Java-based graphical user interface for the MRUI quantitation package. Magnetic Resonance Materials in Physics Biology and Medicine. 2001;12(2–3):141–152. doi: 10.1007/BF02668096. [DOI] [PubMed] [Google Scholar]
- 11.Mountford CE, Stanwell P, Lin A, Ramadan S, Ross B. Neurospectroscopy: The past, present and future. Chemistry Review. 2010 doi: 10.1021/cr900250y. in Press. [DOI] [PubMed] [Google Scholar]
- 12.Mountford CE, Doran S, Lean CL, Russell P. Proton MRS can determine the pathology of human cancers with a high level of accuracy. Chemical Reviews. 2004;104(8):3677–3704. doi: 10.1021/cr030410g. [DOI] [PubMed] [Google Scholar]
- 13.Mountford C, Lean C, Malycha P, Russell P. Proton spectroscopy provides accurate pathology on biopsy and in vivo. Journal of Magnetic Resonance Imaging. 2006;24(3):459–477. doi: 10.1002/jmri.20668. [DOI] [PubMed] [Google Scholar]
- 14.Andronesi OC, Blekas KD, Mintzopoulos D, Astrakas L, Black PM, Tzika AA. Molecular classification of brain tumor biopsies using solid-state magic angle spinning proton magnetic resonance spectroscopy and robust classifiers. International Journal of Oncology. 2008;33(5):1017–1025. doi: 10.3892/ijo_00000000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeener J. Ampere International Summer School. Yugoslavia: Bask Polje; 1971. [Google Scholar]
- 16.Ernst RR. 2-Dimensional Spectroscopy. Chimia. 1975;29(4):179–183. [Google Scholar]
- 17.Aue WP, Karhan J, Ernst RR. Homonuclear Broad-Band Decoupling and 2-Dimensional J-Resolved NMR-Spectroscopy. Journal of Chemical Physics. 1976;64(10):4226–4227. [Google Scholar]
- 18.Jeener J, Meier BH, Bachmann P, Ernst RR. Investigation of Exchange Processes by 2-Dimensional Nmr-Spectroscopy. Journal of Chemical Physics. 1979;71(11):4546–4553. [Google Scholar]
- 19.Balaban RS, Kantor HL, Ferretti JA. Invivo Flux between Phosphocreatine and Adenosine-Triphosphate Determined by Two-Dimensional Phosphorus NMR. Journal of Biological Chemistry. 1983;258(21):2787–2789. [PubMed] [Google Scholar]
- 20.Ryner LN, Sorenson JA, Thomas MA. 3D Localized 2D NMR-Spectroscopy on an MRI Scanner. Journal of Magnetic Resonance Series B. 1995;107(2):126–137. doi: 10.1006/jmrb.1995.1068. [DOI] [PubMed] [Google Scholar]
- 21.Ryner LN, Sorenson JA, Thomas MA. Localized 2d J-Resolved H-1 MR Spectroscopy - Strong-Coupling Effects in-Vitro and in-Vivo. Magn Reson Imaging. 1995;13(6):853–869. doi: 10.1016/0730-725x(95)00031-b. [DOI] [PubMed] [Google Scholar]
- 22.Lange T, Trabesinger AH, Schulte RF, Dydak U, Boesiger P. Prostate spectroscopy at 3 tesla using two-dimensional S-PRESS. Magnetic Resonance in Medicine. 2006;56(6):1220–1228. doi: 10.1002/mrm.21082. [DOI] [PubMed] [Google Scholar]
- 23.Dreher W, Leibfritz D. Detection of homonuclear decoupled in vivo proton NMR spectra using constant time chemical shift encoding: CT-PRESS. Magn Reson Imaging. 1999;17(1):141–150. doi: 10.1016/s0730-725x(98)00156-8. [DOI] [PubMed] [Google Scholar]
- 24.Thomas MA, Yue K, Binesh N, Davanzo P, Kumar A, Siegel B, Frye M, Curran J, Lufkin R, Martin P, Guze B. Localized two-dimensional shift correlated MR spectroscopy of human brain. Magnetic Resonance in Medicine. 2001;46(1):58–67. doi: 10.1002/mrm.1160. [DOI] [PubMed] [Google Scholar]
- 25.McKinnon GC, Bosiger P. Localized Double-Quantum Filter and Correlation Spectroscopy Experiments. Magnetic Resonance in Medicine. 1988;6(3):334–343. doi: 10.1002/mrm.1910060311. [DOI] [PubMed] [Google Scholar]
- 26.Thomas MA, Lange T, Velan SS, Nagarajan R, Raman S, Gomez A, Margolis D, Swart S, Raylman RR, Schulte RF, Boesiger P. Two-dimensional MR spectroscopy of healthy and cancerous prostates in vivo. Magnetic Resonance Materials in Physics Biology and Medicine. 2008;21(6):443–458. doi: 10.1007/s10334-008-0121-7. [DOI] [PubMed] [Google Scholar]
- 27.Ramadan S, Mountford CE. Two-Dimensional Magnetic Resonance Spectroscopy on Biopsy and In vivo; Annual Reports on NMR Spectroscopy; Burlington: Academic Press; 2009. pp. 161–199. [Google Scholar]
- 28.De Graaf RA. In vivo NMR Spectroscopy Principles and Techniques. Chichester, England: John Wiley & Sons Ltd; 2007. [Google Scholar]
- 29.Marjanska M, Henry PG, Bolan PJ, Vaughan B, Seaquist ER, Gruetter R, Ugurbil K, Garwood M. Uncovering hidden in vivo resonances using editing based on localized TOCSY. Magnetic Resonance in Medicine. 2005;53(4):783–789. doi: 10.1002/mrm.20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marjanska M, Henry PG, Ugurbil K, Gruetter R. Editing through multiple bonds: Threonine detection. Magnetic Resonance in Medicine. 2008;59(2):245–251. doi: 10.1002/mrm.21492. [DOI] [PubMed] [Google Scholar]
- 31.Choi IY, Lee SP, Shen J. Selective homonuclear Hartmann-Hahn transfer method for in vivo spectral editing in the human brain. Magnetic Resonance in Medicine. 2005;53(3):503–510. doi: 10.1002/mrm.20381. [DOI] [PubMed] [Google Scholar]
- 32.Andronesi OC, Ramadan S, Ratai EM, Jennings D, Mountford CE, Sorensen AG. Spectroscopic imaging with improved constant adiabaticity gradient modulated pulses on high-field clinical scanners. Journal of Magnetic Resonance. 2010;203(2):283–293. doi: 10.1016/j.jmr.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garwood M, DelaBarre L. The return of the frequency sweep: Designing adiabatic pulses for contemporary NMR. Journal of Magnetic Resonance. 2001;153(2):155–177. doi: 10.1006/jmre.2001.2340. [DOI] [PubMed] [Google Scholar]
- 34.Rance M. Improved Techniques for Homonuclear Rotating-Frame and Isotropic Mixing Experiments. Journal of Magnetic Resonance. 1987;74(3):557–564. [Google Scholar]
- 35.Sorensen OW, Rance M, Ernst RR. Z-Filters for Purging Phase-Distorted or Multiplet-Distorted Spectra. Journal of Magnetic Resonance. 1984;56(3):527–534. [Google Scholar]
- 36.Tannus A, Garwood M. Adiabatic pulses. NMR in Biomedicine. 1997;10(8):423–434. doi: 10.1002/(sici)1099-1492(199712)10:8<423::aid-nbm488>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 37.Kinchesh P, Ordidge RJ. Spin-echo MRS in humans at high field: LASER localisation using FOCI pulses. Journal of Magnetic Resonance. 2005;175(1):30–43. doi: 10.1016/j.jmr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Kupce E, Schmidt P, Rance M, Wagner G. Adiabatic mixing in the liquid state. Journal of Magnetic Resonance. 1998;135(2):361–367. doi: 10.1006/jmre.1998.1607. [DOI] [PubMed] [Google Scholar]
- 39.Peti W, Griesinger C, Bermel W. Adiabatic TOCSY for C,C and H,H J-transfer. Journal of Biomolecular Nmr. 2000;18(3):199–205. doi: 10.1023/a:1026785725363. [DOI] [PubMed] [Google Scholar]
- 40.Braunschweiler L, Ernst RR. Coherence Transfer by Isotropic Mixing - Application to Proton Correlation Spectroscopy. Journal of Magnetic Resonance. 1983;53(3):521–528. [Google Scholar]
- 41.Davis DG, Bax A. Assignment of Complex H-1-NMR Spectra Via Two-Dimensional Homonuclear Hartmann-Hahn Spectroscopy. Journal of the American Chemical Society. 1985;107(9):2820–2821. [Google Scholar]
- 42.Hurd RE. Gradient-Enhanced Spectroscopy. Journal of Magnetic Resonance. 1990;87(2):422–428. doi: 10.1016/j.jmr.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Bottomley PA. Spatial Localization in NMR-Spectroscopy In vivo. Annals of the New York Academy of Sciences. 1987;508:333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 44.Frahm J, Bruhn H, Gyngell ML, Merboldt KD, Hanicke W, Sauter R. Localized High-Resolution Proton Nmr-Spectroscopy Using Stimulated Echoes - Initial Applications to Human-Brain Invivo. Magnetic Resonance in Medicine. 1989;9(1):79–93. doi: 10.1002/mrm.1910090110. [DOI] [PubMed] [Google Scholar]
- 45.Levitt MH, Freeman R, Frenkiel T. Broad-Band Heteronuclear Decoupling. Journal of Magnetic Resonance. 1982;47(2):328–330. [Google Scholar]
- 46.Uhrin D, Barlow PN. Gradient-enhanced one-dimensional proton chemical-shift correlation with full sensitivity. Journal of Magnetic Resonance. 1997;126(2):248–255. [Google Scholar]
- 47.Kover KE, Uhrin D, Hruby VJ. Guadient- and sensitivity-enhanced TOCSY experiments. Journal of Magnetic Resonance. 1998;130(2):162–168. doi: 10.1006/jmre.1997.1309. [DOI] [PubMed] [Google Scholar]
- 48.Ramadan S, Ratai E-M, Andronesi OC, Sorensen AG, Mountford CE. Adiabatic L-COSY at 7T. Proceedings of the 17th ISMRM meeting.2009. [Google Scholar]
- 49.Andronesi OC, Ramadan S, Mountford CE, Sorensen AG. Towards a localized low power adiabatic 2D TOCSY for in-vivo use on clinical platforms. Proceedings of the 18th ISMRM Meeting; Stockolm, Sweeden. 2010. Abstract #3397. [Google Scholar]
- 50.Kupce E, Freeman R. Adiabatic Pulses for Wide-Band Inversion and Broad-Band Decoupling. Journal of Magnetic Resonance Series A. 1995;115(2):273–276. [Google Scholar]
- 51.Haeberlen U, Waugh JS. Coherent Averaging Effects in Magnetic Resonance. Physical Review. 1968;175(2):453. [Google Scholar]
- 52.Levante TO, Baldus M, Meier BH, Ernst RR. Formalized Quantum-Mechanical Floquet Theory and Its Application to Sample-Spinning in Nuclear-Magnetic-Resonance. Molecular Physics. 1995;86(5):1195–1212. [Google Scholar]
- 53.Mangia S, Liimatainen T, Garwood M, Michaeli S. Rotating frame relaxation during adiabatic pulses vs. conventional spin lock: simulations and experimental results at 4 T. Magn Reson Imaging. 2009;27(8):1074–1087. doi: 10.1016/j.mri.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith SA, Levante TO, Meier BH, Ernst RR. Computer-Simulations in Magnetic-Resonance - an Object-Oriented Programming Approach. Journal of Magnetic Resonance Series A. 1994;106(1):75–105. [Google Scholar]
- 55.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR in Biomedicine. 2000;13(3):129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 56.Garwood M, Yong K. Symmetrical Pulses to Induce Arbitrary Flip Angles with Compensation for Rf Inhomogeneity and Resonance Offsets. Journal of Magnetic Resonance. 1991;94(3):511–525. [Google Scholar]
- 57.Ogg RJ, Kingsley PB, Taylor JS. Wet, a T-1-Insensitive and B-1-Insensitive Water-Suppression Method for in-Vivo Localized H-1-NMR Spectroscopy. Journal of Magnetic Resonance Series B. 1994;104(1):1–10. doi: 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- 58.Gruetter R, Tkac I. Field mapping without reference scan using asymmetric echo-planar techniques. Magnetic Resonance in Medicine. 2000;43(2):319–323. doi: 10.1002/(sici)1522-2594(200002)43:2<319::aid-mrm22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 59.van der Kouwe AJW, Benner T, Salat DH, Fischl B. Brain morphometry with multiecho MPRAGE. Neuroimage. 2008;40(2):559–569. doi: 10.1016/j.neuroimage.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conolly S, Nishimura D, Macovski A. A Selective Adiabatic Spin-Echo Pulse. Journal of Magnetic Resonance. 1989;83(2):324–334. doi: 10.1002/mrm.1910180105. [DOI] [PubMed] [Google Scholar]
- 61.Davis AL, Estcourt G, Keeler J, Laue ED, Titman JJ. Improvement of Z Filters and Purging Pulses by the Use of Zero-Quantum Dephasing in Inhomogeneous B1 or B0 Fields. Journal of Magnetic Resonance Series A. 1993;105(2):167–183. [Google Scholar]
- 62.Tran TKC, Vigneron DB, Sailasuta N, Tropp J, Le Roux P, Kurhanewicz J, Nelson S, Hurd R. Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer. Magnetic Resonance in Medicine. 2000;43(1):23–33. doi: 10.1002/(sici)1522-2594(200001)43:1<23::aid-mrm4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 63.Martinez-Ramon M, Gallardo-Antolin A, Cid-Sueiro J, Heileman GL, Yung KT, Zheng WL, Zhao CG, Posse S. Automatic Placement of Outer Volume Suppression Slices in MR Spectroscopic Imaging of the Human Brain. Magnetic Resonance in Medicine. 63(3):592–600. doi: 10.1002/mrm.22275. [DOI] [PubMed] [Google Scholar]
- 64.Thomas MA, Hattori N, Umeda M, Sawada T, Naruse S. Evaluation of two-dimensional L-COSY and PRESS using a 3 T MRI scanner: from phantoms to human brain in vivo. NMR in Biomedicine. 2003;16(5):245–251. doi: 10.1002/nbm.825. [DOI] [PubMed] [Google Scholar]
- 65.Schulte RF, Boesiger P. ProFit: two-dimensional prior-knowledge fitting of J-resolved spectra. NMR in Biomedicine. 2006;19(2):255–263. doi: 10.1002/nbm.1026. [DOI] [PubMed] [Google Scholar]
- 66.Wiggins GC, Triantafyllou C, Potthast A, Reykowski A, Nittka M, Wald LL. 32-Channel 3 tesla receive-only phased-array head coil with soccer-ball element geometry. Magnetic Resonance in Medicine. 2006;56(1):216–223. doi: 10.1002/mrm.20925. [DOI] [PubMed] [Google Scholar]
- 67.Rucker SP, Shaka AJ. Broad-Band Homonuclear Cross Polarization in 2D NMR Using Dipsi-2. Molecular Physics. 1989;68(2):509–517. [Google Scholar]
- 68.Hess A, Andronesi OC, Tisdall MD, Meintjes EM, van der Kouwe AJW. Motion artefact correction in spectroscopic imaging using an EPI navigator and reacquisition. Proceedings of the 18th ISMRM Meeting; Stockolm, Sweeden. 2010. Abstract #3308. [Google Scholar]
- 69.Hess A, Andronesi OC, Tisdall MD, Meintjes EM, van der Kouwe AJW. Real time measurement and correction of B0 field disruption in neuro spectroscopic imaging. Proceedings of the 18th ISMRM Meeting; Stockolm, Sweeden. 2010. Abstract #379. [Google Scholar]
- 70.Adalsteinsson E, Spielman DM. Spatially resolved two-dimensional spectroscopy. Magnetic Resonance in Medicine. 1999;41(1):8–12. doi: 10.1002/(sici)1522-2594(199901)41:1<8::aid-mrm3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 71.Freeman R, Kupce E. New ways to record multidimensional NMR spectra. Current Analytical Chemistry. 2006;2(2):101–105. [Google Scholar]
- 72.Frydman L, Scherf T, Lupulescu A. The acquisition of multidimensional NMR spectra within a single scan. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(25):15858–15862. doi: 10.1073/pnas.252644399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wiggins GC, Polimeni JR, Potthast A, Schmitt M, Alagappan V, Wald LL. 96-Channel Receive-Only Head Coil for 3 Tesla: Design Optimization and Evaluation. Magnetic Resonance in Medicine. 2009;62(3):754–762. doi: 10.1002/mrm.22028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baltes C, Radzwill N, Bosshard S, Marek D, Rudin M. Micro MRI of the mouse brain using a novel 400 MHz cryogenic quadrature RF probe. Nmr in Biomedicine. 2009;22(8):834–842. doi: 10.1002/nbm.1396. [DOI] [PubMed] [Google Scholar]
- 75.Nelson SJ, Vigneron D, Kurhanewicz J, Chen A, Bok R, Hurd R. DNP-hyperpolarized C-13 magnetic resonance metabolic imaging for cancer applications. Applied Magnetic Resonance. 2008;34(3-4):533–544. doi: 10.1007/s00723-008-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]