Abstract
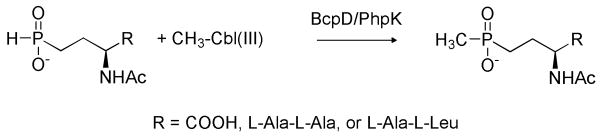
Radical SAM (S-adenosyl-L-methionine), cobalamin-dependent methyltransferases have been proposed to catalyze the methylations of unreactive carbon or phosphorus atoms in antibiotic biosynthetic pathways. To date, none of these enzymes have been purified or shown to be active in vitro. Here we demonstrate the activity of the P-methyltransferase enzyme, PhpK, from the phosalacine producer Kitasatospora phosalacinea. PhpK catalyzes the transfer of a methyl group from methylcobalamin to 2-acetylamino-4-hydroxyphosphinylbutanoate (N-acetyldemethylphosphinothricin or NAcDMPT) to form 2-acetylamino-4-hydroxymethylphosphinylbutanoate (N-acetylphosphinothricin or NAcPT). This transformation gives rise to the only carbon-phosphorus-carbon linkage known to occur in Nature.
Compounds containing a carbon-phosphorus-carbon linkage are often used as neurotransmitter antagonists and enzyme inhibitors.1 However, this bonding sequence is observed in only one naturally-occurring material and its derivatives: 2-amino-4-hydroxymethylphosphinylbutanoate, more commonly known as L-phosphinothricin (L-PT) or glufosinate (Chart S1).2 Due to its structural similarity to L-glutamate, L-PT is a useful inhibitor of bacterial and plant glutamine synthetases.3 L-PT is naturally produced by Streptomyces hygroscopicus, Streptomyces viridochromogenes, and Kitasatospora phosalacinea.4–8 All three species normally produce L-PT as the first amino acid of an exported tripeptide (Chart S1). The Streptomyces tripeptide, L-phosphinothricylalanylalanine or bialaphos, is commercially used as an herbicide.2,3 K. phosalacinea produces L-PT as part of an alternative tripeptide, L-phosphinothricylalanylleucine or phosalacine.
Multiple studies have examined the genetics and mechanistic enzymology of proteins in the two Streptomyces pathways, but no studies have been published specifically investigating the phosalacine pathway.2,9 Little is known about the formation of the unique C-P-C linkage in any of the three organisms. The bcpD gene in S. hygroscopicus (phpK in S. viridochromogenes) was believed to encode the putative P-methyltransferase catalyzing the methylation of the electrophilic phosphinyl group, resulting in the formation of the C-P-C linkage.7,10,11 Experiments with Streptomyces cell extracts indicated that the methyl donor was the vitamin B12 derivative methylcobalamin (CH3Cbl(III)) and the methyl recipient(s) were 2-acetylamino-4-hydroxyphosphinylbutanoate (NAcDMPT) and the corresponding phosphinyl tripeptide, NAcDMPT-L-Ala-L-Ala (Scheme 1).12 The resulting product(s) are NAcPT and NAcPT-L-Ala-L-Ala, respectively. By extension, NAcDMPT and NAcDMPT-L-Ala-L-Leu are the likely substrates for the K. phosalacinea P-methyltransferase.
Scheme 1.
Proposed P-methyl transfer reactions
Sequencing indicated that the P-methyltransferases were related to a pair of putative methyltransferases from the fosfomycin (Fom3) and fortimicin (Fms7) biosynthetic pathways.10,13 Fom3 and Fms7 have been hypothesized to catalyze the transfer of methyl groups from CH3Cbl(III) to unreactive carbon, rather than phosphorus, atoms.13,14 This posed an intriguing mechanistic problem; in nearly all cases studied, biological transfer of a methyl group occurs via an SN2-type nucleophilic substitution reaction.15 Since none of the hypothesized substrates for this family of enzymes were nucleophilic, it appeared likely that these methyltransferases used a different mechanism for catalysis. Transfer of a methyl anion from CH3Cbl(III) was proposed, but such chemistry is unlikely to occur in aqueous solution12.
In 2001, these four proteins were identified as members of the radical SAM superfamily.16 Radical SAM proteins contain three cysteines, typically within a conserved CXXXCXXC motif, which bind a [4Fe-4S] cluster. The reduced +1 state of the cluster donates an electron to SAM, resulting in homolytic cleavage of the carbon-sulfur bond to form the 5′-deoxyadenosyl radical required for catalysis.16 The revelation that these enzymes were members of the radical SAM superfamily suggested another possible mechanism for catalysis: transfer of the methyl group as a radical.9,17 The existence of a chemical precedent for organic radical methylation by CH3Cbl(III) in aqueous solution indicated that the methyl radical mechanism was feasible.18
To gain insight into this group of possible radical SAM methyltransferases, we chose to study the P-methyltransferase from Kitasatospora phosalacinea DSM 43860. Sequencing indicated that phpK from K. phosalacinea is over 99% identical to the published phpK sequence from the bialaphos producer, S. viridochromogenes. 11,19 After overexpression in E. coli and cell lysis, PhpK was not found in the soluble fraction and appeared to express solely in inclusion bodies. To purify the enzyme, we modified a literature procedure for the solubilization and refolding of an iron-sulfur/corrinoid protein.20 All steps were performed in an anaerobic chamber (Coy Laboratory Products). After refolding, PhpK was further purified by anionic exchange chromatography, and the hypothesized [4Fe-4S] clusters were anaerobically reconstituted.
PhpK was dark brown and displayed an ultraviolet-visible spectrum consistent with that of other [4Fe-4S] proteins with a local maximum at 420 nm (Figure S2).21 Iron and sulfide analyses indicated approximately 5.9 moles Fe and 4.4 moles S per mole of PhpK.22–24 EPR spectroscopy was used to verify the presence of the [4Fe-4S] cluster (Figure S3). At 10 K, the EPR spectrum of a sample containing only PhpK and buffer showed a small signal at g = 2.00. Addition of sodium dithionite, a strong chemical reductant, resulted in a new, broad spectral feature at g = 1.93 consistent with an EPR-active, reduced [4Fe-4S]+1 cluster. Addition of all components hypothesized to be required for the reaction (vide infra) resulted in the disappearance of the [4Fe-4S]+1 cluster.
Assaying PhpK for activity was a significant challenge due to the lack of an appropriate ultraviolet or visible handle and significant polarity and/or structural similarities leading to low chromatographic separation. Perhaps most importantly, proposed radical SAM mechanisms can lead to suicide inactivation of PhpK in vitro; use of SAM as a substrate theoretically traps the [4Fe-4S] cluster of the enzyme in an inactive +2 state.9,17 In theory, excess sodium dithionite could reactivate the enzyme by reducing the cluster back to [4Fe-4S]+1. However, in practice, dithionite effected side reactions in the presence of SAM and CH3Cbl(III). Thus, assuming single turnover conditions, the magnitude of the difference between the molecular weights of PhpK (~61 kD) and NAcDMPT (209 Da) implied that large amounts (mg) of enzyme would be needed to observe comparatively small (μg) amounts of product.
We turned to NMR spectroscopy to overcome these experimental limitations. PhpK was incubated with SAM and sodium dithionite, then NAcDMPT (Asischem), CH3Cbl(III), and the enzyme MTAN (to relieve possible product inhibition) were added and anaerobically incubated at ~20 °C.25,26 The reaction mixtures were denatured, and PhpK was removed by ultrafiltration. After partial purification and concentration, the resulting material was dissolved in D2O (Cambridge Isotope Labs). Two-dimensional 1H-31P gHSQC spectra were collected using a coupling constant of 15 Hz. Peak positions varied slightly due to differences in final pH and/or spectrometers. The methylene protons at C-4 of NAcDMPT displayed a strong crosspeak at 1.38 ppm (1H) and 30.0 ppm (31P) due to coupling with 31P (Figures S4, S5, S6, and S7). In the presence of all reaction components above, PhpK catalyzed partial conversion of NAcDMPT to NAcPT as demonstrated by the appearance of a new crosspeak at (1.07, 44.0) ppm corresponding to the methyl group of NAcPT (Figures S5 and S6). Spiking the sample with chemically-synthesized NAcPT increased the intensity of this crosspeak. The amount of NAcPT in the final NMR sample was estimated to be [~100 μM], which corresponds to [~15–20 μM] in the original reaction. Given the initial protein concentration of [~30 μM], under these specific conditions PhpK catalyzes single turnover. In a control lacking dithionite, hypothesized to be required for reduction of the [4Fe-4S] cluster, the NAcPT-associated gHSQC crosspeak is not observed (Figure S4). Thus, our results suggest that PhpK catalyzes the methylation of NAcDMPT in a potentially radical SAM-dependent fashion.
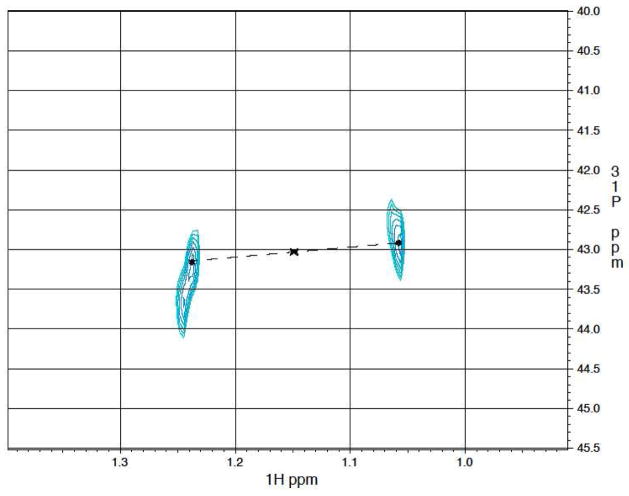
To verify the donor of the methyl group in this reaction, we synthesized 13CH3Cbl(III) and assayed PhpK with either a combination of 12CH3Cbl(III) and 13CH3Cbl(III) or 13CH3Cbl(III) alone.27 The 1H-31P gHSQC spectrum for the mixed-isotope reaction displayed the crosspeak previously observed for the methyl protons of NAcPT at (1.06, 43.8) ppm (Figure S7). Two new crosspeaks appeared at (0.95, 43.7) ppm and (1.16, 44.1) ppm. In the reaction containing only 13CH3Cbl(III), a very clear E.COSY pattern (indicated by the asterisks) of the H-P crosspeaks (centered at (1.15, 43.1) ppm and indicated by the “x”) is observed due to the passive couplings of 1H and 31P to the attached 13C nuclei (Figure 1). This pattern matches the additional crosspeaks observed in the mixed-isotope experiment. The one-bond J coupling of 1JCH is ~127 Hz, which matches the predicted value. Under these conditions, CH3Cbl(III) apparently serves as the sole source for the methyl group of NAcPT. Our data agree with a previous report in which cell-free extracts used only CH3Cbl(III) as a direct methyl donor for the final L-PT antibiotic.12
Figure 1.
H-P gHSQC spectrum of the partially purified 13CH3-Cbl(III) PhpK reaction. The 13CH3 H-P crosspeaks are centered at (1.15, 43.1) ppm (designated by the “x”).
In summary, we have established an active, in vitro system to study a P-methyltransferase, PhpK, that is broadly applicable to related enzymes including Fom3 and Fms7. We are now investigating the hypothesis that PhpK uses a radical SAM mechanism for catalysis.9,17 Isolation of 5′-deoxyadenosine and/or other SAM cleavage products will provide strong evidence supporting a radical mechanism, as would the trapping of Cbl(II). Is NAcDMPT or the tripeptide NAcDMPT-L-Ala-L-Leu the physiological substrate? Another question is whether CH3Cbl(III) is truly the methyl group donor or, as is the case in other CH3Cbl(III)- and methylcorrinoid-dependent enzymes, whether Cbl functions as a coenzyme and CH3Cbl(III) is formed as a methylated intermediate. Previous work in S. hygroscopicus showed that the methyl group of NAcPT originates from CD3-methionine, conserving all three deuterons.28 Taken together, these data suggest that the methyl group is derived from methionine, is transferred to CH3Cbl(III), then is added to NAcDMPT by PhpK. This contrasts with recent reports on the Cfr- and RlmN-catalyzed radical SAM methylation reactions in which one of the three methyl protons is derived from a separate source.29,30 Although methionine is not a methyl donor, it can be converted to the methyl donor SAM by SAM synthetase. PhpK may undergo reductive reactivation with SAM, forming CH3Cbl(III) in a manner similar to that observed in cobalamin-dependent methionine synthase.17 The work presented here sets the stage for the deeper mechanistic investigation of these and other intriguing questions surrounding this fascinating family of enzymes.
Supplementary Material
Acknowledgments
Funding Sources
This study made use of the National Magnetic Resonance Facility at Madison (NMRFAM), which is supported by National Institutes of Health (NIH) grants P41RR02301 (BRTP/NCRR) and P41GM66326 (NIGMS). Additional equipment was purchased with funds from the University of Wisconsin-Madison, the NIH (RR02781, RR08438), the National Science Foundation (NSF) (DMB-8415048, OIA-9977486, BIR-9214394), and the United States Department of Agriculture. The WSU NMR Center equipment was supported by the NIH (RR0631401, RR12948), the NSF (CHE-9115282, DBI-9604689) and the Murdock Charitable Trust. K.D.A. was supported by an NIH training grant (T32GM008336). This research was funded by Washington State University and a Faculty Early Career Development Award (CAREER) from the NSF to S.C.W. (CHE-0953721).
We thank Dr. George Reed for the use of his EPR spectrometers, assistance with spectral collection and interpretation, and insightful discussions; Kenneth Johnson for technical assistance with EPR; Dr. William Hiscox for technical assistance with NMR; Dr. Zhaohui “Sunny” Zhou for the MTAN clone, and Drs. Perry Frey and Christian Whitman for insightful discussions.
ABBREVIATIONS
- SAM
S-adenosyl-L-methionine
- NAcDMPT
N-acetyldemethylphosphinothricin or 2-acetylamino-4-hydroxyphosphinylbutanoate
- NAcPT
N-acetylphosphinothricin
- PT
phosphinothricin
- CH3Cbl(III)
methylcobalamin
- [4Fe-4S]
four-iron, four-sulfur
- EPR
electron paramagnetic resonance
- NMR
nuclear magnetic resonance
- MTAN
5′-methylthioadenosine nucleosidase
- gHSQC
gradient heteronuclear single quantum correlation
- ppm
parts per million
- E.COSY
exclusive correlation spectroscopy
Footnotes
Supporting Information. Experimental procedures and EPR and NMR spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Collinsová M, Jirácek J. Curr Med Chem. 2000;7:629–647. doi: 10.2174/0929867003374831. [DOI] [PubMed] [Google Scholar]
- 2.Thompson CJ, Seto H. Genetics and Biochemistry of Antibiotic Production. 1995. pp. 197–222. [Google Scholar]
- 3.Hoerlein G. Rev Environ Contam Tox. 1994;138:73–145. doi: 10.1007/978-1-4612-2672-7_4. [DOI] [PubMed] [Google Scholar]
- 4.Bayer E, Gugel KH, Hägele K, Hagenmaier H, Jessipow S, König WA, Zähner H. Helv Chim Acta. 1972;55:224–239. doi: 10.1002/hlca.19720550126. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa Y, Tsuruoka T, Inouye S, Niida T. Sci Reports Meiji Seika Kaisha. 1973;13:42–48. [Google Scholar]
- 6.Seto H, Sasaki T, Imai S, Tsuruoka T, Ogawa H, Satoh A, Inouye S, Niida T, Otake N. J Antibiotics. 1982;36:96–98. doi: 10.7164/antibiotics.36.96. [DOI] [PubMed] [Google Scholar]
- 7.Hara O, Murakami T, Imai S, Anzai H, Itoh R, Kumada Y, Takano E, Satoh E, Satoh A, Nagaoka K, Thompson C. J Gen Micro. 1991;137:351–359. doi: 10.1099/00221287-137-2-351. [DOI] [PubMed] [Google Scholar]
- 8.Omura S, Murata M, Hanaki H, Hinotozawa K, Oiwa R, Tanaka H. J Antibiotics. 1984;37:829–835. doi: 10.7164/antibiotics.37.829. [DOI] [PubMed] [Google Scholar]
- 9.Metcalf WW, van der Donk WA. Ann Rev Biochem. 2009;78:65–94. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hidaka T, Hidaka M, Kuzuyama T, Seto H. Gene. 1995;158:149–150. doi: 10.1016/0378-1119(95)00101-b. [DOI] [PubMed] [Google Scholar]
- 11.Blodgett JA, Zhang JK, Metcalf WW. Antimicrob Agents Chemotherap. 2005;49:230–240. doi: 10.1128/AAC.49.1.230-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamigiri K, Hidaka T, Imai S, Murakami T, Seto H. J Antibiotics. 1992;45:781–787. doi: 10.7164/antibiotics.45.781. [DOI] [PubMed] [Google Scholar]
- 13.Kuzuyama T, Seki T, Dairi T, Hidaka T, Seto H. J Antibiotics. 1995;48:1191–1193. doi: 10.7164/antibiotics.48.1191. [DOI] [PubMed] [Google Scholar]
- 14.Woodyer RD, Li G, Zhao H, van der Donk WA. Chem Commun. 2007;4:359–361. doi: 10.1039/b614678c. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee R, Ragsdale SW. Ann Rev Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 16.Sofia HJ, Chen G, Hetzler BG, Reyes-Sindola JF, Miller NE. Nuc Acids Research. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Booker SJ. Curr Opin Chem Biol. 2009;13:58–73. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosimann H, Kräutler B. Angew Chem Int Ed. 2000;39:393–395. doi: 10.1002/(sici)1521-3773(20000117)39:2<393::aid-anie393>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz D, Grammel N, Heinzelmann E, Keller U, Wohlleben W. Appl Environ Microbiol. 2004;70:7093–7102. doi: 10.1128/AEM.70.12.7093-7102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu WP, Schiau I, Cunningham JR, Ragsdale SW. J Biol Chem. 1993;268:5605–5614. [PubMed] [Google Scholar]
- 21.Hagen KS, Watson AD, Holm RH. J Am Chem Soc. 1983;105:3905–3913. [Google Scholar]
- 22.Beinert H. Methods Enzymol. 1978;54:435–445. doi: 10.1016/s0076-6879(78)54027-5. [DOI] [PubMed] [Google Scholar]
- 23.Beinert H. Anal Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy MC, Kent TA, Emptage M, Merkle H, Beinert H, Münck E. J Biol Chem. 1984;259:14463–14471. [PubMed] [Google Scholar]
- 25.Challand MR, Ziegert T, Douglas P, Wood RJ, Kriek M, Shaw NM, Roach PL. FEBS Lett. 2009;583:1358–62. doi: 10.1016/j.febslet.2009.03.044. [DOI] [PubMed] [Google Scholar]
- 26.Farrar CE, Siu KK, Howell PL, Jarrett JT. Biochemistry. 2010;49:9985–96. doi: 10.1021/bi101023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tollinger M, Derer T, Konrat R, Krautler B. J Mol Cat A: Chem. 1997;116:147–155. [Google Scholar]
- 28.Seto H, Imai S, Tsuruoka T, Satoh A, Kojima M, Inouye S, Sasaki T, Otake N. J Antibiotics. 1982;35:1719–1721. doi: 10.7164/antibiotics.35.1719. [DOI] [PubMed] [Google Scholar]
- 29.Yan F, Fujimori DG. Proc Natl Acad Sci USA. 2011;108:3930–3934. doi: 10.1073/pnas.1017781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.