SUMMARY
Biofilms are surface-adhered bacterial communities encased in an extracellular matrix composed of polysaccharides, proteins, and extracelluar (e)DNA, with eDNA required for biofilm formation and integrity. Here we demonstrate that eDNA release is controlled by BfmR, a regulator essential for Pseudomonas aeruginosa biofilm development. Expression of bfmR coincided with localized cell death and DNA release, and could be stimulated by conditions resulting in membrane perturbation and cell lysis. ΔbfmR mutant biofilms demonstrated increased cell lysis and eDNA release suggesting BfmR to suppress, but not eliminate, these processes. Genome-wide transcriptional profiling indicated that BfmR was required for repression of genes associated with bacteriophage assembly and bacteriophage-mediated lysis. Chromatin immunoprecipitation analysis of direct BfmR targets identified the promoter of PA0691, termed here phdA, encoding a previously undescribed homologue of the prevent-host-death (Phd) family of proteins. Lack of phdA expression coincided with impaired biofilm development and increased cell death, a phenotype comparable to ΔbfmR. Expression of phdA in ΔbfmR restored eDNA release, cell lysis, and biofilm formation to wild type levels, with phdA overexpression promoting resistance to the superinfective bacteriophage Pf4, detected only in biofilms. Therefore, we propose that BfmR regulates biofilm development by limiting bacteriophage-mediated lysis and thus, eDNA release, via PhdA.
Keywords: Pseudomonas aeruginosa, prevent-host-death protein, bacteriophage Pf4, extracellular DNA, biofilm development, cell death
INTRODUCTION
The opportunistic pathogen Pseudomonas aeruginosa is responsible for a wide range of acute and chronic infections (Costerton et al., 1999). The transition to chronic infections is accompanied by physiological changes in the bacteria favoring the formation of biofilm communities. Biofilms are complex communities of microorganisms attached to surfaces and embedded in a self-produced extracellular matrix (Costerton et al., 1995). The extracellular polymeric substance (EPS) matrix can constitute up to 90 % of the biofilm biomass (Flemming et al., 2000). The EPS contains one or more polysaccharides that provide a hydrated scaffolding to stabilize and reinforce the structure of the biofilm, mediate cell-cell and cell-surface interactions, and provide protection from biocides and antimicrobial agents (Mah & O'Toole, 2001, Stewart & Costerton, 2001, Flemming et al., 2007). In P. aeruginosa, three polysaccharides have been identified, namely alginate, Pel, and Psl (Jackson et al., 2004, Friedman & Kolter, 2004, Ryder et al., 2007). In addition to polysaccharides, EPS has been demonstrated to contain proteins, DNA, and other macromolecules (Sutherland, 2001). Their relative importance as structural components may depend on the environmental conditions, on the age of the biofilm, and on the particular strain of P. aeruginosa forming the biofilm.
The importance of extracellular genomic DNA (eDNA) as a structural component of biofilms was first demonstrated in P. aeruginosa. Whitchurch et al. demonstrated that P. aeruginosa PAO1 biofilm formation was attenuated under static growth conditions and significantly reduced under flowing growth conditions by the presence of DNase in the growth medium (Whitchurch et al., 2002). However, the contribution of eDNA appeared to be temporal, as 2-day-old biofilms readily dispersed upon addition of DNase I, while mature biofilms did not. The findings suggested that the cells in young PAO1 biofilms are held together by eDNA whereas the cells in more mature PAO1 biofilms are held together primarily by components other than eDNA (Matsukawa & Greenberg, 2004, Whitchurch et al., 2002). The importance of eDNA in biofilm EPS has subsequently been demonstrated for clinical P. aeruginosa isolates, as well as for a variety of bacterial species, through analyses of biofilm formation by lysis-defective mutants and of DNA removal from the biofilm matrix (Izano et al., 2008, Nemoto et al., 2003, Steinberger & Holden, 2005, Thomas et al., 2009). For instance, eDNA was also found to be essential for the initial phase of biofilm development by Staphylococcus epidermidis, under both static and flowing conditions (Qin et al., 2007).
Microscopic investigation of flow chamber-grown wild-type P. aeruginosa biofilms stained with different nucleic acid stains suggested that the eDNA is located primarily in the stalks of mushroom-shaped multicellular structures called microcolonies, with a high concentration especially in the outer part of the stalks forming a border between the stalk-forming bacteria and the cap-forming bacteria (Allesen-Holm et al., 2006). The findings indicated that eDNA was organized in distinct patterns in P. aeruginosa biofilms and suggested a specific mechanism for eDNA release. In P. aeruginosa, eDNA has been described to be released in a PQS quorum sensing dependent manner and released due to cell lysis (Spoering & Gilmore, 2006, Yang et al., 2007, D'Argenio et al., 2002, Allesen-Holm et al., 2006). It has been suggested that PQS quorum sensing-regulated DNA release might be linked to phage induction in biofilms causing cell lysis (Allesen-Holm et al., 2006, Webb et al., 2003).
Goedeke et al. recently demonstrated a link between eDNA release and bacteriophage activity by demonstrating that phage-induced lysis enhanced biofilm formation in Shewanella oneidensis MR-1, while a mutant devoid of the prophages was severely impaired in biofilm formation through all stages of development due to reduced eDNA release (Godeke et al., 2010). However, the contribution of phage-mediated cell lysis to biofilm formation appears to vary from species to species. For instance, deletion of phage genes in E. coli coincided with increased biofilm formation (Wang et al., 2009). Similarly, Zegans et al. demonstrated that lysogenic infection of P. aeruginosa PA14 with bacteriophage DMS3 inhibits biofilm formation and swarming motility, both important bacterial group behaviors (Zegans et al., 2009). In contrast, deletion of Pf4 genes resulted in reduced biofilm formation by P. aeruginosa (Rice et al., 2009), while the presence of Pf4 has been linked to both cell death and lysis of P. aeruginosa PAO1 cells in biofilms (Webb et al., 2004, Webb et al., 2003). However, little is known about the regulation of filamentous prophage excision in P. aeruginosa.
While it is apparent that phage-mediated lysis and eDNA release play an important role in biofilm formation and architectural differentiation, little is known about the mechanism regulating the temporal and spatial activation of prophage genes in P. aeruginosa biofilms and thus, biofilm development. Here, we demonstrate that the novel two-component regulator BfmR, which was previously demonstrated to be required for P. aeruginosa biofilms to transition to maturation-1 biofilm developmental stage (Petrova & Sauer, 2009), characterized by cell clusters exceeding 10 μm in diameter (Sauer et al., 2002), regulates biofilm development through the suppression of premature release of eDNA by controlling bacteriophage-mediated lysis via PhdA (PA0691), a homologue of the Phd (prevent host death) family of proteins.
RESULTS
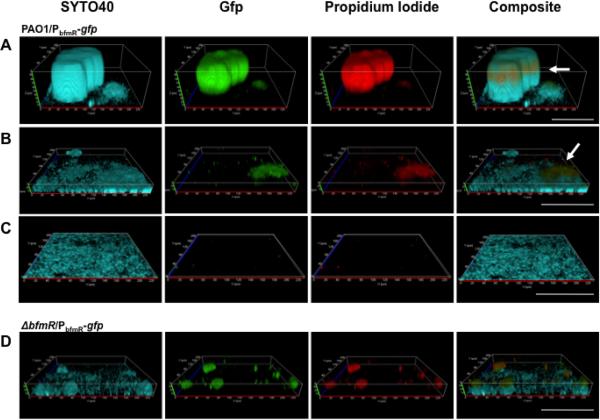
To elucidate the mechanism by which the novel two-component system (TCS) BfmSR regulates the transition of P. aeruginosa biofilms to the maturation-1 stage, we first monitored bfmR expression within P. aeruginosa PAO1 biofilms by employing chromosomal transcriptional gfp-fusion constructs (PbfmR-gfp). In addition, biofilms were stained with propidium iodide (PI) to visualize the presence of extracellular DNA (eDNA), and counterstained with SYTO 40. SYTO 40 is a member of the SYTO family of membrane-permeable nucleic acid stains that do not act exclusively as nuclear stains in live cells. While both PI and SYTO 40 stains are capable of binding nucleic acids, PI has a higher affinity for eDNA and displaces SYTO 40. The expression of bfmR was monitored in three different areas of PAO1/PbfmR-gfp biofilms: within microcolonies (Fig. 1A), areas harboring small clusters (Fig. 1B) and those only showing a thin layer of cells attached to the substratum (Fig. 1C). Fluorescent image composites indicated that in PAO1/PbfmR-gfp biofilms, GFP indicative of bfmR expression was detected predominantly within microcolonies and small clusters (Fig. 1A–B), while bfmR expression in the substratum layer was negligible (Fig. 1C). Furthermore, expression of the PbfmR-gfp reporter was only detectable in areas of the biofilm that also stained with PI (Fig. 1A–B).
Figure 1. Detection of bfmR expression in P. aeruginosa biofilms.
P. aeruginosa PAO1 reporter strain PbfmR-gfp was grown for 144 hr in flow cells after which time the biofilms were stained with propidium iodide (PI) and SYTO 40. PI was used to visualize eDNA, while SYTO 40 was used as a counterstain to stain all cells present in the biofilm. gfp was used to monitor bfmR expression. Three different areas of PAO1/PbfmR-gfp biofilms were visualized by confocal microscopy: (A) areas harboring microcolonies, (B) small clusters, (C) and those only showing a thin layer of cells attached to the substratum. Biofilms were grown in 1/20 diluted LB. Arrows point to location of eDNA on the outside of a microcolony, as well as eDNA deposition on top of a small cluster. (D) Visualization of PbfmR-gfp reporter activity in ΔbfmR biofilms. White bar = 100 μm.
The microscopic analysis of the composites furthermore suggested the presence of a `ring' or `disk' composed of cells/biomass stained by PI and expressing GFP/bfmR, which was detectable on the outside of microcolonies of PAO1/PbfmR-gfp (Fig. 1A, arrow). Moreover, cells and material stained by PI and expressing GFP/bfmR were also detectable in smaller cell clusters where it appeared to be deposited on top of the aggregates (Fig. 1B, arrow). Strain PAO1/Tn7-gfp, which constitutively expresses gfp, was used as a negative control. No correlation between PI-stainable material and gfp expression was noted when composites of PAO1/Tn7-gfp were analyzed (not shown).
The observation of a ring/disk surrounding microcolonies is consistent with previous findings indicating the presence of eDNA between the stalk and the cap of microcolonies where the DNA not only functions as a cell–cell interconnecting compound but also as a structural component of biofilms (Allesen-Holm et al., 2006). Moreover, the microscopic analysis suggests that bfmR expression occurs predominantly in areas of biofilms that are stained by PI.
Biofilms inactivated in bfmR demonstrate higher levels of PI-stainable biofilm biomass
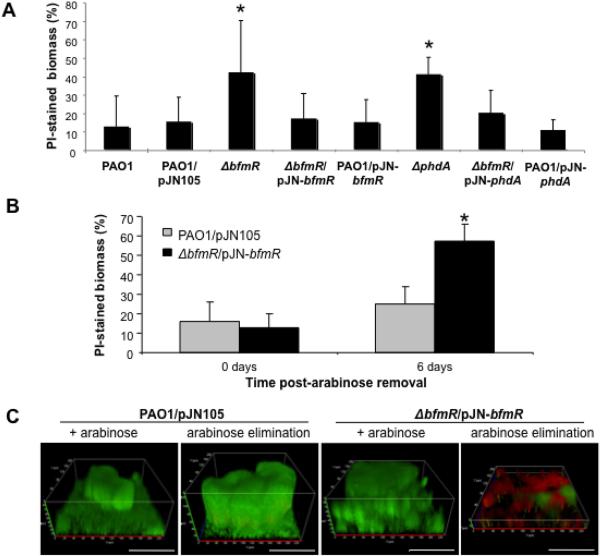
As bfmR transcription was predominantly detectable in areas of biofilms that also stained with PI, we asked whether BfmR is associated with the regulation of cell death and DNA release. Visual observation of PAO1 ΔbfmR flow cell-grown biofilms indicated increased levels of bacterial cells stainable with PI and the presence of PI-stainable material that was not associated with bacterial cells (Fig. 1D, Fig. S1). This finding indicated not only that bfmR expression occurs in areas of biofilms that are predominantly stained by propidium iodide, but also that inactivation of bfmR coincides with increased PI stainable biofilm biomass. We subsequently quantified the percentage of PI stainable material in wild type and ΔbfmR biofilms following staining with the LIVE/DEAD BacLight viability kit and visualization by confocal microscopy (see Fig. S2). In PAO1 biofilms, 35.47±10.05% of the cells present in microcolonies exhibited PI fluorescence, while areas devoid of microcolonies only exhibited 3.38% (±5.48) PI fluorescence (Fig. S2A). This is consistent with previous findings demonstrating the presence of eDNA and the occurrence of cell death within localized regions in microcolonies of P. aeruginosa biofilms (Allesen-Holm et al., 2006, Yang et al., 2007, Whitchurch et al., 2002, Webb et al., 2003). Not considering architectural differences, the total PI-stained biomass in PAO1 biofilms was found to be 12.55±16.34% (Fig. 2A). In contrast, inactivation of bfmR coincided with higher levels of PI staining throughout the thin biofilm, with PI-stained biomass accounting for 41.79±31.6% of the total ΔbfmR biofilm biomass (Fig. 2A, C). Complementation (ΔbfmR/pJN-bfmR) restored the proportion of PI-stained biomass to wild type levels (Fig. 2A, C, Fig. S2B), while overexpression of bfmR (PAO1/pJN-bfmR) significantly reduced regions of cell death and potential DNA release within microcolonies (Fig. S3).
Figure 2. Quantitation of propidium iodide stainable biofilm material in P. aeruginosa PAO1 and mutant biofilms inactivated in or overexpressing bfmR and phdA.
(A) Quantitation of propidium iodide stainable material (reported as percent of the total detectable biomass) in biofilms following 144 hrs of growth under flowing conditions. (B) Quantitation of propidium iodide stainable biofilm biomass in P. aeruginosa PAO1/pJN105 and ΔbfmR biofilms expressing bfmR following arabinose removal. Wild type PAO1 bearing the empty pJN105 vector and complemented ΔbfmR bearing pJN-bfmR were grown in the presence of arabinose for 144 hrs, at which point, arabinose was removed in order to inactivate transcription from the PBAD promoter. Biofilms were allowed to grow for an additional 72 hrs before confocal images were acquired for COMSTAT analysis. (C) Confocal images of PAO1/pJN105 and ΔbfmR/pJN-bfmR prior and 72 hrs post arabinose removal. Prior to image acquisition by confocal microscopy, biofilms were stained with SYTO 9 and propidium iodide (PI) using the LIVE/DEAD BacLight viability kit. + arabinose, biofilms were grown for 144 hrs in the presence of arabinose. Arabinose elimination, biofilms 72 hrs post arabinose removal. Biofilms were grown in VBMM medium. Images showing separate SYTO9 and propidium iodide staining are shown in Fig. S2B–C. White bar = 100 μm. Error bars indicate standard deviation. *, significantly different from PAO1 (p < 0.05).
To determine whether the detected increase in PI stainable biofilm biomass was due to impaired biofilm formation, we tested two additional mutants, ΔbfiS and ΔmifR, which were previously reported to also be arrested in biofilm development (Petrova & Sauer, 2009). Inactivation of bfiS and mifR did not result in an increase in PI-stainable biofilm biomass (Fig. S4A).
Since decreasing the expression of bfmR in developed biofilms resulted in biomass disaggregation and architectural collapse (Petrova & Sauer, 2009), we next asked whether decreasing bfmR expression in already established, mature biofilms would also result in elevated levels of PI fluorescence. Following 144 hr of growth under inducing conditions, arabinose-inducible expression of bfmR was abrogated in ΔbfmR via a switch to an arabinose-free medium, with the wild type strain PAO1 containing the empty vector used as a control. The wild type biofilms demonstrated an approximately 1.6-fold increase in the PI fluorescence (from 15.26±13.01% to 25.23±11.92% of total biomass) following 72 hr post arabinose removal (Fig. 2B–C, Fig. S2B–C). In contrast, arabinose removal brought about a 4-fold increase (from 12.75±11.22% to 51.36±16.52% of total biomass) in the PI-stainable material and biomass of the ΔbfmR/pJN-bfmR strain (Fig. 2B–C, Fig. S2B–C) with biofilm biomass disaggregation noticeable within 24 hr post arabinose removal (Fig. S2D). In contrast, deactivation of bfiS and mifR in complemented ΔbfiS and ΔmifR biofilms and disaggregation of these biofilms did not result in similar increases in the proportion of PI fluorescence (Fig. S4B–C). In fact, these biofilms exhibited 1.89- and 1.49-fold reductions in the proportion of the PI-staining biomass/material, respectively, demonstrating that general deactivation of biofilm-specific regulators and biomass disaggregation in mature biofilms does not result in cell lysis or increased eDNA release (Fig. S4C).
Inactivation of bfmR results in increased DNA release and cell death in biofilms
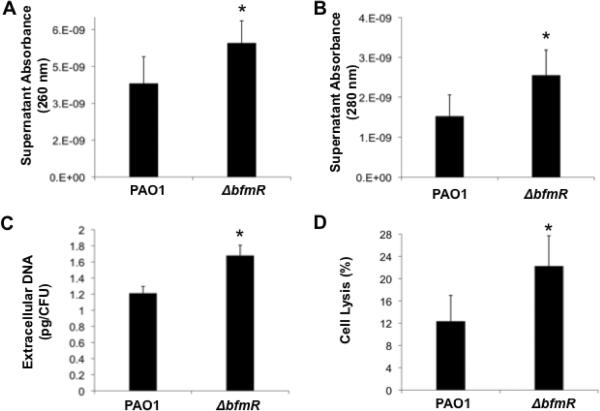
To determine whether increased PI-stainable biofilm biomass correlated with increased DNA release and cell lysis in ΔbfmR biofilms, we first compared the concentration of DNA and proteins released into the supernatants of 72-hr-old ΔbfmR and PAO1 biofilms grown under continuous flow conditions spectrophotometrically and standardized to total viable biofilm cells (CFUs). The analysis indicated a significant increase in the DNA and protein concentration in supernatants obtained from ΔbfmR biofilms (43% and 68%, respectively) when compared to PAO1 biofilm supernatants (Fig. 3A–B). Similar results were obtained using a propidium iodide (PI) DNA binding assay confirming the spectrophotometric based findings of ΔbfmR biofilms containing more released DNA than PAO1 biofilms (Fig. 3C). In contrast, no difference in eDNA was observed when supernatants of ΔbfmR and PAO1 grown planktonically were compared (not shown).
Figure 3. Relative comparison of DNA and protein content of wild type PAO1 and ΔbfmR biofilm supernatants.
The absorbance at 260 (A) and 280 (B) nm of supernatants from 72-hr-old PAO1 and ΔbfmR biofilms was determined as a relative measure of DNA and protein content, respectively. The absorbance was normalized with respect to CFU ml−1. (C) Determination of extracellular DNA present in 72-hr-old PAO1 and ΔbfmR biofilms using the propidium iodide binding assay. (D) Determination of cell lysis levels in wild type and ΔbfmR biofilm supernatants. Relative cell lysis levels in 72-hr-old PAO1 and ΔbfmR biofilms were calculated as the ratio of the concentrations of released (supernatant) and total (supernatant and cellular) DNA in the biofilms. Absolute DNA concentrations were measured via qPCR amplification of the housekeeping mreB gene and calculated with a standard curve established using known concentrations of genomic DNA. Error bars indicate standard deviation. *, significantly different from PAO1 (p < 0.05).
To determine whether the presence of eDNA was a result of cell lysis, a qPCR-based approach was used to quantify the relative levels of cell lysis in PAO1 and ΔbfmR biofilms (Ma & Wood, 2009). The analysis revealed significantly higher levels of lysis in ΔbfmR biofilms (22.20±5.50%) when compared to the wild type (12.29±4.71%, Fig. 3D). The findings strongly suggest that increased levels of eDNA in ΔbfmR biofilms were due to increased cell lysis. Taken together, these findings underscore the importance of BfmR in the regulation of eDNA cell lysis in PAO1 biofilms undergoing maturation.
Genome-wide transcriptional profiling of wild-type P. aeruginosa and an isogenic bfmR mutant
We next addressed the question of how BfmR controls the amount of DNA release and/or cell death by assessing the full range of genes regulated by BfmR in a biofilm. We therefore compared the transcriptome of P. aeruginosa ΔbfmR to wild-type P. aeruginosa PAO1 under biofilm growth conditions. Among the 5678 genes surveyed, 1246 genes showed significantly altered (either reduced or increased) expression in P. aeruginosa ΔbfmR as compared to its isogenic wild-type parent (Table S1–S2).
Many of the genes exhibiting BfmR-dependent changes in expression encode proteins involved in transcriptional regulation, secretion/export, membrane proteins, genes related to phages, catabolism and energy metabolism (Fig. S5A). For instance, multiple genes/operons encoding phage-related genes including genes encoding defective phages (PA0616-PA0648) and the filamentous Pf1-like phage (PA0715-PA0728) were significantly upregulated in a ΔbfmR mutant biofilm (Fig. S5). In parallel with bfmR-mediated induction of prophage gene expression, we also noted 20 genes involved in DNA repair including helicases, recombinases, ligases, and endonucleases to be significantly upregulated (Table S1). Similarly, genes encoding secreted factors and the type II and III secretion systems were significantly increased in the bfmR mutant strain (Table S1). In addition, genes encoding the PhoPQ- and PmrAB-regulated cationic antimicrobial peptide (CAP) resistance operon PA3552–PA3559 (arnBCADTEF-ugd) were found to be significantly upregulated (Table S1). The CAP operon has been demonstrated to be induced by sub-inhibitory concentrations of eDNA (Mulcahy et al., 2008) supporting the finding of increased presence of eDNA in ΔbfmR biofilms (Fig. 3). Taken together, the common feature of all of these genes is that they are associated with bacteriophage, DNA release, and potential defense mechanisms in response to phage-mediated lysis.
Conversely, a number of genes encoding factors associated with the propagation and spread of bacteriophages were repressed in P. aeruginosa ΔbfmR biofilms, including many genes associated with the expression of type IV pili and flagella. The bfmR mutant strain showed significant downregulation of 16 genes encoding the type IV pili apparatus and 11 genes involved in flagellar driven motility (Fig. S5B). Furthermore, each of the 38 structural and regulatory genes of the translation machinery (30S and 50S ribosomes) were significantly downregulated (Fig. S5B). In parallel with bfmR-mediated repression of protein translation, we also observed significant downregulation of structural genes encoding the DNA directed polymerase, rne (ribonuclease E), ribonucleoside reductases, and tsf (elongation factor Tsf). Furthermore, 13 genes involved in protein folding (e.g. chaperones) were down-regulated (Table S2). These included groEL, hslVU, dnaK-grpE, and tig-clpPL.
We also identified proteins controlled in a BfmR-dependent manner using a proteomic approach. Proteins obtained from PAO1 and the isogenic bfmR mutant following 24, 72, and 144 hr of biofilm growth were analyzed by 2D/PAGE, and differentially produced proteins were subsequently identified by LC-MS-MS. Consistent with the transcriptome analysis, proteins absent in ΔbfmR biofilms were chaperones including HslU, several proteases including the Lon protease (PA1803), and ribonuclease E (rne) (Table 1). In contrast, proteins involved in secretion and export were increased in production (Table 1).
Table 1.
Proteins demonstrating altered expression in biofilms following bmfR inactivation.
| Locus | Biofilm sample | Protein | ||||
|---|---|---|---|---|---|---|
| PAO1 | ΔbfmR | |||||
| 24 hr | 72 hr | 144 hr | 144 hr | |||
| PA4498 | √ | √ | √ | - | probable metallopeptidase | |
| PA1803 | lon | √ | √ | √ | - | Lon protease |
| PA4502 | √ | √ | √ | - | probable binding component of ABC transporter | |
| PA4175 | piv | √ | √ | √ | - | protease IV, “probable endoproteinase Arg-C precursor PvdS-regulated endoprotease, lysyl class” |
| PA5054 | hslU | √ | √ | √ | - | heat shock protein HslU |
| PA2976 | rne | √ | √ | √ | - | ribonuclease E |
| PA1800 | tig | √ | √ | - | - | trigger factor |
| PA2385 | pvdQ | √ | - | - | √ | 3-oxo-C12-homoserine lactone acylase PvdQ |
| PA3471 | √ | - | - | √ | probable malate dehydrogenase | |
| PA2023 | galU | √ | - | - | √ | UTP--glucose-1-phosphate uridylyltransferase |
| PA1585 | sucA | - | - | - | √ | 2-oxoglutarate dehydrogenase (E1 subunit) |
| PA5210 | - | - | - | √ | probable secretion pathway ATPase | |
| PA4938 | purA | - | - | - | √ | adenylosuccinate synthetase |
| PA0266 | gabT | - | - | - | √ | 4-aminobutyrate aminotransferase |
√ Protein detected on a silver stained 2D-PAGE gel;
- Protein not detected on a silver stained 2D-PAGE gel.
BfmR regulates biofilm development through the prevent-host-death family protein PA0691 (PhdA)
The transcriptome analysis strongly suggested that BfmR suppressed the expression of bacteriophage genes and genes involved in DNA repair and defense. We next addressed the question of how BfmR controls the expression of bacteriophage-mediated lysis and, thus, DNA release by investigating the direct regulatory target(s) of BfmR. This was done by analyzing the DNA binding sites of this regulator during biofilm growth via chromatin immunoprecipitation (ChIP) analysis, followed by cloning and sequencing of the respective DNA binding sites.
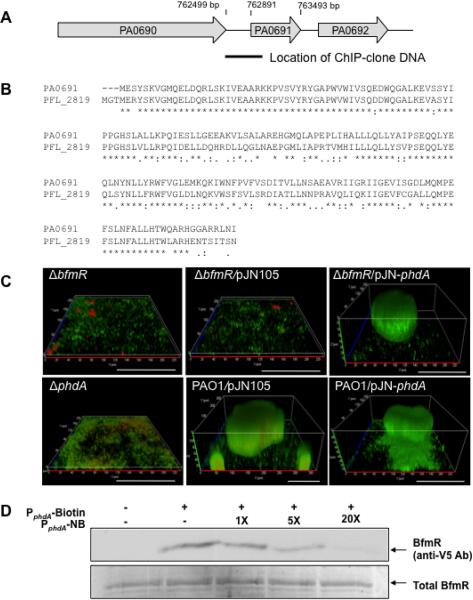
The analysis resulted in the identification of a sequence upstream of the gene PA0691 (Fig. 4A). Although www.pseudomonas.com indicates that the encoded protein PA0691 does not contain any known conserved domains, it is 84% similar and 72% identical to the P. fluorescens prevent-host-death (Phd) family protein PFL_2819 (Paulsen et al., 2005) (Fig. 4B). Phd proteins have been previously shown to prevent bacteriophage-mediated cell lysis (Garcia-Contreras et al., 2008, Lehnherr & Yarmolinsky, 1995). Due to the high similarity to the Phd family protein PFL_2819, we named PA0691 prevent-host-death protein A (PhdA).
Figure 4. BfmR activates a homologue of the prevent host death protein, PA0691 (PhdA), which is required for normal biofilm development by P. aeruginosa.
(A) Location of DNA that co-purified with BfmR via ChIP-cloning from 24-hr-old PAO1 biofilms was identified via sequencing as a fragment upstream of the gene PA0691 (phdA). The sequenced region corresponded to the 762651-763061 bp region of the PAO1 genome. (B) Alignment of PA0691 (PhdA) and P. fluorescens PFL_2819 prevent-host-death family protein. *, denotes identical amino acids;: and. denote similar amino acids. (C) Complementation of ΔbfmR with phdA restores biofilm formation to wild type levels. Confocal images of were acquired following 144 hr of growth. White bar = 100 μm. (D) Streptavidin magnetic bead binding assay demonstrating binding of V5/6×His-tagged BfmR protein to 2.5 pmol biotinylated PphdA. Non-biotinylated PphdA (PphdA-NB) was used as specific competitor DNA in 1-, 5-, and 20-fold excess. BfmR binding to PphdA was detected by immunoblot analysis using anti-V5 antibodies. The lower panel shows total BfmR present in the streptavidin binding assays. +, indicates presence of PphdA-Biotin or PphdA -NB; −, indicates absence of PphdA-Biotin or PphdA-NB. All experiments were carried out at least in triplicate.
To confirm that the ΔbfmR biofilm phenotype was linked to phdA expression, we expressed phdA in a ΔbfmR mutant and quantified cell death and eDNA release in the resulting mutant biofilm. Expression of phdA resulted in a significant reduction of PI-stainable biofilm biomass to a level comparable to that observed in wild type biofilms (Fig. 2A, Fig. S2A). More importantly, expression of phdA in a ΔbfmR mutant restored the biofilm architecture to wild type levels (Fig. 4C, Table 2). Overexpression of phdA in PAO1 had a similar effect on PI-stainable biofilm biomass (Fig. 2A). Moreover, overexpression of phdA resulted in increased biofilm biomass accumulation (Fig. 4A, Table 2). In contrast, inactivation of phdA resulted in biofilms that were not only architecturally comparable to those formed by ΔbfmR (Fig. 4C), but also exhibited PI staining levels similar to those of ΔbfmR biofilms (Fig. 2A).
Table 2.
COMSTAT analysisa of P. aeruginosa wild type and mutant biofilm structure post 144 hrs of growth as a biofilm.
| Strains | Total biomass (μm3/μm2) | Substratum coverage (%) | Average thickness (μm) | Maximum thickness (μm) | Roughness coefficient (dimensionless) |
|---|---|---|---|---|---|
| PAO1 | 10.88 (±3.43) | 25.17 (±9.38) | 12.09 (±5.24) | 62.51 (±12.60) | 1.26 (±0.32) |
| Δ bfmR | 1.70 (±0.79)* | 15.34 (±11.88)* | 1.41 (±0.74)* | 9.06 (±5.73)* | 1.54 (±0.23)* |
| ΔbfmR/pJN-phdA | 11.21(±3.04) | 54.02 (±19.75)* | 11.09 (±3.15) | 48.5 (±14.40) | 1.17 (±0.25) |
| PAO1/pJN-phdA | 19.47 (±13.30)* | 41.03 (±14.24)* | 20.30 (±14.79)* | 58.50 (±29.91) | 0.96 (±0.38) |
COMSTAT analysis was carried out from biofilms grown in triplicate using at least 6 images per replicate.
Significantly different from 144-hr-old PAO1; p ≤ 0.05 as determined by ANOVA and SigmaStat.
The binding of BfmR to the sequence upstream of phdA was verified by streptavidin magnetic bead DNA binding assays, with BfmR binding to the sequence present in all identified phdA ChIP clones (−240 to +0 relative to the translational start site, PphdA) (Fig. 4D). In addition, phdA expression was found to be BfmR-dependent, with deletion of bfmR resulting in a 3-fold reduction of phdA transcript abundance in ΔbfmR biofilms as compared to wild type biofilms (Table 3). Overall, these findings strongly suggest that BfmR is directly involved in coordinating biofilm formation through PhdA by possibly suppressing bacteriophage-mediated cells lysis and thus, the amount of released DNA.
Table 3.
Expression of phdA and selected genes in P. aeruginosa ΔbfmR mutant biofilms relative to P. aeruginosa PAO1 biofilms as determined by qRT-PCR.
| Transcript | ΔbfmR biofilms (relative to P. aeruginosa PAO1 biofilms, %) |
|---|---|
| phdA | 36.75 (±11.7)* |
| pqsA | 97.37 (±29.0) |
| pqsL | 127.80 (±18.8) |
| lasB | 97.33 (±15.0) |
| rhlA | 111.96 (±30.3) |
| phoP | 233.83 (±52.9)* |
| PA3553 (arnC) | 321.50 (±25.5)* |
Significantly different from PAO1; p ≤ 0.05 as determined by ANOVA and SigmaStat.
Inactivation of bfmR coincides with increased bacteriophage-mediated lysis during biofilm development
Phd proteins have been previously shown to prevent bacteriophage-mediated lysis of E. coli (Garcia-Contreras et al., 2008, Lehnherr & Yarmolinsky, 1995). Furthermore, the filamentous prophage Pf4 has been implicated in bacterial apoptosis and biofilm dispersion in P. aeruginosa biofilms (Rice et al., 2009, Webb et al., 2003). We, thus, assessed whether bacteriophage release, cell lysis, and eDNA accumulation in P. aeruginosa biofilms is dependent on BfmR and PhdA by isolating and quantifying bacteriophages from wild type and ΔbfmR biofilms, ΔbfmR biofilms complemented with bfmR and phdA (ΔbfmR/pJN-bfmR, ΔbfmR/pJN-phdA), and wild type biofilms overexpressing phdA (PAO1/pJN-phdA) (Fig. 5A).
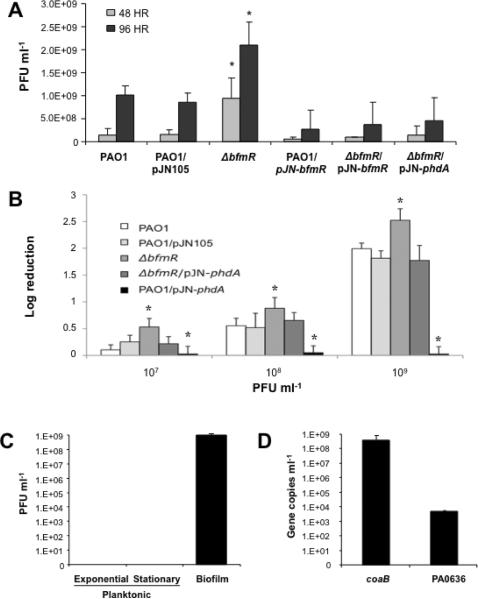
Figure 5. Inactivation of bfmR coincides with increased release of bacteriophages Pf4 in biofilms and increased susceptibility of ΔbfmR grown planktonically to biofilm-isolated phages.
(A) Inactivation of bfmR coincides with increased release of bacteriophages following 48 and 96 hrs of biofilm growth compared to wild type PAO1 biofilms. Release of phages is restored to wild type levels following expression of bfmR and phdA in ΔbfmR biofilms. (B) ΔbfmR grown planktonically is more susceptible to biofilm-isolated phages than wild type PAO1, while overexpression of phdA renders P. aeruginosa more resistant. (C) Plaque formation is only detectable when using biofilm, but not planktonic, supernatants. (D) Determination of coaB and PA0636 gene copy numbers in biofilm supernatants following DNase treatment. All experiments were performed in triplicate. Error bars indicate standard deviation. *, significantly different from PAO1 biofilm of same age (p ≤ 0.05), as determined by ANOVA and Sigma Stat.
We were able to detect and directly isolate bacteriophages capable of infecting and lysing the P. aeruginosa parental strain in the fluid supernatants and effluents from wild type and ΔbfmR biofilms. Contrary to previous reports indicating the appearance of phage only following extended periods of growth (4–10 days) of P. aeruginosa biofilms (Webb et al., 2003, Rice et al., 2009), bacteriophages were already evident in our tube reactor system following 48 hr of biofilm growth (Fig. 5A). On average, 1.5×108 ±1.3 ×108 plaque forming units (PFU) ml−1 were detected in 48-hr-old wild type biofilms, which increased to 1.0×109 ±1.9 ×108 PFU ml−1 in 96-hr-old biofilms. In contrast, 48-hr-old ΔbfmR biofilms were characterized by a 6.3-fold increase in PFU ml−1 compared to the wild type. The increase in PFU in ΔbfmR biofilms was consistent with the higher levels of lysis observed for ΔbfmR biofilms (see Fig. 3D). Interestingly, overexpression of bfmR significantly reduced the numbers of PFUs in 48- and 96-hr-old biofilms to below 6.0 ×107 and 3.0 ×108 PFU ml−1, respectively (Fig. 5A). Similarly, complementation of the ΔbfmR mutant by expressing bfmR (ΔbfmR/pJN-bfmR) or phdA (ΔbfmR/pJN-phdA) restored the release of bacteriophages to wild type levels (Fig. 5A). Overexpression of phdA had a similar effect (not shown). Taken together, these findings suggest that BfmR plays a role via PhdA in suppressing (but not abolishing) the release of bacteriophages and phage-mediated cell lysis during biofilm development.
To determine whether inactivation of bfmR renders cells more sensitive to phage-mediated lysis, viability upon exposure to 107, 108, and 109 biofilm-isolated phage particles per mL in the growth medium was assessed. Both wild type and ΔbfmR strains were susceptible to biofilm-isolated bacteriophages, with the ΔbfmR strain being more susceptible to phage-mediated lysis as compared to the wild type (Fig. 5B). Complementation of ΔbfmR with phdA reduced the susceptibility to wild type levels, while overexpression of phdA rendered P. aeruginosa (PAO1/pJN-phdA) resistant to biofilm-isolated bacteriophages (Fig. 5B).
Bacteriophage Pf4 isolated from cell-free biofilm supernatants causes plaques
It is generally accepted that filamentous phages do not kill host cells as they can be extruded from the cell. Recent evidence, however, suggests that bacteriophage Pf1 (renamed to Pf4) may convert or mutate to a superinfective form during biofilm development (Webb et al., 2003). Plaque formation was only observed when using supernatants from biofilms but not from planktonic cells grown to exponential and late stationary phase (Fig. 5C). To determine whether plaque formation was due to bacteriophage Pf4, we made use of a quantitative PCR-based approach to detect the coaB gene, encoding the coat B protein of bacteriophage Pf4. Amplification of the genes mreB, mexA, and 16S rDNA was used to determine the presence and amount of genomic DNA in cell-free biofilm supernatants and were found to be detectable at low copy numbers. In the case of mreB, only 2500 copies were detected which dropped to below detection levels (< 80 gene copies, Ct > 40 cycles) following DNase treatment (Fig. S6). In contrast, cell-free biofilm supernatants, which were shown to give rise on average to 108 PFU ml−1, contained a total of 1.2×109 copies of the coaB gene per ml (Figs. 5D, S6). Following repeated DNase treatments, 3.8 ×108 copies of the coaB gene remained detectable indicating that these genes were protected from DNase treatment (Fig. 5D). Similar results were obtained for the detection of coaB in ΔbfmR biofilm supernatants (not shown). In contrast, supernatants of planktonic cells harbored less than 1×105 copies of the coaB gene (Fig. S6). To determine whether bacteriocins that resemble bacteriophage tail-like structures (pyocin) play a role in plaque formation, gene copy numbers of PA0636 encoding the minor tail protein of the defective phage were analyzed as well, as pyocin DNA has been shown to be associated with the pyocins (Lee et al., 1999). However, no correlation between the presence of PA0636 and PFU was found. For both growth conditions, the PA0636 copy numbers were comparable to those of mreB following DNase treatment (Fig. S6) and more than 4 magnitudes lower than coaB and the number of plaques detected in biofilm supernatants (Fig. 5C–D).
These findings are in agreement with bacteriophage Pf4 being capable of switching from a nonlytic to a superinfective, lytic phenotype (Webb et al., 2003). Moreover, our findings correlate PFU with the presence of DNAseI-protected bacteriophage Pf4 coaB in biofilm supernatants suggesting bacteriophages to be the cause of plaque formation.
Expression of bfmR is induced by cell lysis and membrane perturbing conditions
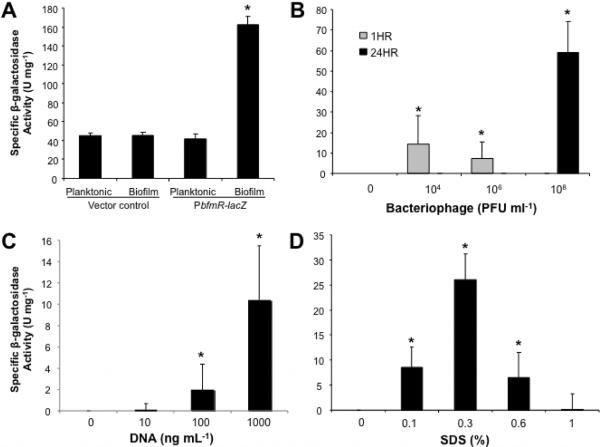
Since BfmR was found to activate the expression of a potential Phd homolog in biofilms resulting in the reduction of DNA release, presumably by preventing phage-mediated cell lysis, we asked whether comparable physiological conditions (e.g. extracellular DNA, exposure to phage particles) would result in activation of the bfmR promoter under planktonic growth conditions. Consequently, a chromosomal lacZ transcriptional fusion construct (PbfmR-lacZ) in PAO1 was employed. Under biofilm growth conditions, a specific activity of 162 U mg−1 was detected (Fig. 6A). Under planktonic conditions, however, the PbfmR reporter activity was comparable to a lacZ reporter construct lacking a promoter (vector control, Fig.6A), indicating that bfmR transcription was biofilm-specific. The reporter construct was subsequently used to determine β-galactosidase activity of planktonic cells in response to various physiological conditions.
Figure 6. bfmR transcription is activated by SDS, extracellular DNA, and exposure to bacteriophage Pf4.
(A) PbfmR-lacZ reporter activity is biofilm-specific. P. aeruginosa PAO1-PbfmR-lacZ was grown planktonically and as a 3-day old biofilm. A chromosomal lacZ reporter construct lacking a promoter was used as vector control. For all subsequent experiments (B–D), the β-galactosidase activity shown has been corrected by subtracting the β-galactosidase activity of the vector control. PAO1-PbfmR-lacZ was grown planktonically to exponential phase and exposed to increasing biofilm-isolated PFU ml−1 (B), increasing concentrations of salmon sperm DNA (C), and SDS (D), after which time the specific β-galactosidase activity (U mg−1) was determined. All experiments were performed in triplicate. Error bars indicate standard deviation. *, significantly different from untreated PAO1 (p ≤ 0.05), as determined by ANOVA and Sigma Stat.
Exposure to 104 and 106 phage particles for 1 hr resulted in detectable PbfmR reporter activity (14 and 7 U mg−1, respectively), while no activity was detected upon exposure to 108 phage particles for 1 hr (Fig. 6B). In contrast, specific β-galactosidase activity increased to 59 U mg−1 upon exposure for 24 hr to 108 biofilm-isolated phage particles per mL (Fig. 6B). To determine whether the increase in reporter activity upon exposure to phage particles was due to cell lysis or released DNA, planktonic cells were exposed to spent supernatants of cells grown to stationary phase, varying concentrations of DNA and the detergent SDS. Spent culture supernatant had no effect on bfmR transcription (not shown). In contrast, addition of salmon sperm DNA resulted in increased bfmR transcription at elevated DNA concentrations (Fig. 6C). Similar results were obtained when genomic DNA isolated from P. aeruginosa was tested (not shown), indicating that exposure to extracellular DNA at elevated concentrations, regardless of the origin, affects bfmR expression. Since eDNA has previously been demonstrated to cause membrane perturbation and cell lysis (Mulcahy et al., 2008), bfmR expression was also determined in the presence of the detergent SDS. Treatment with SDS ranging from 0.1–0.6% positively affected bfmR transcription, with the highest β-galactosidase activity (27 U mg−1) noted upon treatment with 0.3% SDS (Fig. 6D). Higher SDS concentrations had little or no effect on PbfmR reporter activity (Fig. 6D).
Taken together, the findings indicate that conditions resulting in cell lysis/membrane perturbation, in particular exposure to biofilm-isolated bacteriophages, result in increased bfmR transcription, further supporting the notion that BfmR is involved in limiting cell lysis.
Inactivation of bfmR increases expression of CAP resistance genes but does not alter expression of quorum sensing related genes
In P. aeruginosa biofilms, eDNA functioning as a cell–cell interconnecting compound and structural component has been shown to be regulated via quorum sensing, in particular via the PQS quorum-sensing system (Allesen-Holm et al., 2006, Yang et al., 2007). We, therefore, sought to assess any potential changes in quorum sensing signaling by comparing the levels of quorum sensing-regulated transcripts in wild type and ΔbfmR biofilms. Compared to wild type biofilms, no significant difference in pqsA and pqsL transcript abundance was detected in ΔbfmR biofilms (Table 3). Furthermore, no bfmR-dependent effects on the Las and Rhl quorum sensing systems were detected as revealed by rhlA and lasB transcript abundance analysis (Table 3), suggesting that BfmR functions independently of quorum sensing signaling.
Genes encoding the PhoPQ- and PmrAB-regulated cationic antimicrobial peptide (CAP) resistance operon PA3552–PA3559 (arnBCADTEF-ugd) were found to be upregulated in ΔbfmR biofilms as revealed by DNA microarray analysis. Upregulation of PA3553 (arnC) and phoP was confirmed by qRT-PCR. The finding confirms that increased presence of eDNA in ΔbfmR biofilms resulted in increased cation chelation and thus, activation of the PhoPQ and increased expression of the CAP operon (Table 3).
DISCUSSION
Extracelluar DNA (eDNA) has been shown to be essential for the development, integrity, architecture and antimicrobial resistance of P. aeruginosa biofilms (Allesen-Holm et al., 2006, Whitchurch et al., 2002, Yang et al., 2007, Barken et al., 2008, Mulcahy et al., 2008, Nemoto et al., 2003), with such a role requiring complex temporal and spatial regulation of eDNA release. Evidence suggests that the PQS quorum sensing-regulated DNA release might be linked to phage induction in biofilms causing cell lysis (Webb et al., 2004, Allesen-Holm et al., 2006). However, little is known about how phage-mediated lysis is regulated in P. aeruginosa biofilms to accomplish such a temporal and spatial release of eDNA.
Here, we present evidence that bacteriophage-mediated lysis and subsequent DNA release are linked to biofilm development via the two-component regulator BfmR, which is required for the transitioning of P. aeruginosa biofilms from the initial attachment stage to the maturation-1 stage of biofilm development (Petrova & Sauer, 2009). To our knowledge, this is the first description of a regulatory program linking the regulation of bacteriophage and DNA release to the stage-specific biofilm development by P. aeruginosa. This is accomplished by BfmR suppressing the premature release of eDNA by controlling bacteriophage-mediated lysis via the transcription of PhdA (PA0691). Bacteriophage mediated lysis was biofilm specific and correlated with the presence of the DNAse I-protected bacteriophage Pf4 coaB gene in biofilm supernatants. The detection of the bacteriophage Pf4 coaB gene and the absence of plaque-forming units in the supernatants of planktonic cells are consistent with previous results by Webb et al. indicating that bacteriophage Pf4 can switch to a superinfective form capable of lysing P. aeruginosa upon transitioning to surface associated growth (Webb et al., 2003).
While the genetic basis of this switch remains to be elucidated, superinfective bacteriophages were detectable in flow cell grown P. aeruginosa biofilms following 4–10 days of growth, primarily within microcolonies prior to dispersion (Rice et al., 2009, Webb et al., 2004, Webb et al., 2003). Our findings suggested an earlier switch to superinfectivity by Pf4, which was exacerbated in a ΔbfmR mutant. It is likely that the difference in timing is due to differences in growth conditions, especially considering that a ΔbfmR mutant displays phenotypes opposite to a bacteriophage Pf4-deficient mutant, which was shown to only form biofilms composed of significantly smaller microcolonies compared to the wild type biofilm and to be unable to undergo lysis (Rice et al., 2009). The reduced size of microcolonies in a bacteriophage Pf4-deficient mutant supports a role for bacteriophages prior to dispersion to enable cluster and microcolony formation. Moreover, these findings indicate a requirement for balanced bacteriophage activity and bacteriophage-mediated lysis to enable P. aeruginosa biofilms to progress past the maturation-1 stage of biofilm development, with both increases and decreases in lytic activity resulting in arrest of biofilm development. The data are furthermore consistent with the role of eDNA as an architectural element of P. aeruginosa biofilms. While the absence of eDNA resulted in inhibition of biofilm formation, here we demonstrated that excess or uncontrolled cell lysis and eDNA release arrest biofilm formation at a similar stage (Fig. 2C, 4C, Fig. S1) (Petrova & Sauer, 2009).
In P. aeruginosa biofilms, restriction of cell lysis is accomplished via the BfmR-dependent activation of PhdA, a homologue of the prevent host death protein PFL_2819 belonging to the antitoxin family. While little is known about the prevent host death protein PFL_2819 with the exception of it matching a protein family harboring the TIGR01552 domain (Paulsen et al., 2005), members of the Phd family have been shown to prevent bacteriophage-mediated lysis by countering the action of toxin or death-on-curing (Doc) proteins (Garcia-Contreras et al., 2008, Lehnherr & Yarmolinsky, 1995). This is consistent with our finding of phdA overexpression resulting in increased resistance to bacteriophages (Fig. 5B). The best-characterized member of this family of proteins is the plasmid-borne Phd protein of E. coli bacteriophage P1, the antidote partner of the Doc protein, which is controlled by the ClpXP serine protease (Lehnherr & Yarmolinsky, 1995). While the mechanism by which PhdA is controlled remains to be explored, inactivation of bfmR resulted in the differential expression of several proteases including the tig-clpPX operon (PA1800–1802) and the Lon protease (PA1803) (Tables 1, S1–S2).
Analysis of the phage genome obtained by extracting the RF plasmid from Pf4-infected cells recently indicated the presence of a putative ORF downstream of PA0728 encoding an integrase and 9 bp upstream of PA0729 a hypothetical protein harboring a ParE plasmid stabilization system domain (Webb et al., 2004). This putative ORF with homology to the Phd antitoxin protein of Pseudomonas syringe, was believed to be part of a toxin/anti-toxin system comprising PA0729. However, evidence for this ORF is lacking from the current P. aeruginosa genome website (pseudomonas.com) and BLAST searches using the intergenic region did not reveal homologies to Phd-like proteins (not shown). Since the locus was not further investigated, it is not clear whether this intergenic region is indeed part of a toxin/anti-toxin system or whether the Phd-like protein is similar in function or sequence to PhdA.
In E. coli, the genes encoding Phd and Doc are organized in an operon (Lehnherr et al., 1993). No Doc homologue was identified in this study. In P. aeruginosa, phdA is predicted to be encoded as a single gene operon, which is not juxtapositioned to the bacteriophage Pf4 operon. Moreover, BLAST searches did not result in the identification of a Doc homologue (not shown). However, Doc expression has been linked to rapid cell-growth arrest accompanied by inhibition of translation without significant perturbation of transcription or replication (Liu et al., 2008), a phenotype not unlike the one observed for the ΔbfmR mutant (Fig. S5, Tables S1–S2). These data suggest the presence of a yet unidentified Doc-like protein in P. aeruginosa.
Transcriptomic and proteomic analysis indicated differential expression of a number of genes and proteins associated with bacteriophage release and potential defense mechanisms in response to phage-mediated lysis. In addition to bacteriophage Pf4 related genes, helicases, recombinases, ligases, and endonucleases were significantly upregulated (Table S1), which may indicate activation of DNA repair mechanisms. Filamentous-like prophage excision has been linked to the emergence of small-colony variants (SCVs) in P. aeruginosa. While the genetic basis of the emergence of these SCVs is largely unknown, bacteriophage Pf4 has been suggested to be the genetic cause for SCV formation (Webb et al., 2004). However, it is as likely that the respective gene products assist in phage excision. Increased expression of virulence factors and genes involved in secretion/toxin delivery further suggested a response by the bacterial cells to limit the spread of bacteriophages. Moreover, the microarray analysis provided possible insight into phage propagation. Allesen-Holm et al. suggested that DNA release might occur via induction of a prophage in a few cells followed by flagella/pili-dependent phage propagation and lysis of a subpopulation of the cells (Allesen-Holm et al., 2006), with type IV pili also binding DNA (Aas et al., 2002, van Schaik et al., 2005). Here, we observed reduction of pili and flagella gene expression but increased expression of cup genes in ΔbfmR biofilms (Fig. S5), indicating that bacteriophage Pf4 may make use of the cup fimbriae rather than type IV pili and flagella in ΔbfmR biofilms to propagate.
Moreover, our findings suggested the notion of BfmR being activated upon exposure to bacteriophages and sensing signals such as membrane perturbing conditions, probably via its cognate sensor BfmS. Exposure to increasing numbers of phage particles indicated an immediate and a delayed response to bacteriophages. One can speculate that the immediate response is accomplished via the direct action of PhdA, by repressing the transcription of prophage genes and genes encoding cup fimbriae (Fig. S5). Similarly, the E. coli antitoxin YbaJ has been demonstrated to repress the transcription of two prophage gene clusters as well as the production of fimbriae (Garcia-Contreras et al., 2008). The delayed response may be the consequence of genes that indirectly result in persistence to bacteriophages (e.g. repression of genes involved in protein translation and those having chaperone functions, Fig. S5, Table 1) or alternatively, due to BfmSR-activating conditions such as exposure to eDNA considering that eDNA has been shown to result in perturbation of both the outer and inner membrane as well as cell lysis (Mulcahy et al., 2008).
In conclusion, our data suggest that BfmR plays an essential role in biofilm development by P. aeruginosa by controlling or suppressing bacteriophage-mediated lysis via the previously undescribed PhdA, a homologue of the prevent host death proteins. Activation of BfmR and BfmR-dependent suppression of bacteriophage and eDNA release appear to be dependent on biofilm architecture, since expression of bfmR was only detectable in clusters and microcolonies but not in cells attached to the substratum (Fig. 1) or planktonic cells (Fig. 6). Activation of PhdA, probably in response to sensing membrane perturbation and eDNA, thus results in fine tuning of cell lysis and DNA release soon after initial attachment by P. aeruginosa to enable biofilm formation. Once activated, BfmR, through the action of PhdA, suppresses lytic phage activity while restoring or promoting normal growth functions including transcription and translation, chaperone function, and expression of genes encoding appendages (e.g. pili). To our knowledge, this is the first demonstration of a two-component regulatory system essential for modulating the extent of cell lysis and eDNA release via regulation of bacteriophage release in biofilms. A more thorough understanding of how bacteria activate BfmSR to regulate cell lysis may be key for developing novel strategies to target biofilms.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids, media, and culture conditions
All bacterial strains and plasmids used in this study are listed in Table S3. P. aeruginosa strain PAO1 was used as the parental strain. All planktonic cultures were grown in VBMM medium containing citrate as sole carbon source (Schweizer, 1991) or in Luria-Bertani (LB) broth in shake flasks at 220 rpm. Biofilms were grown as described below at 22°C in 1/20 diluted LB or VBMM medium containing citrate as sole carbon source supplemented with arabinose.
Biofilm formation
Biofilms were grown using a once-through continuous flow tube reactor system for biofilm sample collection and in flow cells (BioSurface Technologies) for the analysis of biofilm architecture as previously described (Sauer et al., 2002, Sauer et al., 2004, Petrova & Sauer, 2009). Quantitative analysis of confocal scanning laser microscopy (CSLM) images of flow cell-grown biofilms was performed using COMSTAT (Heydorn et al., 2000).
Reporter activity
To monitor bfmR expression in biofilms, the PbfmR-gfp reporter constructs were used. Biofilms were stained using propidium iodide (PI) capable of binding either released DNA or DNA within cells with compromised membrane integrity only, and SYTO-40, a member of the SYTO family of cell-permeable nucleic acid stains that do not act exclusively as nuclear stains in live cells (Invitrogen). Specific β-galactosidase activity (U mg−1) of strains harboring the bfmR promoter reporter construct (PbfmR-lacZ) was determined using the Miller assay (Miller, 1972) essentially as previously described (Petrova & Sauer, 2010) using an extinction coefficient for o-nitrophenyl-β-galactoside cleavage at 420 nm of 4500 nl nmol−1 cm−1.
Quantification of DNA release and cell lysis
Extracellular DNA and protein concentrations of biofilms and planktonic cultures were initially assessed by measuring the absorbance at 260 and 280 nm of cell-free biofilm supernatants, collected from biofilms at indicated ages via centrifugation (5 min at 16000xg, 4°C) and subsequent filtration through a 0.2 μm filter. To quantify the amount of DNA release in wild type and mutant cells grown as biofilms, a propidium iodide (PI) DNA binding assay was used essentially as previously described (Allesen-Holm et al., 2006). To quantify the differences in cell lysis as indicated by PI staining of biofilms by confocal microscopy, biofilms were first stained with the LIVE/DEAD BacLight stain and then, separate images were acquired for SYTO 9- and PI-stained biofilm cells by using two distinct fluorescence channels. The resulting images were analyzed by COMSTAT. Moreover, using the qPCR-based assay described by Ma and Wood (Ma & Wood, 2009), the percent lysis in ΔbfmR and PAO1 biofilms was determined as a ratio of released (supernatant) to total (supernatant and cells) DNA concentrations.
RNA isolation and preparation for Affymetrix GeneChip analysis
The samples were prepared identically as previously described (Morici et al., 2007). For biofilm growth experiments, three independent replicates of P. aeruginosa strains PAO1 and ΔbfmR were grown as biofilms in a flow-through system as described above. Cells were treated with RNAprotect (Qiagen) and total RNA was extracted using an RNeasy mini purification kit (Qiagen) per the manufacturer's instructions. RNA quality and the presence of residual DNA were checked on an Agilent Bioanalyzer 2100 electrophoretic system pre- and post-DNase treatment. Ten micrograms of total RNA was used for cDNA synthesis, fragmentation, and labeling according to the Affymetrix GeneChip P. aeruginosa genome array expression analysis protocol. Briefly, random hexamers (Invitrogen) were added (final concentration, 25 ng μl−1) to the 10 μg of total RNA along with in vitro-transcribed Bacillus subtilis control spikes (as described in the Affymetrix GeneChip P. aeruginosa genome array expression analysis protocol).
cDNA was synthesized using Superscript II (Invitrogen) according to the manufacturer's instructions under the following conditions: 25°C for 10 min, 37°C for 60 min, 42°C for 60 min, and 70°C for 10 min. RNA was removed by alkaline treatment and subsequent neutralization. The cDNA was purified with use of the QIAquick PCR purification kit (Qiagen) and eluted in 40 μl of buffer EB (10 mM Tris-HCl, pH 8.5). The cDNA was fragmented by DNase I (0.6 U μg−1 of cDNA; Amersham) at 37°C for 10 min and then end labeled with biotin-ddUTP with use of the Enzo BioArray Terminal Labeling kit (Affymetrix) at 37°C for 60 min. Proper cDNA fragmentation and biotin labeling were determined by gel mobility shift assay with NeutrAvadin (Pierce) followed by electrophoresis through a 5% polyacrylamide gel and subsequent DNA staining with SYBR Green I (Roche).
Microarray data analysis
Microarray data were generated using Affymetrix protocols as previously described (Frisk et al., 2004, Lizewski et al., 2004, Morici et al., 2007). Absolute expression transcript levels were normalized for each chip by globally scaling all probe sets to a target signal intensity of 500. Three statistical algorithms (detection, change call, and signal log ratio) were then used to identify differential gene expression in experimental and control samples. The detection metric (presence, absence, or marginal) for a particular gene was determined using default parameters in MAS software (version 5.0; Affymetrix). Batch analysis was performed in MAS to make pairwise comparisons between individual experimental and control GeneChips in order to generate change calls and a signal log ratio for each transcript. These data were imported into Data Mining Tools (version 3.0; Affymetrix). Transcripts that were absent under both control and experimental conditions were eliminated from further consideration. Statistical significance of signals between the control and experimental conditions (P < 0.05) for individual transcripts was determined using the t test. We defined a positive change call as one in which greater than 50% of the transcripts had a call of increased (I) or marginally increased (MI) for up-regulated genes and decreased (D) or marginally decreased (MD) for down-regulated genes. Finally, the mean value of the signal log ratios from each comparison file was calculated. Only those genes that met the above criteria and had a mean signal log ratio of greater than or equal to 1 for up-regulated transcripts and less than of equal to 1 for down-regulated transcripts were kept in the final list of genes. Signal log ratio values were converted from log2 and expressed as fold changes. The microarray data are available on the GEO (Gene Expression Omnibus) website at http://www.ncbi.nlm.nih.gov/projects/geo (GEO accession no., pending).
ChIP-cloning
In order to identify the in vivo DNA binding sites of the response regulator BfmR, 24-hr-old biofilms of P. aeruginosa PAO1/pJN-bfmR, bearing the 6XHis/V5-tagged bfmR, were subjected to chromatin immunoprecipitation (ChIP) analysis essentially as previously described (Petrova & Sauer, 2010). Reactions containing no antibody or no 6XHis/V5-tagged BfmR were used as controls.
Isolation of bacteriophages, PFU determination, and detection of coaB in cell free biofilm supernatants by quantitative PCR (qPCR)
Cell-free biofilm supernatants collected following 48 and 96 hr of biofilm growth were used for PFU determination essentially as described by (Webb et al., 2003), by spotting dilutions in SM buffer on top agar (0.8%) seeded with P. aeruginosa cells. In addition, cell-free supernatants of cells grown planktonically to exponential and stationary phase were used. Bacteriophage stocks were prepared as previously described (Webb et al., 2003). Cell-free planktonic culture and biofilm supernatants were used as a source of DNA to quantitate the coaB and PA0636 genes before and after repeated DNase treatment by qPCR using the Eppendorf Mastercycler® ep realplex (Eppendorf AG, Hamburg, Germany). Amplification of the genes mreB, mexA, and 16SrDNA were used as control for the presence of genomic DNA. The number of gene copies was determined with standard curves established using genomic DNA as follows: NmreB = 2.718−(Ct-41.626)/1.525; NcoaB = 2.718−(Ct-43.527)/1.302.; NPA0636 = 2.718−(Ct-44.345)/1.6095.
Quantitative reverse transcriptase PCR (qRT-PCR)
Isolation of mRNA and cDNA synthesis was carried out as previously described (Petrova & Sauer, 2010). qRT-PCR was performed using the Eppendorf Mastercycler® ep realplex and the KAPA SYBR FAST qPCR Kit (KAPABIOSYSTEMS), with oligonucleotides listed in Table S4. mreB were used as controls. Relative transcript quantitation was accomplished using the ep realplex software (Eppendorf AG) by first normalizing transcript abundance (based on Ct-value) to the control genes followed by determining transcript abundance ratios. Melting curve analyses were employed to verify specific single product amplification.
Protein production analysis by 2D/PAGE and protein identification
Preparation of crude protein extracts and protein determination was carried out as previously described (Southey-Pillig et al., 2005). The resulting proteins were subsequently separated by 2D/PAGE and analyzed using the 2D ImageMaster software (GE Healthcare) (Sauer & Camper, 2001, Sauer et al., 2002). Differentially produced proteins were identified essentially as previously described using a QStarXL mass spectrometer (Applied Biosystems) (Petrova & Sauer, 2009).
Streptavidin Magnetic bead DNA Binding Assay
BfmR binding to the putative phdA promoter was confirmed using the streptavidin magnetic bead DNA binding assay as described by Chandrachud and Gal (Chandrachud & Gal, 2009) with the following modifications. Briefly, biotinylated target DNA fragment PphdA (−240 to 0 relative to translational start site, based on the sequence in identified phdA ChIP clones) was amplified using the primer pair PphdA_for/PphdA_rev (Table S4). A total of 2.5 pmol of target DNA was incubated for 30 min at room temperature with 5 pmol of purified His/V5-tagged BfmR in 25mM Tris-Cl, pH 8, 5mM MgCl2, 0.5mM dithiothreitol, 1mM EDTA, and 50ng/uL poly(dI-dC) as nonspecific competitor DNA. For specific competition, non-biotinylated target DNA (0–50 pmol) was used. Streptavidin magnetic beads (Thermo Scientific, 100 μg) were used to capture biotinylated DNA. Following three washes, the proteins co-purified with the biotinylated DNA were separated by 11% SDS/PAGE and assessed by immunoblot analysis for the presence of BfmR using anti-V5 antibodies (Invitrogen). An aliquot prior to the addition of streptavidin magnetic beads was used to determine total BfmR present in each DNA binding assay.
Statistical analysis
A Student's t-test was performed for pair-wise comparisons of groups, and multivariant analyses were performed using a 1-Way ANOVA followed by a post-priori test using Sigma Stat software. All experiments were carried out at least in triplicate.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by a grant from the National Institutes of Health (2R15 HL073835-02).
REFERENCES
- Aas FE, Wolfgang M, Frye S, Dunham S, Løvold C, Koomey M. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Molecular Microbiology. 2002;46:749–760. doi: 10.1046/j.1365-2958.2002.03193.x. [DOI] [PubMed] [Google Scholar]
- Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, Givskov M, Whitchurch CB, Engel JN, Tolker-Nielsen T. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- Chandrachud U, Gal S. Three assays show differences in binding of wild-type and mutant p53 to unique gene sequences. Technol Cancer Res Treat. 2009;8:445–453. doi: 10.1177/153303460900800606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annual Review of Microbiology. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H-C, Neu TR, Wozniak DJ. The EPS matrix: the “house of biofilm cells”. J. Bacteriol. 2007;189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J, Griegbe T, Mayer C. Physicochemical properties of biofilms. In: Evans LV, editor. Biofilms: Recent advances in their study and control. Harwood Academic Publishers; Amsterdam: 2000. pp. 19–34. [Google Scholar]
- Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- Frisk A, Schurr JR, Wang G, Bertucci DC, Marrero L, Hwang SH, Hassett DJ, Schurr MJ. Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect. Immun. 2004;72:5433–5438. doi: 10.1128/IAI.72.9.5433-5438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Contreras R, Zhang X-S, Kim Y, Wood TK. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS ONE. 2008;3:e2394. doi: 10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeke J, Paul K, Lassak J, Thormann KM. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 2010 doi: 10.1038/ismej.2010.153. In Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 2008;74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FKN, Dudas KC, Hanson JA, Nelson MB, LoVerde PT, Apicella MA. The R-type pyocin of Pseudomonas aeruginosa C is a bacteriophage tail-like particle that contains single-stranded DNA. Infect. Immun. 1999;67:717–725. doi: 10.1128/iai.67.2.717-725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnherr H, Maguin E, Jafri S, Yarmolinsky MB. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. Journal of Molecular Biology. 1993;233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- Lehnherr H, Yarmolinsky MB. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang Y, Inouye M, Woychik NA. Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. Proceedings of the National Academy of Sciences. 2008;105:5885–5890. doi: 10.1073/pnas.0711949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizewski SE, Schurr JR, Jackson DW, Frisk A, Carterson AJ, Schurr MJ. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J. Bacteriol. 2004;186:5672–5684. doi: 10.1128/JB.186.17.5672-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Wood TK. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ Microbiol. 2009;11:2735–2746. doi: 10.1111/j.1462-2920.2009.02000.x. [DOI] [PubMed] [Google Scholar]
- Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends in Microbiology. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2004;186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor; N.Y.: 1972. pp. 352–355. [Google Scholar]
- Morici LA, Carterson AJ, Wagner VE, Frisk A, Schurr JR, zu Bentrup KH, Hassett DJ, Iglewski BH, Sauer K, Schurr MJ. Pseudomonas aeruginosa AlgR represses the Rhl quorum-sensing system in a biofilm-specific manner. J. Bacteriol. 2007;189:7752–7764. doi: 10.1128/JB.01797-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto K, Hirota K, Murakami K, Taniguti K, Murata H, Viducic D, Miyake Y. Effect of Varidase (streptodornase) on biofilm formed by Pseudomonas aeruginosa. Chemotherapy. 2003;49:121–125. doi: 10.1159/000070617. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GSA, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, Dodson RJ, Durkin AS, Brinkac LM, Daugherty SC, Sullivan SA, Rosovitz MJ, Gwinn ML, Zhou L, Schneider DJ, Cartinhour SW, Nelson WC, Weidman J, Watkins K, Tran K, Khouri H, Pierson EA, Pierson LS, Thomashow LS, Loper JE. Complete genome. 2005 [Google Scholar]
- Petrova OE, Sauer K. Pseudomonas aeruginosa biofilm development. PLoS Pathogens. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. Journal of Bacteriology. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, Hauser A, McDougald D, Webb JS, Kjelleberg S. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. Isme J. 2009;3:271–282. doi: 10.1038/ismej.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Current Opinion in Microbiology. 2007;10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 2001;183:6579–6589. doi: 10.1128/JB.183.22.6579-6589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer HP. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa. J. Bacteriol. 1991;173:6798–6806. doi: 10.1128/jb.173.21.6798-6806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey-Pillig CJ, Davies DG, Sauer K. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2005;187:8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering AL, Gilmore MS. Quorum sensing and DNA release in bacterial biofilms. Current Opinion in Microbiology. 2006;9:133–137. doi: 10.1016/j.mib.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Steinberger RE, Holden PA. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 2005;71:5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. The Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Sutherland IW. The biofilm matrix - an immobilized but dynamic microbial environment. Trends in Microbiology. 2001;9:222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Molecular Microbiology. 2009;72:1022–1036. doi: 10.1111/j.1365-2958.2009.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik EJ, Giltner CL, Audette GF, Keizer DW, Bautista DL, Slupsky CM, Sykes BD, Irvin RT. DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J. Bacteriol. 2005;187:1455–1464. doi: 10.1128/JB.187.4.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Wood TK. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J. 2009;3:1164–1179. doi: 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Lau M, Kjelleberg S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2004;186:8066–8073. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2003;185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology. 2007;153:1318–1328. doi: 10.1099/mic.0.2006/004911-0. [DOI] [PubMed] [Google Scholar]
- Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 2009;191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.