Abstract
Bacterial type III secretion system (T3SSs) chaperones pilot substrates to the export apparatus in a secretion-competent state, and are consequently central to the translocation of effectors into target cells. Chlamydia trachomatis is a genetically intractable obligate intracellular pathogen that utilizes T3SS effectors to trigger its entry into mammalian cells. The only well-characterized T3SS effector is TARP (translocated actin recruitment protein), but its chaperone is unknown. Here we exploited a known structural signature to screen for putative type III secretion chaperones encoded within the C. trachomatis genome. Using bacterial two-hybrid, co-precipitation, cross-linking, and size exclusion chromatography we show that Slc1 (SycE-like chaperone 1; CT043) specifically interacts with a 200 amino acid residue N-terminal region of TARP (TARP1–200). Slc1 formed homodimers in vitro, as shown in crosslinking and gel filtration experiments. Biochemical analysis of an isolated Slc1-TARP1–200 complex was consistent with a characteristic 2:1 chaperone-effector stoichiometry. Furthermore, Slc1 was co-immunoprecipitated with TARP from C. trachomatis elementary bodies. Also, co-expression of Slc1 specifically enhanced host cell translocation of TARP by a heterologous Yersinia enterocolitica T3SS. Taken together, we propose Slc1 as a chaperone of the C. trachomatis T3SS effector TARP.
INTRODUCTION
Bacterial virulence factors designed to modulate host cell functions are translocated from the bacteria into the host cell where they target and subvert key host processes to promote bacterial infection and replication (Cornelis & Van Gijsegem, 2000, Ghosh, 2004). Effector translocation, requires a specialized machinery called a Type III secretion system (T3SS) that serves as a conduit that connects the bacterial cytosol and the host cell cytoplasm (Creasey et al., 2003, Elliott et al., 1999, Matsumoto & Young, 2009, Page et al., 2001, Page & Parsot, 2002, Thomas et al., 2005). Most T3SS Class I chaperones are small (14–18 kDa), have an acidic isoelectric point, and share a secondary structural α-β-β-β-α-β-β motif (Akeda & Galan, 2005, Birtalan & Ghosh, 2001, Singer et al., 2004). In general, T3SS chaperones also dimerize (Schubot et al., 2005, Birtalan & Ghosh, 2001), and bind their cognate effector with a stoichiometry of two chaperone protomers (i.e. a dimer) to one effector molecule (Buttner et al., 2005). Chaperone dimers bind to the N-terminal regions of their effectors and are proposed to pilot the effector to the T3SS (Spaeth et al., 2009), stabilize the effector (Elliott et al., 1999, Matsumoto & Young, 2009), maintain the effector in a translocation-competent conformation (Rodgers et al., 2008, Stebbins & Galan, 2001), and prevent premature effector homo- and hetero-oligomerisation or aggregation in the bacterial cytosol (Letzelter et al., 2006).
The best-studied T3SS-directed process in Chlamydia is invasion (Clifton et al., 2005, Clifton et al., 2004, Jamison & Hackstadt, 2008). Being obligate intracellular pathogens, chlamydial access to their intracellular niche is paramount to their survival. Uniquely, chlamydiae exist in two distinct developmental forms, a small robust elementary body (EB) and a vegetative reticulate body (RB). The extracellular EBs that initiate infection are compact and spore-like in structure, and are apparently metabolically inert (Fields & Hackstadt, 2002). Nevertheless, EBs bind and force their own entry into non-phagocytic mammalian target cells (Byrne, 1976). Upon internalization, pathogen-containing vacuoles are diverted from the normal host endolysosomal maturation pathway, enabling the formation of a conditioned intracellular niche or ‘inclusion’ in which chlamydiae differentiate and replicate (Fields & Hackstadt, 2002).
One of the only well-characterized T3SS effectors in Chlamydia is the translocated actin recruiting protein (TARP), which initiates the remodelling of the actin cytoskeleton directly or by stimulating cellular signalling to indirectly activate the host actin-nucleating Arp2/3 complex (Clifton et al., 2004, Jewett et al., 2006, Lane et al., 2008). Actin remodelling is critical for Chlamydia invasion (Carabeo et al., 2002), so timely T3SS-dependent TARP translocation is key to the survival of the obligate intracellular bacteria.
All chlamydiae spp encode a type III secretion system (T3SS). However, studying the activities of the chlamydial T3SS system is compounded by the genetic intractability of these bacteria. Nevertheless, chlamydia infection of cultured cells is inhibited by known chemical inhibitors of other T3SSs (Muschiol et al., 2006, Wolf et al., 2006), suggesting that T3SS effectors are pivotal in manipulating host cell processes throughout the lifecycle. Putative translocated effectors have been identified, primarily from the use of surrogate bacterial systems (Subtil et al., 2005). Perhaps the best characterized is the entry-associated effector TARP.
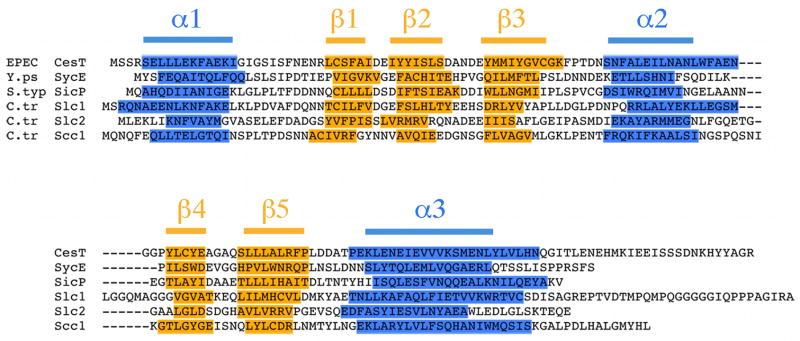
The T3SS chaperone for TARP has not been identified. It is clear that substrate-specific chaperones are necessary for the efficient translocation of effectors in disparate T3SSs, so it is likely that chlamydiae spp are no exception. Shotgun proteomic studies of purified EBs suggested three candidate proteins of unknown function with chaperone-like characteristics – CT043, CT088/Scc1, and CT663 – which are apparently present in EBs together with TARP (Skipp et al., 2005). Indeed, although alignment of the putative chaperones, CT043, CT663, and CT088/Scc1 encoded within the C. trachomatis serovar D genome revealed very limited sequence similarity at the amino acid level, direct comparison of their predicted secondary structure revealed more significant homology to known type III export chaperones (Beeckman & Vanrompay, 2010, Spaeth et al., 2009, Pallen et al., 2005, Kim, 2001). The α-β-β-β-α-β-β signature common to bacterial class 1 T3SS chaperones was readily identified in each of the three putative chlamydial chaperones when aligned with CesT (enteropathogenic Escherichia coli, EPEC), SycE (Yersinia enterocolitica) and SicP (Salmonella enterica serovar typhimurium) (Figure 1) (Page & Parsot, 2002, Luo et al., 2001, Birtalan & Ghosh, 2001). Based on this close structural relationship with SycE, we propose the names Slc1 and Slc2 (SycE-like chaperone) for CT043 and CT663, respectively.
Figure 1.
Three C. trachomatis proteins share predicted secondary structural features characteristic of known type III export chaperones. Primary amino acid sequence alignment of known type III export chaperones CesT (enteropathogenic Escherichia coli, EPEC), SycE (Yersinia enterocolitica) and SicP (Salmonella enterica serovar typhimurium) with the putative export chaperones Slc1 (CT043), Slc2 (CT663) and Scc1 (CT088) of C. trachomatis. Secondary structure was predicted using Jpred and sequences aligned manually to according to the α-β-β-β-α-β-β-α signature characteristic of some type III export chaperones. Residues predicted to form α-helices and β-strands are highlighted blue and yellow, respectively.
Full-length TARP is translocated by the Yersinia enterocolitica T3SS, and the first 200 residues of TARP are sufficient for secretion in the same heterologous T3SS, suggesting the possible presence of a binding site for chaperones within this region (Clifton et al., 2004, Clifton et al., 2005). Here we screened for interactions between the first 200 amino acids of TARP (TARP1–200) and each of the putative chaperones using multiple independent assays. We show that homodimeric Slc1 specifically interacts with TARP1–200. When co-expressed with TARP in laboratory E.coli, a TARP1–200-Slc1 complex could be isolated. Furthermore, the apparent molecular weight of this complex is consistent with a 2:1 ratio of chaperone to TARP. Furthermore, Slc1 co-immunoprecipitated with TARP from C. trachomatis EB lysates. Translocation of TARP by the heterologous Yersinia enterocoilitica T3SS is specifically enhanced in the presence of co-expressed Slc1. We propose that Slc1 is the T3SS chaperone for the C. trachomatis invasion effector TARP, and that the first 200 amino acids of TARP contain the information sufficient for Slc1 binding and translocation.
RESULTS
The N-terminal 200 residues of TARP interact specifically with the putative chlamydial chaperone Slc1
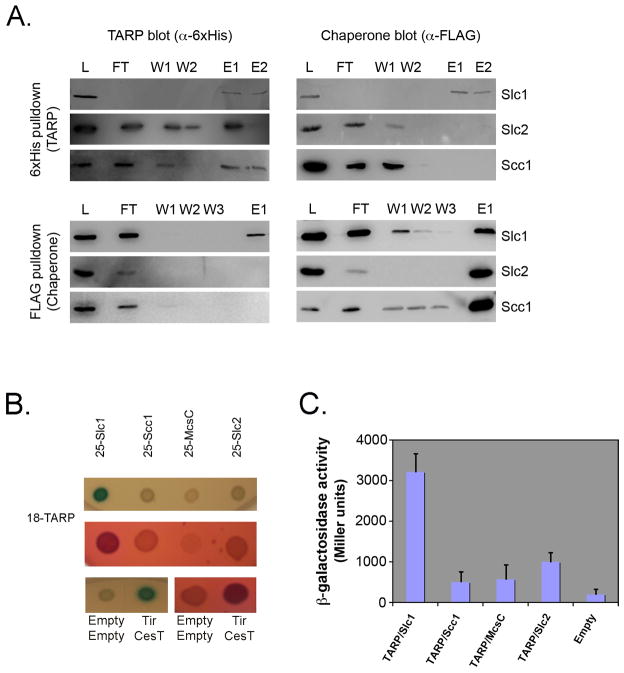
As the first 200 amino acids of TARP are sufficient for secretion by the Yersinia T3SS (Clifton et al., 2004), we used co-precipitation and bacterial two-hybrid analyses to investigate whether TARP1–200 could interact with any of the three putative chlamydial chaperones Slc1, Slc2, or Scc1 highlighted by our sequence analysis. For the co-precipitation experiments, His-tagged TARP1–200 was co-expressed in laboratory E.coli with the FLAG-tagged version of Slc1, Slc2, or Scc1, and the chaperones captured using anti-FLAG agarose. His6-TARP1–200 and FLAG-chaperones were monitored in the lysate, unbound (flow-through, FT), wash (W) and elution (E) fractions by immunoblotting. His6-TARP1–200 was specifically co-precipitated with Slc1-FLAG in the elution fraction, but not with Slc2-FLAG or Scc-FLAG under equivalent conditions (Figure 2).
Figure 2.
The N-terminal 200 amino acids of Tarp interact with Slc1. (A). Co-precipitation of Slc1-FLAG, but not Slc2-FLAG or Scc1-FLAG by His6-Tarp1–200 (top) and co-immunoprecipitation of His6-Tarp1–200 by Scl1-FLAG, but not Slc2-FLAG or Scc1-FLAG from laboratory E.coli lysates following co-expression of Tarp and chaperone. Samples are of flowthrough (FT), washes (W) and elution (E) fractions. (B). LB-X-Gal-IPTG (upper) and McConkey-maltose (lower) agar plates indicating positive blue and fuchsia colonies, respectively. In this bacterial two-hybrid assay, partner interaction reconstitutes toxin activity, which is reported by the colour change in colonies on appropriate indicator plates. Positive control for interaction was EPEC Tir with its cognate chaperone CesT. Empty vectors represented the negative control (C). β-galactosidase activity of E.coli DHM1 strains co-expressing Cya18-Tarp1–200 and Cya25-Slc1, Cya25-Slc2, Cya25-Scc1 or Cya25-Mcsc. Activity is expressed as Miller Units. Data in (B) and (C) are representative of three independent trials.
To confirm the specific interaction between Slc1-FLAG and His6-TARP1–200, a second reciprocal precipitation was performed whereby His6-TARP1–200 was captured using His6-binding Ni-NTA magnetic beads from an E.coli lysate following co-expression of the FLAG-tagged version of Slc1, Slc2, or Scc1. Slc1-FLAG was specifically co-precipitated by immobilized His6-TARP1–200 (Figure 2). As expected, neither Slc2-FLAG nor Scc1-FLAG was captured from lysates following equivalent co-expression with His6-TARP1–200.
Next, we exploited the bacterial two-hybrid system to independently validate these co-precipitation data. In these assays, interaction is reported by in trans reassembly of active Bordetella pertussis adenylate cyclase (Cya) from two constituent fragments (termed 18- and 25-), which are fused to the target and bait proteins, respectively. Target-bait interaction allows Cya-dependent conversion of ATP to cAMP, reported as either lac-dependent β-galactosidase (β-gal) production allowing cleavage of the colorimetric substrate X-Gal to yield blue bacterial colonies, or a mal-dependent pH-induced colour change of phenol red on McConkey agar indicator plates (Karimova et al., 1998). Cya18-TARP1–200 was co-expressed in cya-deficient E.coli DHM1 together with individual Cya25-chaperone fusions (Cya25-Slc1, Cya25-Slc2, or Cya25-Scc1), and additionally the recently described Mcsc that has a chaperone-like activity (Cya25-Mcsc) (Spaeth et al., 2009). Cya18-TARP1–200 and Cya25-Slc1 co-expression induced blue and fuchsia colonies on X-Gal and McConkey plates, respectively (Figure 2B), similar to the positive control Cya18-Tir and Cya25-CesT co-transformants, with CesT being the cognate chaperone of Tir. In contrast, bacteria co-expressing Cya18-TARP1–200 and Cya25-Slc2, Cya25-Scc1 or Cya25-Mcsc induced no significant colour changes on either indicator media.
These qualitative observations were quantified by measuring β-galactosidase-dependent conversion of ONPG to the colorimetric molecule ONP, standardized as Miller Units as previously described (Griffith & Wolf, 2002). Only Cya18-TARP1–200/Cya25-Slc1 co-expressing bacteria exhibited activity that differed significantly from the negative control strain, whereas the other Tarp-chaperone pairs exhibited only minimal levels of β-galactosidase activity (Figure 2C). Taken together, these combined data from the co-precipitation and bacterial two-hybrid experiments support a specific interaction between the first 200 residues of the C. trachomatis TARP effector and the putative Slc1 chaperone.
Purified Slc1 forms homodimers in solution
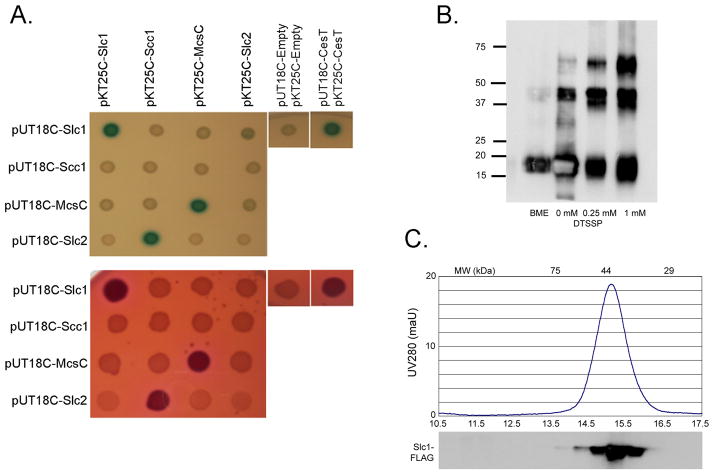
Class I T3SS chaperones typically function as a homodimer, although chaperone heterodimerization has been suggested in Chlamydia (Spaeth et al., 2009, Fields et al., 2005). We therefore investigated whether Slc1 could interact with itself, Slc2 or Scc1 in addition to TARP1–200 using a similar bacterial two-hybrid approach. Chaperone-chaperone interactions were systematically evaluated using E.coli 2-hybrid analyses by screening all the possible pair-wise combinations. Interactions were detected between Cya18-Slc1:Cya25-Slc1, Cya18-Mcsc:Cya25-Mcsc, and Cya18-Slc2:Cya25-Scc1 as indicated by blue and fuchsia colonies on X-gal and McConkey indicator plates, respectively (Figure 3A). This screen therefore identified both a novel interaction (Slc1-Slc1) and confirms those suggested previously by a recent two-hybrid study (Mcsc:Mcsc and Slc2:Scc1) (Spaeth et al., 2009, Fields et al., 2005). No other pair-wise interaction partners were identified. Intriguingly, while Cya18-Slc2:Cya25-Scc1 demonstrated interaction, the reverse pairing Cya18-Scc1 and Cya25-Slc2 did not, a disparity possibly related to conformational differences in the Cya18 and Cya25 fusions.
Figure 3.
Detection of chaperone:chaperone interactions. (A) LB + X-gal + IPTG (top) or McConkey + maltose (bottom) agar plates indicate interaction between co-expressed Cya15 or Cya18 fused chlamydial chaperones in the cya-deficient E. coli strain DHM1. Upregulation of the lactose and maltose operons results from fusion-protein interaction and is indicated by blue or fuchsia colonies. Data in (A) are representative of multiple trials. (B) An immunoblot of β-mercaptoethanol (βME), mock-treated (−), or 0.25 mM DTSSP crosslinked (+) purified Slc1 was probed with αFLAG antibody following separation in 12% SDS-PAGE gel under non-reducing conditions. (C) The relative abundance of native M2-purified Slc1 complexes were detected and eluted in 500 ul fractions by gel filtration chromatography. An immunoblot of trichloracetic acid (TCA)-precipitated fractions was detected with α-FLAG antibody. Molecular weight standards are indicated at the top of the graph.
While Class I T3SS chaperones are predominantly homodimeric, trimeric and tetrameric states have been reported in other organisms (Fraser et al., 2003). Although the two-hybrid analyses reported Slc1:Slc1 interaction, this technique cannot distinguish the underlying oligomerization state. When analyzed by SDS-PAGE, purified Slc1-FLAG migrated predominantly at ~18kDa, although weak SDS-resistant species at ~40kDa were also present, which could be restored to monomers under reducing conditions (Figure 3B). Crosslinking with DTSSP substantially stabilized the ~40kDa species and a higher-order species at ~60kDa (Figure 3B). These in vitro data again show that Slc1 forms homooligomers. While it is tempting to speculate that the preferential stabilization of the ~40kDa species would also indicate that dimers predominate in solution, our chemical cross-linking also captured a minor higher order species, which may either reflect a cross-linking artefact or be of relevance. To investigate this further, we next employed gel filtration chromatography to independently analyze the native oligomeric state of Slc1. Purified Slc1-FLAG was loaded onto a calibrated Superdex 200 10/300 GL column with an in-line UV detector, developed in phosphate-buffered saline. In parallel, elution fractions were collected and Slc1-FLAG detected by immunoblotting. A single monodispersed Slc1 species was always observed (Figure 3C). Comparison of the peak fraction to a series of standards indicated a molecular weight of ~40kDa, consistent with the expected size of an Slc1 dimer. These combined data strongly suggest that purified Slc1 forms homodimers in solution.
Characterisation of the Slc1-TARP complex
To gain a more detailed insight into the composition of the Slc1-TARP complex, we continued to employ gel filtration chromatography to compare the apparent molecular weights of purified His6-TARP1–200 and the His6-TARP1–200:Slc1-FLAG complex. Purified His6-TARP1–200 alone eluted as a single species when subjected to gel filtration, corresponding to a molecular weight ~25kDa (Figure 4, TARP in blue). This is consistent with His6-TARP1–200 exclusively forming monomers in solution. Following co-expression of Slc1-FLAG and His6-TARP1–200, the His6-TARP1–200:Slc1-FLAG complex was again isolated from lysates using Ni-NTA resin. The complex was analyzed by gel filtration chromatography and each component detected in the resultant fractions using anti-His6 conjugate (TARP) and anti-FLAG antibodies (Slc1). The dominant peak, correlating with a molecular weight >75kDa, contained both Slc1-FLAG and His6-TARP1–200 (Figure 4, Slc1 and TARP in red). This represents a significant shift in the elution profile of Slc1 in the presence of TARP1–200 (compare Figure 4 to Slc1-FLAG alone in Figure 3C). A second peak, corresponding to monomeric His6-TARP1–200 was also observed (Figure 4, compare TARP traces in red and blue), the relative amount of which varied between trials. This would be expected since any excess His6-TARP1–200 in the lysate would also be captured by the Ni-NTA resin used to bind the complex, the amount of which would depend upon the relative expression of His6-TARP1–200 and Slc1-FLAG in each individual experiment. Interestingly, the His6-TARP1–200 present in the complex was more readily detected upon immunoblotting than the monomeric species, perhaps indicative of increased exposure of the His6 tag in the chaperone-bound state. Nevertheless, these were the only two peaks observed, illustrating that His6-TARP1–200 always engaged Slc1-FLAG in a complex of defined stoichiometry. The size of this preferential single complex is consistent with the typical 2:1 chaperone:effector stoichiometry. Furthermore, co-expression of His6-TARP1–200 and Slc1-FLAG 1-was apparently necessary for complex formation, as co-incubation of pre-purified His6-TARP1–200 and Slc1-FLAG isolated from separate E.coli lysates failed to efficiently reconstitute this complex (Supplementary Figure 1).
Figure 4.
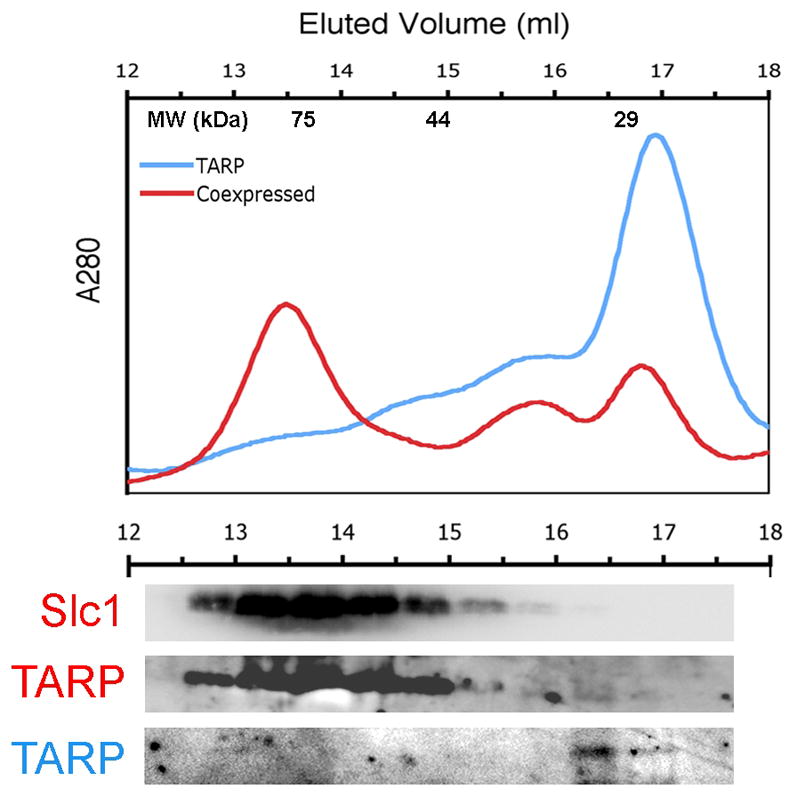
Separation and detection of co-expressed Slc1: Tarp complexes. The relative abundances of native NiNta-purified Tarp1–200 complexes alone (blue trace) or co-expressed with Slc1 (red trace) were detected and eluted in 500 ul fractions (top) by gel filtration chromatography. Molecular weight standards are indicated at the top of the graph. Immunoblots of the TCA-precipitated elution fractions were detected with α6XHIS (TARP) and αFLAG (Slc1) antibodies (bottom).
Isolation of the Slc1-Tarp complex from C. trachomatis elementary bodies
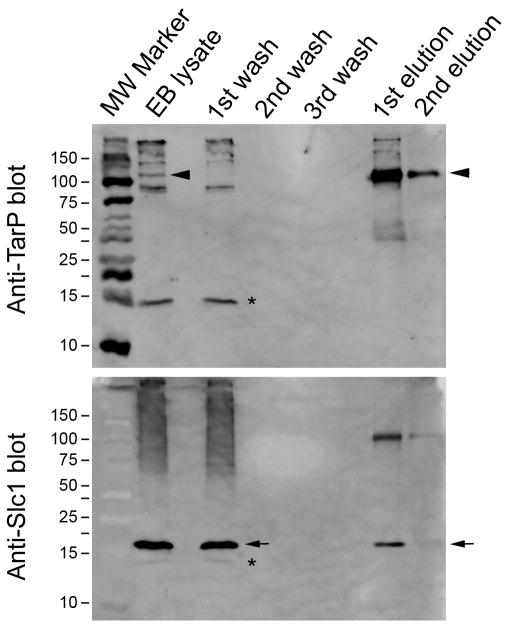
Given the apparent specific interaction of Slc1 with TARP in laboratory E.coli and the co-temporal expression of these proteins in C. trachomatis EBs, we investigated whether Slc1 and TARP form a complex in EBs. TARP was immunoprecipitated from lysates of density gradient-purified C. trachomatis serovar D EBs using a rabbit polyclonal anti-TARP antibody. The eluate contained a major species at >100 kDa (Figure 5, arrowheads), which was also present in the EB lysate, but absent in the wash fractions. The size is consistent with the predicted molecular weight of the unmodified (i.e. unphosphorylated) form of TARP. The blot was stripped and reprobed with an anti-Slc1 rabbit polyclonal antibody, which detected a prominent band of ~18 kDa in size in the lysate and wash fractions (Figure 5, arrows). More importantly, the same band was present in the eluate fractions. The >100 kDa species present on the anti-Slc1 blot were likely residual TARP from inefficient stripping, rather than cross-reactivity of the Slc1 antibody and TARP, since the complex would also have been present in the EB lysate. This observation of Slc1-TARP interaction in C. trachomatis EBs validated the co-precipitation and bacterial two-hybrid experiments using laboratory E.coli and also demonstrate an interaction between Slc1 and full-length TARP.
Figure 5.
Slc1 interacts with TARP in C. trachomatis elementary bodies. Immunoprecipitation of TARP (top blot) from EB lysates pulled down Slc1 (bottom blot) indicating interaction of the chaperone with TARP in Chlamydia EBs. Lanes from left to right are molecular weight markers (MW), lysate (Lys), wash (W) and elution (E) fractions. Anti-TARP blot was stripped and reprobed with anti-Slc1 antibody. The 103 kDa TARP and the 18 kDa Slc1 bands are indicated by the arrow and arrowheads, respectively. Asterisks indicate the background band to differentiate it from the Slc1 band.
TARP translocation by the Yersinia T3SS is specifically enhanced by Slc1
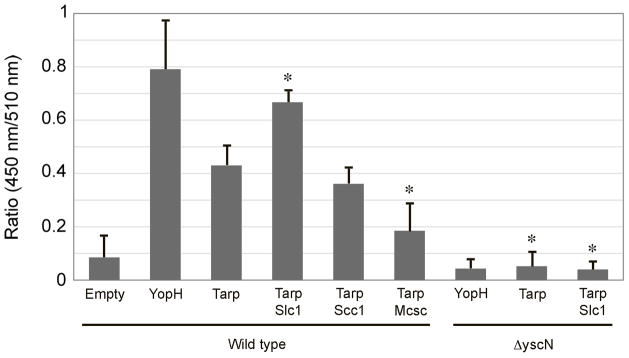
In the absence of a genetic system to mutate C. trachomatis slc1, we adopted a surrogate T3SS to investigate whether Slc1 plays a role in TARP translocation. The Yersinia enterocolitica T3SS is closely related to the putative Chlamydia T3SS. This system has been used previously both to screen for chlamydial substrates and to demonstrate that TARP1–200 is sufficient for TARP secretion via the T3SS (Clifton et al., 2005, Clifton et al., 2004). Furthermore, full-length TARP from C. trachomatis serovar L2 could be translocated into HeLa cells and tyrosine phosphorylated (Clifton et al., 2005). We used a reporter system to monitor TARP translocation into HeLa cells in the presence and absence of co-expressed Slc1. In this assay, T3SS effectors are fused to β-lactamase, which upon translocation into the target cell will cleave the β-lactam ring that brings in close proximity two fluorophores in the reporter molecule CCF2-AM. Induced cleavage leads to a loss of fluorescence resonance energy transfer (FRET) between the intramolecular reporter fluorophores, an activity proportional to the amount of effector delivered (Charpentier & Oswald, 2004). Using this assay, translocation of the Yersinia enterocolitica T3SS effector YopH and C. trachomatis TARP1–200 into HeLa cells was clearly detected, although as expected TARP translocation was less efficient than that of endogenous YopH (Figure 6, wild-type TARP1–200 and YopH). The observed basal level of Tarp translocation confirms previous findings. However, translocation of TARP1–200 was significantly and reproducibly enhanced upon co-expression of Slc1 (Figure 6, wild-type TARP1–200 and TARP1–200 + Slc1), but not with Scc1 or Mcsc. The differences in translocation were observed under conditions of similar multiplicity of infection (MOI) and similar levels of TARP-β-lactamase fusion protein expression (Supplemental data 2). Translocation of both TARP and YopH reported by loss-of-FRET was negligible when analogous experiments were performed in a Y. enterocolitica ΔyscN deletion mutant that lacks the ATPase essential for the function of the T3SS. These findings demonstrate that the chaperone Slc1 specifically enhances the translocation of the cognate effector TARP via the surrogate Yersinia T3SS, and that the N-terminal 200 amino acids of TARP contain the necessary information for translocation.
Figure 6.
Enhancement of TARP1–200-β-lactamase fusion translocation by Slc1 in HeLa cells. Translocation of TARP fused to -β-lactamase in the absence or presence of expressed Slc1, Mcsc, or Slc2 were monitored in a Yersinia pseudotuberculosis Type III wild-type or “null” background. The level of translocation is indicated by the 450 nm:510 nm ratio. 450 nm values are indicators of a loss of FRET due to cleavage of CCF2/AM, and hence translocation. The data were obtained from four independent experiments each in triplicate, and each set adjusted by normalizing the values for the “EMPTY” group. Data are represented as mean ± S.D. Asterisks indicate groups that were statistically significantly different from the “TARP/wild type” group (p < 0.001, Student’s T-test).
DISCUSSION
Host cell invasion by the obligate intracellular bacterium Chlamydia is essential for its survival and subsequent replication. This process is driven by the bacteria, which deploy effector proteins via a T3SS to force their own uptake (Clifton et al., 2004). Delivered TARP rapidly triggers actin reorganization in the host cell both directly and indirectly (Carabeo et al., 2002, Jewett et al., 2006, Lane et al., 2008), although other effectors also likely contribute to this critical process (Hower et al., 2009). In other well-studied T3SSs, including the archetypal flagellum assembly pathway, substrate export and virulence protein translocation require the activity of cognate chaperones. In general, these chaperones are substrate-specific, although a few are more promiscuous. Here we show that CT043/Slc1, a previously uncharacterized protein in C. trachomatis, encodes a T3SS chaperone that binds to the N-terminal region of the effector TARP. Slc1 shares a structural signature reminiscent of class I T3SS chaperones, does not detectably interact with other putative chaperones identified from the chlamydial genome and forms homodimers in solution. Additionally, our further biochemical analysis showed that Slc1 dimers likely engage a single protomer of a TARP derivative that encompasses the first 200 residues (TARP1–200), a complex that only forms efficiently upon co-expression of TARP1–200 and Slc1 in E.coli. To extend these findings, we demonstrated by co-immunoprecipitation that Slc1 exists in a complex with TARP in Chlamydia elementary bodies. While TARP1–200 is sufficient for secretion by the Y. enterocolitica T3SS, we additionally demonstrate that it also directs TARP translocation into HeLa cells, an activity significantly enhanced by Slc1. On the basis of these combined features and their similarity to other known examples, we propose that the previously anonymous Slc1 is a T3SS chaperone for TARP.
Our notion that Slc1 is a T3SS a chaperone is also consistent with available transcriptome and proteome data. The Chlamydia slc1 gene is expressed throughout the infection cycle (Belland et al., 2003), possibly indicative of an essential function. The Slc1-Tarp complex in EBs would enable TARP to be primed in an export-competent state, ready for efficient translocation upon EB contact with the target cell. Indeed, TARP translocation does occur rapidly both upon cell contact and when induced artificially in vitro (Clifton et al., 2004). There is precedent for such activity; for example, the rapid translocation of Yersinia YopE upon contact with macrophages is dependent upon its chaperone SycE, as effector Yops are similarly pooled in a primed state in the bacterial cytosol (Lee et al., 1998).
Structural conservation amongst T3SS chaperones suggests that related interfaces govern chaperone dimerization and effector interactions (Luo et al., 2001, Yip & Strynadka, 2006). As with Slc1-TARP1–200, chaperone-binding regions lie within the N-terminal regions of other effectors. High-resolution crystal structures of Salmonella SicP-SptP35–139 (Stebbins & Galan, 2001) and SycE-YopE17–85 (Birtalan & Ghosh, 2001) show similar contacts even though the binding regions of the effectors share little primary sequence similarity. Chaperone-effector interactions occur between secondary structural elements of the effector and four hydrophobic patches located on the chaperone dimer (Birtalan & Ghosh, 2001, Luo et al., 2001, Stebbins & Galan, 2001). However, these contacts do not confer specificity, which arises from additional hydrogen bonds formed between partner residues outside these regions (Luo et al., 2001). Examination of the N-terminal region of TARP does not reveal any obvious motifs involved in chaperone binding, but it is clear that there must be conserved features as TARP from C. caviae serovar GPIC also interacts with Slc1 from C. trachomatis serovar L2. A closer analysis of these divergent N-terminal regions reveal a series of hydrophobic amino acid residues conserved between TARP sequences from other chlamydial species, which might be involved in chaperone interactions (Brinkworth & Carabeo, unpublished observations). This series of hydrophobic residues, along with the structural conservation of many T3SS chaperones possibly contribute to TARP translocation in the absence of Slc1. Interestingly, the expression of Mcsc, which does not interact with TARP modestly inhibited Slc1-independent TARP translocation. Mcsc participates a multi-protein interaction hub in Chlamydia (Spaeth et al., 2009), so could potentially interfere with essential protein-protein interactions required for translocation in Yersinia due to its incompatibility with other heterologous T3SS components.
We consistently recovered chaperone-effector complexes at a greater efficiency when both partners were co-expressed together in E.coli. This was particularly evident in our co-precipitation and size exclusion chromatography experiments (data not shown). This might reflect a requirement for co-translational binding of the effector by a pre-synthesised chaperone pool to prime the effector for delivery to the T3SS and subsequent translocation. Such interaction might additionally prevent premature effector oligomerization. While our biochemical analysis of TARP1–200 is consistent with the exclusive formation of monomers in the absence of Slc1, direct actin modulation requires TARP oligomerization. It therefore remains possible that Slc1 binding also impedes self-assembly of full-length TARP, ensuring that effector activity is constrained to the host cell.
A peculiarity of the chlamydial T3SS seems to be the apparent formation of chaperone heterodimers. While our bacterial two-hybrid assays also detect Slc2/CT088 heterodimerization, the majority of interactions seem to be self-self, for example Slc1/Slc1 and Mcsc2/Mcsc2. When quantified using β-galactosidase activity, these homooligomeric interactions were significant and well above the background; hence the lack of heterodimerization of Slc1 and Mcsc with other chaperones is unlikely to reflect limitations in our experimental system. Furthermore Slc1 homooligomerization is observed when the purified protein is analyzed in vitro. Interestingly, homodimerization of Slc1 and Mcsc was not detected in a recent yeast two-hybrid screen (Spaeth et al., 2009), although the obvious explanation would be the use of two different heterologous systems. To date, despite an equivalently large effector cohort, fewer T3SS chaperones have been identified in Chlamydia than in other systems. Intuitively, heterodimerization might represent a mechanism to compensate for this numeric reduction, by permitting increased promiscuity using a smaller number of components. However, it remains possible that there are additional chaperones encoded within the chlamydial genome that have yet to be discovered. Regardless, our data show that the Slc1 homodimer is the form that binds TARP, and that this interaction is sufficient to enhance T3SS-dependent TARP translocation in a heterologous system. Nevertheless, given the emerging number of effectors, it is clear that more chaperones must be present, and that further work is required to study the possible roles of chaperone heterodimerization. By identifying the cognate chaperone for one of the earliest identified effectors, we hope to contribute further to understanding the chlamydial T3SS and how deployed effectors mediate interaction with the host.
MATERIALS AND METHODS
Bacterial strains and cell culture
Chemically competent Top10 and BL21star E. coli strains from Invitrogen were used for plasmid propagation and protein expression, respectively. The adenylate cyclase (cya) deficient DHM1 E. coli strains (Charpentier & Oswald, 2004) were grown at 30°C on indicator plates (LB + 40 μg/ml X-gal + 1mM IPTG or McConkey + 1% maltose) for 48 h or in Luria-Bertani broth (LB) at 37°C. All other E. coli strains were grown at 37°C in LB broth or agar plates. The Yersinia enterocolitica WT strain MRS40 or ΔYscN E40 (pMSl41) strain ((Sory et al., 1995, Sory & Cornelis, 1994); gifts from Professor Guy Cornelis) were grown in LB or Brain-Heart-Infusion (BHI) broth at 26°C. Media was supplemented with 100 μg/ml carbenicillin (carb), 60 μg/ml kanamycin (kan), 12.5 μg/ml tetracycline (tet), or 35 μg/ml Nalidixic Acid, when appropriate. Hela cells were cultivated at 37°C with 5% CO2 in DMEM supplemented with10% FBS and 2 mM L-glutamine. Cultivation and harvesting of C. trachomatis serovar D, strain UW-3/Cx was as described previously (Caldwell et al. 1981).
Plasmid construction
slc1, scc1, mcsC, slc2 genes and a fragment of tarp encoding the N-terminal 200 residues were amplified from Chlamydia trachomatis genomic DNA (gDNA), CesT and the N-terminal 600 nt of Tir from enteropathogenic E. coli gDNA, and YopH from Y. enterocolitica MRS40 gDNA by polymerase chain reaction (PCR) using the primers listed in Table 3. Cloning into pET101, pET200, pBAD-TOPO and pENTR-D/SD-TOPO utilized the endogenous topoisomerase activity of Invitrogen directional cloning vectors. The PCR fragments CACC-CT043-FLAG-stop and CACC-CT663-FLAG-stop were cloned into pET101/D-TOPO, resulting in the plasmids pET101-Slc1-FLAG and pET101-Slc2-FLAG, respectively. The PCR fragment CACC-TARP200 was cloned into pET200/D-TOPO, resulting in the plasmid pET200-TARP200. All of the constructed plasmids were transformed into TOP10 cells, recovered in SOC medium for 1h at 37° C, and plated on LB agar supplemented with appropriate antibiotics. The resulting colonies were re-streaked and grown overnight. Presence of the correct plasmid inserts was confirmed by colony PCR and sequencing. The PCR fragments CACC-CT043 and CACC-CT088-FLAG were cloned into pENTR/SD/D-TOPO, resulting in the plasmids pENTR-Slc1 and pENTR-Scc1-FLAG, respectively. These new entry vectors were recombined with pET-56-DEST using LR recombinase II reaction mix (Invitrogen), transformed into TOP10, and selected for on LB+carb agar overnight. The resulting colonies were re-streaked on carbenicillin plates and grown overnight. The presence of the correct plasmids was determined by colony PCR and sequencing. Plasmid DNA was purified from the TOP10 genetically-modified strains by using Qiaprep Spin Miniprep Kit.(Qiagen). The BL21star expression strain was transformed or co-transformed with pET200-TARP200, pET101-Slc1, pET101-Slc2, and pET56-Scc1-FLAG, as appropriate. For the expression of chlamydial proteins in Yersinia, slc1 and scc1 were subcloned from pET101-Slc2 and pET101-Scc1 using XbaI and ScaI-HF (NEB). These digested inserts were then ligated into the corresponding XbaI and ScaI sites of pBAD18 ((Guzman et al., 1995); American Tissue Culture Collection). mcsc and slc1 were cloned into the pBAD-TOPO vector. SLIM PCR was used to generate a KpnI sited downstream of tarp in the vector pET200-tarp200 to form pET200-tarp200-kpnI. This vector was then digested with NdeI and KpnI and the resulting NdeI-Tarp200-KpnI fragment was subcloned into the corresponding sites of pCX340 (provided by Prof. Gadi Frankel, Imperial College). For co-expression in the bacterial 2-hybrid system, PCR fragments were generated to include flanking XbaI and KpnI sites and were cloned into the same sites in pUT18C and/or pKT25 (kind gifts from Dr. Daniel Ladant, Institut Pasteur).
Protein expression, lysate preparation, and protein purification
Overnight cultures of BL21star strains were diluted 1:20 in LB + antibiotic, and grown to OD600 of 0.6. Expression of recombinant proteins was induced by addition of 0.5 mM isopropyl-β-D thiogalactopyranoside (IPTG) and incubated for 3 h 37°C with shaking. Bacteria were centrifuged (8000g, 20min, 4°C), and the resulting pellet resuspended in 1/10 culture volume of lysis buffer (50mM NaH2PO4, 300mM NaCl, 0.05% Tween, 0.1 mg/ml Dnase I, 1 mg/ml lysozyme, Halt Protease Inhibitor Cocktail (Pierce)). For preparation of lysates that were added to NiNta Magnetic Beads, 25 mM imidazole was added to the lysis buffer. Bacteria were incubated in lysis buffer for 30 min at room temperature, sonicated to shear DNA, and stored at −80°C. Purifications of Slc1-FLAG and His6-TARP1–200 were performed using M2-Anti-FLAG agarose (Sigma) and Ni-Nta Agarose (Qiagen), respectively. Lysozyme was excluded from the lysis buffer and a French pressure cell was used prior to clarification (15,000g, 30 min, 4°C). Purified proteins were analyzed by 12% SDS-PAGE gels stained with a 0.2% Coomassie Blue R250.
Coprecipitation assays and immunodetection
Bacterial lysates were thawed and clarified (4000g 15 min, 4°C) prior to addition to M2 Anti-FLAG Agarose (Sigma) or NiNta Magnetic Beads (Qiagen). 30 mM imidazole was added to 500 μl lysates prior to incubation with the magnetic beads on an end-over-end rotator overnight at 4°C. The flow-through fraction or supernatant was separated from the beads with a magnetic separator, and the beads were washed five times with NiNta Native Wash Buffer (50 mM Na2HPO4, 300 mM NaCl, 20 mM imidazole, 0.05% Tween 20, PH 8.0). The wash fractions were retained for analysis, and the native protein complexes eluted from the beads by incubation with 100 μl NiNta Native Elution Buffer (50 mM Na2HPO4, 300 mM NaCl, 300 mM imidazole, 0.05% Tween 20, pH 8.0) at 4°C for 4 h. For each co-precipitation experiment with M2 Anti-FLAG agarose, 1 ml of bacterial lysate was added to 100 μl of packed agarose resin and incubated on an end-over-end rotator at 4°C overnight. The flow-through fractions were collected (3000g, 1min, 4°C), and the resin washed five times with Tris-buffered saline (TBS), prior to elution of the native protein complexes by incubation with 100 μl of 200 ng/ml 3XFLAG peptide (Sigma) in TBS at 4°C for 4 hours-overnight. Precipitation fractions were incubated 1:1 with Laemmli sample buffer (Biorad) + 0.5 % β-mercaptoethanol at 95°C for 10 min prior to separation by SDS-PAGE. Proteins were transferred to polyvinylidine fluoride (PVDF) membrane and detected by western blot probed with 6XHIS (ab18184), DDDDK (ab21536, ab49763), and β-lactamase (ab12251) - specific antibodies from Abcam. HRP-conjugated goat-anti-mouse (Millipore 12–349) or HRP-goat-anti-rabbit (Pierce #185415) were used as secondary antibodies when necessary, followed by development with Millipore Immobilin Western Substrate.
Bacterial 2-hybrid
pUT18C and pKT25 vectors containing putative chlamydial chaperones or TARP1–200 were co-transformed into the adenylate cyclase deficient E.coli strain DHM1. DHM1 strains co-expressing T18 and T25-fused chlamydial proteins were grown at 30°C on indicator plates (LB + 40 μg/ml X-gal + 1mM IPTG or McConkey + 1% maltose) for 48 h or in Luria-Bertani broth (LB) at 37°C. Briefly, 5 μl of co-expressing overnight culture was plated onto indicator plates (LB+X-gal and McConkey agar) and incubated for 2 days at 30°C to allow for color development. A modified Miller assay was followed. Briefly, co-expressing DHM1 strains were grown overnight in LB+antibiotic at 37°C followed by a 1:50 subculture and growth in fresh LB + antibiotic to an OD600 of 0.5–0.8. The OD600 of the culture was recoded and 20 μl of culture was added to 80 μl of permeabilization buffer (20 mM KCl, 2 mM MgSO4, 0.8 mg/mL CTAB (hexadecyltrimethylammonium bromide), 0.4 mg/mL sodium deoxycholate, 5.4 μL/mL beta-mercaptoethanol) and incubated at 30° C for 30 minutes. The time was recorded as 600 μl of pre-warmed substrate solution (60 mM Na2HPO4, 40 13 mM NaH2PO4, 1 mg/mL o-nitrophenyl-β-D-galactoside (ONPG), 2.7 μL/mL β-mercaptoethanol) was added to each permeabilized sample and incubated at 30 °C. Upon developing a yellow color, the samples were stopped with 700 μl of 1 M Sodium Carbonate (Na2CO3) and the stop time was noted.
Co-immunoprecipitation from Chlamydia elementary bodies
5 × 109 infectious C. trachomatis serovar D elementary bodies were pelleted by centrifugation, and the SPG storage buffer aspirated. Pellets were resuspended in 500 μl lysis buffer (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP-40, 5% glycerol; pH 7.4, Halt Protease Inhibitor Cocktail (Pierce)). The lysate was incubated overnight at 4°C with 10 μl of rabbit polyclonal anti-TARP antibody crosslinked to Protein A/G - agarose beads (Pierce). Conjugation by crosslinking of 10 μg of anti-TARP antibodies to the Protein A/G agarose beads was performed according to the manufacturer’s instructions. TARP-Slc1 complexes were eluted by incubation with a denaturing, but non-reducing sample buffer. TARP and Slc1 contents of the lysate, flow-through, wash, and elution fractions were analysed by Western blotting with anti-TARP and anti-Slc1 antibodies, respectively. The antibodies were generously provided by Dr. Raphael Valdivia (Duke University Medical Center, USA).
Translocation Assay
HeLa cells were plated in black, flat, glass-bottomed 96-well plates at a density of 1 × 104 cells per well and allowed to grow for 24 hours prior to infection. One day prior to infection TEM-1 fusion protein (pCX340) and pBAD-chaperone expressing Y. enterocolitica strains were inoculated into 3 ml of BHI broth + tetracycline ± carbenicillin and grown at 26°C overnight. The following day, the cultures were diluted 1:20 in BHI + antibiotics and grown at 26°C for 1 hr. 1 mM IPTG and 13 mM L-Arabinose were added to the cultures and were grown for 2 more hours at 26°C. OD600 values were determined for each culture and adjusted to 0.1 prior to infection of HeLa cells. Monolayers were washed twice with HBSS, and infected with 10 μl of cultures (OD600 = 0.1) of Y. enterocolitica. After 1 hr of infection at 37°C, the monolayer was washed twice and fresh DMEM was added. The infection progressed for 2 more hours at 37°C in 5% CO2. Infected cells were washed 3 times with HBSS and to which 100 μl of HBSS was added. The 1x CCF2/AM substrate was prepared as per the manufacturer’s manual (Invitrogen) and 20 μl was added to each well. Infected cells were incubated at RT for 1.5 hrs in the dark prior to fluorescence detection at 450 and 510 nm with a fluorimeter (excitation at 410 nm).
Supplementary Material
Supplementary Figure 1. Separation and detection of additive Slc1: Tarp complexes. The relative abundance of native purified Slc1 alone migrating as a dimer, purified Tarp1–200 alone, or the addition of singly purified dimeric Slc1 and Tarp1–200 complexes were detected and eluted in 500 ul fractions (top) by gel filtration chromatography.
Supplementary Figure 2. TARP1–200-β-lactamase fusion are expressed at similar levels in the different Yersinia transformants as indicated. Aliquots of cultures used to infect HeLa cells were lysed and subjected to immunoblotting with an anti-β-lactamase monoclonal antibody.
Table 1.
Plasmids used
| Plasmid | Description | Reference |
|---|---|---|
| pET101 | Directional TOPO expression vector | Invitrogen |
| pET200 | Directional TOPO expression vector | Invitrogen |
| pENTR-D/SD- TOPO | Directional TOPO entry vector for LR recombination | Invitrogen |
| pET56-DEST | Bacterial expression destination vector (LR) | Novagen |
| pSlc1-FLAG | pET101 expressing Slc1-FLAG | This study |
| pSlc2-FLAG | pET101 expressing Slc2-FLAG | This study |
| pTARP200 | pET200 expressing N-term 200aa of L2 TARP | This study |
| p56-Scc1 | pDEST-56 expressing Scc1-FLAG | This study |
| p56-Slc1 | pDEST-56 expressing Slc1 | This study |
| pUT18C | E.coli 2-hybrid vector - cya fragment 18 fusion | Ref. 35 |
| pKT25 | E.coli 2-hybrid vector - cya fragment 25 fusion | Ref. 35 |
| p18C-TARP | E2H -cya-18 fragment fused to Nterm 200aa TARP | This study |
| p25-CesT | E2H -cya-18 fragment fused to Nterm 200aa Tir | This study |
| p18C-Tir | E2H -cya-18 fragment fused to CT043 | This study |
| p18C-CT043 | E2H -cya-18 fragment fused to CT088 | This study |
| p18C-CT088 | E2H -cya-18 fragment fused to CT260 | This study |
| p18C-CT260 | E2H -cya-18 fragment fused to CT663 | This study |
| p18C-CT663 | E2H -cya-25 fragment fused to CT043 | This study |
| p25-CT043 | E2H -cya-25 fragment fused to CT088 | This study |
| p25-CT088 | E2H -cya-25 fragment fused to CT260 | This study |
| p25-CT260 | E2H -cya-25 fragment fused to CT663 | This study |
| p25-CT663 | E2H -cya-25 fragment fused to CesT | This study |
| pCX340 | TEM1 fusion, IPTG inducible | Ref. 39 |
| pBAD-TOPO | Arabinose-inducible promoter | Invitrogen |
| pBAD18 | Arabinose-inducible promoter | Ref. 46 |
| pCX340-Tarp200 | N-term 200aa of TARP fused to TEM1 | This study |
| pCX340-YopH | N-term 200aa of YopH fused to TEM1 | This study |
| pBAD-CT043 | pBAD-TOPO with CT043-FLAG insert | This study |
| pBAD-CT088 | pBAD18 with CT088-FLAG insert | This study |
| pBAD-CT260 | pBAD-TOPO with CT260-FLAG insert | This study |
| pBAD-CT663 | pBAD18 with CT663-FLAG insert | This study |
Table 2.
Primers used
| Target gene | Plasmid vector | Sequence |
|---|---|---|
| Slc1 (incorp. FLAG-Tag) | pET101 | (AJB1 F) CACCATGTCCAGGCAGAATGCTGA (AJB1 R) TCACTTGTCATCGTCGTCCTTGTAGTAGCACGGATTCCTGCGGTA |
| Slc2 (incorp. FLAG-Tag) | pET101 | (AJB2 F) CACCATGTTGGAAAAATTGATAAAGAATTTTGTGGCG (AJB2 R) TTACTCCTGCTCCGTTTTACTTAGTCTAAATC |
| Scc1 (incorp. FLAG-Tag) | pET56-DEST | (AJB3 F) CACCATGCAAAATCAATTTGAACAACTC (AJB3 R) ATCGATTTACTTGTCGTCATCGTCCTTGTAGTCCAGGTGATACAT |
| TARP200aa | pET200 | (AJB4 F) CACCATGACGAATTCTATATCAGGTGATC (AJB4 R) TTATCCTACGGTATCAATCAGTGAGCT |
| Slc1 | pET56-DEST | (AJB5 R) TTATGCACGGATTCCTGTGGAGG |
| pET200-TARP-Kpn1 (SLIM-PCR) | pET200-TARP | (AJB6 FT) CCTTCACCAAGGGTACCTCAACGATCCGGCTGCTAACAAAGCC (AJB6 FS) CCGGCTGCTAACAAAGCC (AJB6 RT) TCGTTGAGGTACCCTTGGAGGAGCCTCTTAGAGA (AJB6 RS) CTTGGAGGAGCCTCTTAGAGA |
| pET200-TARP-Δ6xHis (SLIM-PCR) | pET200-TARP | (AJB7 FT) CGATTAAATAAGGAGGAATAACATATGACGAATTCTATATCAGGTGATC (AJB7 RS) ACGAATTCTATATCAGGTGATCAACCTA (AJB7 RT) CATATGTTATTCCTCCTTATTTAATCGCGATACATTAATATATACCTCT (AJB7 RS) CGATACATTAATATATACCTCTTTAATTTTTAATAATAAAG |
| Slc1 | pBAD18-Cm | (AJB8 F) CTATTTCTAATACTAATGGTACCGAAGGAAGATTCCCTCATGTCCAGGCAGAATGCT (AJB8 R) AATGTTAATGATATGTTGTCGACTTATTAGTCCTTATCGTCATCGTCCTTGTAATCAGCACGGATTCCTGCTGG |
| Scc1 | pBAD18-Cm | (AJB9 F) CTATTTCTAATACTAATGGTACCGAAGGAAGATTCCCTCATGCAAAATCAATTTGAACAAC (AJB9 R) AATGTTAATGATATGTTGTCGACTTATTAGTCCTTATCGTCATCGTCCTTGTAATCCAGGTGATACATACCTAGA |
| TARP200 | pCX340 | (AJB10 F) CTGTACGCATATGACGAATTCTATATCAGG (AJB10 R) AGCACAAGTGACCCAGGTACCTGAATCA |
| YopH | pCX340 | (AJB12 F) CGTAGGACGCATATGAACTTATCATTAAGCGAT (AJB12 R) TGAATCTCAGGTACCGCGGCTGCTGAGTTC |
| TARP200 | pUT18C | (AJB13 F) TAGCATCGGGTACCGATGACGAATTCTATATCAG (AJB13 R) CGTAGCTATCGATGAGCCACTTGTGCTAGA |
| Tir | pUT18C | (AJB14 F) GTTACGATCTAGAGATGCCTATTGGTAACCTTGGTAATA (AJB14 R) CTAGTGCAGGTACCTTACGTATCAACGTCCTT |
| Slc1 | pUT18C | (AJB15 F) TTACGTATTCTAGAGATGTCCAGGCAGAATGCT (AJB15 R) GCATTAGTGGTACCCTTATGCACGGATTCCTGC |
| Scc1 | pUT18C | (AJB16 F) TGTACCGTATTCTAGAGATGCAAAATCAATTTGAAC (AJB16 R) TGCTATCGGTACCTTACAGGTGATACATACCT |
| Mcsc | pUT18C | (AJB17 F) TTACGTATTCTAGAGATGACAACGTGGACTTTG (AJB17 R) GCATTAGTGGTACCCCTAAGGCTCTAGCTGATC |
| Slc2 | pUT18C | (AJB18 F) GGACGGACTCTAGAGATGTTGGAAAAATTGATA (AJB18 R) GCATTAGTGGTACCCTTACTCCTGCTGCGTTTT |
| CesT | pKT25 | (AJB19 F) TAACAGTAGGTACCTTATCTTCCGGCGTA (AJB19 R) TTACGTATTCTAGAGATGTCATCAAGATCTGAAC |
| Slc1 | pKT25 | (AJB20 F) CATTATTCTAGAGATGTCCAGGCAGAATGCTGAG (AJB20 R) TTGCTAGTACCCGTTATGCACGGATTCCTGCTGG |
| Scc1 | pKT25 | (AJB21 F) CATTATTCTAGAGATGCAAAATCAATTTGAACAA (AJB21 R) TTGCTAGTACCCGTTACAGGTGATACATACCTAG |
| Mcsc | pKT25 | (AJB22 F) CATTATTCTAGAGATGACAACGTGGACTTTGAAT (AJB22 R) TTGCTAGTACCCGTTAAGGCTCTAGCTGATCGGA |
| Slc2 | pKT25 | (AJB23 F) CATTATTCTAGAGATGTTGGAAAAATTGATAAAG (AJB23 R) TTGCTAGTACCCGTTACTCCTGCTCCGTTTTCTT |
Acknowledgments
The authors acknowledge the generous contributions of antibodies by Dr. Raphael Valdivia (Duke University), bacterial strains and/or plasmids by Prof. Guy Cornelis (Biozentrum), Dr. Daniel Ladant (Institut Pasteur), and Prof. Gadi Frankel (Imperial College). We would also like to thank Drs. Gunnar Schroeder, Robert Fagan, Scot Ouellette, Alex Wong, Diana Munera, and members of the Carabeo lab for technical advice. The work was supported by the Medical Research Council and the National Institutes of Health. RDH is a Royal Society University Research Fellow.
References
- Akeda Y, Galan JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–915. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- Beeckman DSA, Vanrompay DCG. Bacterial Secretion Systems with an Emphasis on the Chlamydial Type III Secretion System. Current Issues in Molecular Biology. 2010;12:17–41. [PubMed] [Google Scholar]
- Belland RJ, Nelson DE, Virok D, Crane DD, Hogan D, Sturdevant D, Beatty WL, Caldwell HD. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc Natl Acad Sci U S A. 2003;100:15971–15976. doi: 10.1073/pnas.2535394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtalan S, Ghosh P. Structure of the Yersinia type III secretory system chaperone SycE. Nat Struct Biol. 2001;8:974–978. doi: 10.1038/nsb1101-974. [DOI] [PubMed] [Google Scholar]
- Buttner CR, Cornelis GR, Heinz DW, Niemann HH. Crystal structure of Yersinia enterocolitica type III secretion chaperone SycT. Protein Sci. 2005;14:1993–2002. doi: 10.1110/ps.051474605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne GI. Requirements for ingestion of Chlamydia psittaci by mouse fibroblasts (L cells) Infect Immun. 1976;14:645–651. doi: 10.1128/iai.14.3.645-651.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabeo RA, Grieshaber SS, Fischer E, Hackstadt T. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect Immun. 2002;70:3793–3803. doi: 10.1128/IAI.70.7.3793-3803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier X, Oswald E. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J Bacteriol. 2004;186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton DR, Dooley CA, Grieshaber SS, Carabeo RA, Fields KA, Hackstadt T. Phosphorylation of the chlamydial effector protein Tarp is species specific and not required for the recruitment of actin. Infect Immun. 2005;73:3860–3868. doi: 10.1128/IAI.73.7.3860-3868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annual Review of Microbiology. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- Creasey EA, Delahay RM, Bishop AA, Shaw RK, Kenny B, Knutton S, Frankel G. CesT is a bivalent enteropathogenic Escherichia coli chaperone required for translocation of both Tir and Map. Mol Microbiol. 2003;47:209–221. doi: 10.1046/j.1365-2958.2003.03290.x. [DOI] [PubMed] [Google Scholar]
- Elliott SJ, Hutcheson SW, Dubois MS, Mellies JL, Wainwright LA, Batchelor M, Frankel G, Knutton S, Kaper JB. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol Microbiol. 1999;33:1176–1189. doi: 10.1046/j.1365-2958.1999.01559.x. [DOI] [PubMed] [Google Scholar]
- Fields KA, Fischer ER, Mead DJ, Hackstadt T. Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J Bacteriol. 2005;187:6466–6478. doi: 10.1128/JB.187.18.6466-6478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields KA, Hackstadt T. The chlamydial inclusion: escape from the endocytic pathway. Annu Rev Cell Dev Biol. 2002;18:221–245. doi: 10.1146/annurev.cellbio.18.012502.105845. [DOI] [PubMed] [Google Scholar]
- Fraser GM, Gonzalez-Pedrajo B, Tame JR, Macnab RM. Interactions of FliJ with the Salmonella type III flagellar export apparatus. J Bacteriol. 2003;185:5546–5554. doi: 10.1128/JB.185.18.5546-5554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev. 2004;68:771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith KL, Wolf RE., Jr Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun. 2002;290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hower S, Wolf K, Fields KA. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol. 2009;72:1423–1437. doi: 10.1111/j.1365-2958.2009.06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison WP, Hackstadt T. Induction of type III secretion by cell-free Chlamydia trachomatis elementary bodies. Microb Pathog. 2008;45:435–440. doi: 10.1016/j.micpath.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett TJ, Fischer ER, Mead DJ, Hackstadt T. Chlamydial TARP is a bacterial nucleator of actin. Proc Natl Acad Sci U S A. 2006;103:15599–15604. doi: 10.1073/pnas.0603044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JF. Revisiting the chlamydial type III protein secretion system: clues to the origin of type III protein secretion. Trends in Genetics. 2001;17:65–69. doi: 10.1016/s0168-9525(00)02175-2. [DOI] [PubMed] [Google Scholar]
- Lane BJ, Mutchler C, Al Khodor S, Grieshaber SS, Carabeo RA. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 2008;4:e1000014. doi: 10.1371/journal.ppat.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VT, Anderson DM, Schneewind O. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol Microbiol. 1998;28:593–601. doi: 10.1046/j.1365-2958.1998.00822.x. [DOI] [PubMed] [Google Scholar]
- Letzelter M, Sorg I, Mota LJ, Meyer S, Stalder J, Feldman M, Kuhn M, Callebaut I, Cornelis GR. The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J. 2006;25:3223–3233. doi: 10.1038/sj.emboj.7601202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Bertero MG, Frey EA, Pfuetzner RA, Wenk MR, Creagh L, Marcus SL, Lim D, Sicheri F, Kay C, Haynes C, Finlay BB, Strynadka NC. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat Struct Biol. 2001;8:1031–1036. doi: 10.1038/nsb717. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Young GM. Essential Role of the SycP Chaperone in Type III Secretion of the YspP Effector. Journal of Bacteriology. 2009;191:1703–1715. doi: 10.1128/JB.01021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschiol S, Bailey L, Gylfe A, Sundin C, Hultenby K, Bergstrom S, Elofsson M, Wolf-Watz H, Normark S, Henriques-Normark B. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A. 2006;103:14566–14571. doi: 10.1073/pnas.0606412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AL, Fromont-Racine M, Sansonetti P, Legrain P, Parsot C. Characterization of the interaction partners of secreted proteins and chaperones of Shigella flexneri. Mol Microbiol. 2001;42:1133–1145. doi: 10.1046/j.1365-2958.2001.02715.x. [DOI] [PubMed] [Google Scholar]
- Page AL, Parsot C. Chaperones of the type III secretion pathway: jacks of all trades. Mol Microbiol. 2002;46:1–11. doi: 10.1046/j.1365-2958.2002.03138.x. [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Beatson SA, Bailey CM. Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: a Darwinian perspective. FEMS Microbiol Rev. 2005;29:201–229. doi: 10.1016/j.femsre.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Rodgers L, Gamez A, Riek R, Ghosh P. The type III secretion chaperone SycE promotes a localized disorder-to-order transition in the natively unfolded effector YopE. J Biol Chem. 2008;283:20857–20863. doi: 10.1074/jbc.M802339200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubot FD, Jackson MW, Penrose KJ, Cherry S, Tropea JE, Plano GV, Waugh DS. Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J Mol Biol. 2005;346:1147–1161. doi: 10.1016/j.jmb.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Singer AU, Desveaux D, Betts L, Chang JH, Nimchuk Z, Grant SR, Dangl JL, Sondek J. Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure. 2004;12:1669–1681. doi: 10.1016/j.str.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Skipp P, Robinson J, O’Connor CD, Clarke IN. Shotgun proteomic analysis of Chlamydia trachomatis. Proteomics. 2005;5:1558–1573. doi: 10.1002/pmic.200401044. [DOI] [PubMed] [Google Scholar]
- Sory MP, Boland A, Lambermont I, Cornelis GR. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci U S A. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sory MP, Cornelis GR. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- Spaeth KE, Chen YS, Valdivia RH. The Chlamydia Type III Secretion System C-ring Engages a Chaperone-Effector Protein Complex. PLoS Pathog. 2009;5:e1000579. doi: 10.1371/journal.ppat.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- Subtil A, Delevoye C, Balana ME, Tastevin L, Perrinet S, Dautry-Varsat A. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol Microbiol. 2005;56:1636–1647. doi: 10.1111/j.1365-2958.2005.04647.x. [DOI] [PubMed] [Google Scholar]
- Thomas NA, Deng W, Puente JL, Frey EA, Yip CK, Strynadka NC, Finlay BB. CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol Microbiol. 2005;57:1762–1779. doi: 10.1111/j.1365-2958.2005.04802.x. [DOI] [PubMed] [Google Scholar]
- Wolf K, Betts HJ, Chellas-Gery B, Hower S, Linton CN, Fields KA. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol Microbiol. 2006;61:1543–1555. doi: 10.1111/j.1365-2958.2006.05347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip CK, Strynadka NC. New structural insights into the bacterial type III secretion system. Trends Biochem Sci. 2006;31:223–230. doi: 10.1016/j.tibs.2006.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Separation and detection of additive Slc1: Tarp complexes. The relative abundance of native purified Slc1 alone migrating as a dimer, purified Tarp1–200 alone, or the addition of singly purified dimeric Slc1 and Tarp1–200 complexes were detected and eluted in 500 ul fractions (top) by gel filtration chromatography.
Supplementary Figure 2. TARP1–200-β-lactamase fusion are expressed at similar levels in the different Yersinia transformants as indicated. Aliquots of cultures used to infect HeLa cells were lysed and subjected to immunoblotting with an anti-β-lactamase monoclonal antibody.