Abstract
Many flaviviruses are significant human pathogens. No effective antiviral therapy is currently available for treatment of flavivirus infections. Development of antiviral treatment against these viruses is urgently needed. The flavivirus methyltransferase (MTase) responsible for N-7 and 2'-O methylation of the viral RNA cap has recently been mapped to the N-terminal region of nonstructural protein 5. Structural and functional studies suggest that the MTase represents a novel antiviral target. Here we review current understanding of flavivirus RNA cap methylation and its implications for development of antivirals. The 5' end of the flavivirus plus-strand RNA genome contains a type 1 cap structure (m7GpppAmG). Flaviviruses encode a single MTase domain that catalyzes two sequential methylations of the viral RNA cap, GpppA-RNA→m7GpppA-RNA→m7GpppAm-RNA, using S-adenosyl-L-methionine (SAM) as the methyl donor. The two reactions require different viral RNA elements and distinct biochemical assay conditions. Despite exhibiting two distinct methylation activities, flavivirus MTase contains a single binding site for SAM in its crystal structure. Therefore, substrate GpppA-RNA must be re-positioned to accept the N-7 and 2'-O methyl groups from SAM during the two methylation reactions. Structure-guided mutagenesis studies indeed revealed two distinct sets of amino acids on the enzyme surface that are specifically required for N-7 and 2'-O methylation. In the context of virus, West Nile viruses (WNV) defective in N-7 methylation are non-replicative; however, WNVs defective in 2'-O methylation are attenuated and can protect mice from subsequent wild-type WNV challenge. Collectively, the results demonstrate that the N-7 MTase represents a novel target for flavivirus therapy.
Keywords: Flavivirus NS5, Methyltransferase, Flavivirus replication, Antiviral therapy, West Nile virus, dengue virus, yellow fever virus, Japanese encephalitis virus, tick-borne encephalitis virus
1. Introduction
Many members of the genus Flavivirus in the family Flaviviridae are arthropod-borne, and cause significant human disease (Gubler et al., 2007). The four serotypes of dengue virus (DENV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), West Nile virus (WNV), and tick-borne encephalitis virus (TBEV) are all emerging or reemerging pathogens. Vaccines for humans are currently available only for YFV, JEV, and TBEV; no clinically approved vaccine or antiviral therapy for humans is available for WNV or DENV (Kramer et al., 2007, Gould et al., 2008, Leyssen et al., 2008, Monath, 2008). Development of a vaccine for DENV has been challenging, principally because of the need to simultaneously immunize and induce long-lasting protection against all four DENV serotypes; an incomplete immunization of the four serotypes may give rise to dengue hemorrhagic fever or dengue shock syndrome, upon subsequent natural DENV infection. Furthermore, due to the sporadic nature of outbreaks of many flaviviruses (such as WNV), it may not be cost-effective to vaccinate the general population, even if vaccines become available. These complications have underscored the importance of developing effective therapies for flaviviral diseases. The goal of this review is to highlight recent progress in understanding flavivirus RNA cap methylation. The advances clearly suggest that flavivirus methyltransferase (MTase) is a valid antiviral target.
2. Flavivirus RNA cap formation
The flaviviral genome is a single-stranded, plus-sense RNA approximately 11 kb in length. It consists of a 5'-untranslated region (UTR), a single long open reading frame, and a 3'-UTR (Fig. 1A). The single open reading frame encodes three structural proteins (capsid, premembrane or membrane, and envelope) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Lindenbach et al., 2007). Two of the ten viral proteins have enzymatic activities (Fig. 1A). NS3 acts as a serine protease (with NS2b as a cofactor), 5'-RNA triphosphatase (RTPase), nucleoside triphosphatase (NTPase), and helicase (Wengler and Wengler, 1991, Warrener et al., 1993, Wengler and Wengler, 1993, Li et al., 1999, Bartelma and Padmanabhan, 2002, Benarroch et al., 2004). NS5 functions as an RNA-dependent RNA polymerase (RdRp) (Tan et al., 1996, Ackermann and Padmanabhan, 2001, Guyatt et al., 2001) and a MTase involved in methylation of the 5'-cap structure of genomic RNA (Koonin, 1993, Egloff et al., 2002, Ray et al., 2006, Zhou et al., 2007). Flavivirus replication occurs on the cellular endoplasmic reticulum membrane without viral entry into the nucleus, although some viral proteins can be detected in the nucleus in infected cells (Lindenbach et al., 2007).
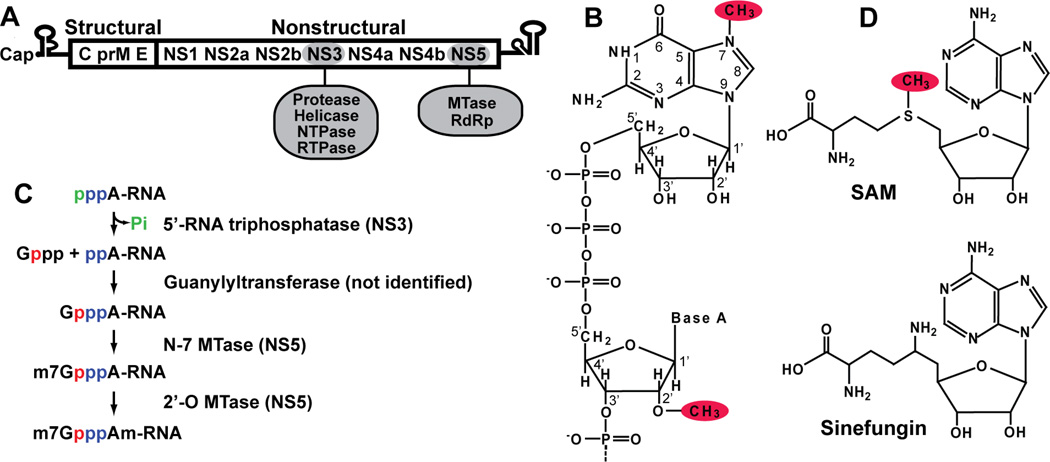
FIG. 1. Flavivirus genome and 5' cap formation.
(A) Flavivirus genome structure. Flavivirus genomic RNA consists of a 5'-UTR, a single open reading frame (ORF), and a 3'-UTR. The single ORF encodes three structural and seven nonstructural proteins. The enzymatic activities of NS3 and NS5 are indicated. (B) Flavivirus 5' cap structure. The cap structure is formed by connection of a guanosine to the first nucleotide adenosine of RNA through a 5'-to-5' triphosphate bridge. The GpppA cap is methylated at the guanine N-7 position, and the first nucleotide adenosine is methylated at the ribose 2'-O position. The N-7 and 2'-O methyl groups are shaded in red. (C) Flavivirus 5' cap formation. Four enzymatic modifications are required for cap formation. The 5'-RNA triphosphatase has been mapped to the viral NS3, while the N-7 and 2'-O MTase activities are performed by NS5; the guanylyltransferase remains to be identified. Phosphates from different molecules are colored individually to indicate their sources. (D) Structures of S-adenosyl-L-methionine (SAM) and sinefungin.
The 5' end of the flavivirus genome contains a type 1 cap structure, followed by the conserved dinucleotide sequence AG (m7GpppAmG; Fig. 1B) (Cleaves and Dubin, 1979, Wengler, 1981). The cap structure of most eukaryotic and viral mRNAs is critical for mRNA stability and efficient translation (Furuichi and Shatkin, 2000). In general, RNA capping consists of four enzymatic modifications (Fig. 1C): (i) the 5'-triphosphate end of the nascent RNA transcript is hydrolyzed to a 5'-diphosphate by an RNA triphosphatase; (ii) the GMP moiety of GTP is transferred to the 5'-diphosphate of RNA by an RNA guanylyltransferase; (iii) the N-7 position of guanine is methylated by an RNA (guanine-N(7))-MTase (N-7 MTase); and (iv) the first nucleotide of the RNA transcript is further methylated at the ribose 2'-OH position by a RNA (nucleoside-2'-O)-MTase (2’-O MTase), resulting in a type 1 cap (m7GpppAm) (Fig. 1C). S-adenosyl-L-methionine (SAM; Fig. 1D) is the methyl donor for both the N-7 and 2'-O methylations, generating S-adenosyl-L-homocysteine (SAH) as a by-product.
Since host mRNA capping occurs in the nucleus, viruses that replicate in the cytoplasm encode their own RNA capping machineries. For flaviviral cap formation, viral NS3 functions as an RNA triphosphatase (Wengler and Wengler, 1993), and NS5 functions as both a N-7 MTase and a 2'-O MTase (Egloff et al., 2002, Ray et al., 2006); the guanylyltransferase, however, remains to be identified. The capping mechanisms of some viral mRNAs are different from those of the host mRNA (Furuichi and Shatkin, 2000). For example, the minus-strand RNA vesicular stomatitis virus (VSV) transfers GDP, rather than GMP, to the 5'-monophosphate of the RNA (Abraham et al., 1975). The plus-strand RNA alphaviruses methylate GTP prior to the transfer of m7GMP to the 5'-diphosphate of the RNA (Ahola and Kaariainen, 1995). The differences between the host and viral cap formation processes can potentially be exploited for development of antiviral therapy.
3. Identification of NS5-mediated N-7 and 2'-O methylations of flavivirus RNA cap
The flavivirus MTase resides in the N-terminal one-third of NS5, while the C-terminal two-thirds of NS5 contain an RNA-dependent RNA polymerase (RdRp). The NS5 MTase domain was initially predicted by a sequence alignment (Koonin, 1993). Using short RNA GpppA(C)5 or m7GpppA(C)5 as substrates, Egloff and co-workers (2002) first showed that recombinant DENV-2 MTase (containing the N-terminal 296 amino acids of NS5) has a 2'-O methylation activity. The 2'-O methylation activity was later confirmed using recombinant MTase and full-length NS5 of WNV. In addition to the 2'-O methylation, the WNV MTase domain can perform N-7 cap methylation. Furthermore, the two cap methylation events are sequential, in the order GpppA-RNA→m7GpppA-RNA→m7GpppAm-RNA (Ray et al., 2006). Using authentic RNA substrates representing the first 190 nucleotides of the flavivirus genome, Dong and co-workers (2007) showed that NS5 MTases from four serocomplexes of flaviviruses (DENV-1, YFV, and WNV, plus Powassan virus as a TBEV representative) also perform N-7 and 2'-O cap methylations sequentially. The discrepancy between the results of the earlier (Egloff et al., 2002) and the later studies (Ray et al., 2006, Dong et al., 2007, Zhou et al., 2007) could have arisen from differences in the assay conditions, including buffer components and RNA substrates. As detailed in the following two sections, the buffer conditions (especially the pH) and the RNA substrate (sequence and structure) are critical for detection of robust flavivirus RNA cap methylations.
3.1. Distinct assay conditions for optimization of flavivirus N-7 and 2'-O methylation
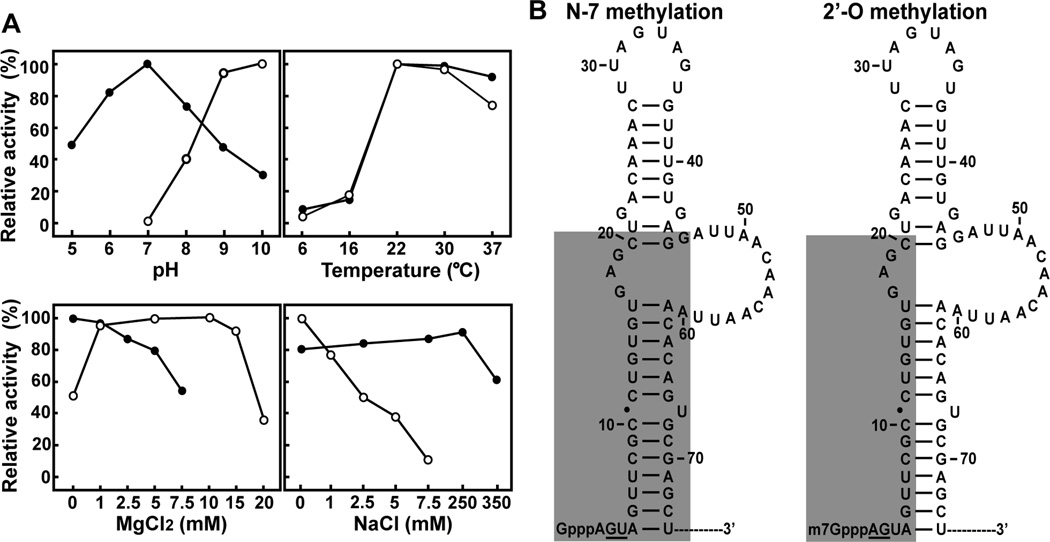
Optimization of assay conditions using WNV MTase showed that N-7 and 2'-O cap methylations require distinct assay parameters (Fig. 2A). Both activities reached maximum when performed at 22°C; however, the optimal pH and the concentrations of MgCl2 and NaCl were different (Zhou et al., 2007). N-7 methylation required neutral pH (pH 7.0) and 50–100 mM NaCl; MgCl2 inhibited the activity. In contrast, 2'-O methylation required high pH (pH 10) and 5–10 mM MgCl2; NaCl inhibited the activity. Similar results were obtained for the DENV-1 and YFV MTases. These data demonstrate that optimal N-7 and 2'-O methylations of flavivirus cap require distinct biochemical conditions, suggesting that the two methylations occur through different mechanisms. The distinct pH requirements for the N-7 and 2'-O methylations give us an experimentally useful means to isolate the two reactions. Specifically, the requirement of a high pH for the 2'-O methylation allows us to selectively perform the N-7 methylation of GpppA-RNA→m7GpppA-RNA, without the subsequent 2'-O methylation, by incubating the reactions in a neutral pH buffer. The 2'-O methylation of m7GpppA-RNA→m7GpppAm-RNA can be efficiently accomplished if the reaction is performed in a high-pH buffer.
FIG. 2. Optimal assay conditions and minimal RNA requirement for two WNV cap methylations.
(A) Optimal assay conditions for the N-7 (●) and 2'-O (○)methylations. Activities were measured on radioactively labelled substrates. The N-7 and 2'-O methylations achieve conversion of G*pppA-RNA→m7G*pppA-RNA and m7G*pppA-RNA→m7G*pppAm-RNA, respectively. Symbol * indicates that the following phosphate is 32P-labeled. The optimal conditions were determined by individual titrations of pH, temperature, MgCl2 and NaCl, while other parameters were held constant at optimal levels. The reaction mixtures were then treated with nuclease P1, analyzed on thin layer chromatography plates, and quantified by PhosphorImager. For each parameter, relative activities are presented with the optimal level set at 100%. For details, see (Zhou et al., 2007). (B) Minimal RNA requirement for WNV cap methylation. The stem-loop structure formed by the 5'-terminal 74 nucleotides of the WNV genome was predicted by the Mfold program. The shaded regions are important for N-7 (left) and 2'-O (right) methylation. Essential wild-type nucleotides are underlined.
3.2. Requirement of viral RNA for flavivirus cap methylations
The specific requirement for viral RNA elements for methylation of the flavivirus cap was initially made clear by the finding that WNV MTase can methylate the cap of the viral RNA, but not the cap of a plasmid-derived RNA of the same length (Ray et al., 2006). Systematic mutagenesis of the 5'-stem-loop structure in the WNV genome showed that distinct RNA elements are required for the N-7 and the 2'-O methylations (Fig. 2B). The N-7 methylation requires wild-type nucleotides at the 2nd (G) and 3rd (U) positions, and a 5'-stem-loop structure; in contrast, 2'-O methylation requires wild-type nucleotides at the 1st (A) and 2nd (G) positions, and a minimum of 20 nucleotides of viral RNA. Cap analogues, GpppA and m7GpppA, are not active substrates for WNV MTase. Footprinting experiments using GpppA-RNA and m7GpppA-RNA suggested that the 5' termini of RNA substrates interact with NS5 during both methylation reactions. An antisense oligomer with a sequence complementary to the first 20 nucleotides of the WNV genome can inhibit WNV cap methylation (Dong et al., 2007).
3.3. A K-D-K-E motif as an active site for 2'-O methylation
A K-D-K-E tetrad is conserved among various RNA MTases, including flavivirus MTases (Egloff et al., 2002). The tetrad residues are distant from one another in the primary sequence, but clustered together in the three-dimensional structures. The K-D-K-E motif was previously shown to be the active site for the 2'-O MTases of vaccinia virus VP39 and RrmJ (a heat shock-induced 2'-O MTase) (Schnierle et al., 1994, Hager et al., 2002). Mutagenesis of the K61-D146-K182-E218 tetrad (depicted on the right panel in Fig. 3C) of WNV MTase showed that the enzyme requires distinct amino acids for the N-7 and 2'-O methylations. The entire K61-D146-K182-E218 motif is essential for 2'-O methylation, whereas N-7 methylation requires only D146. The other three amino acids of the tetrad facilitate, but are not essential for, N-7 methylation (Ray et al., 2006). Structural alignment of the WNV MTase with the tertiary complex of vaccinia virus VP39 (MTase, short RNA with cap, and SAH) showed that the K-D-K-E tetrad is nearly superimposable between one enzyme and the other, suggesting, by analogy, that K182 within the WNV tetrad directly participates in the deprotonation of ribose 2'-OH (Egloff et al., 2002, Zhou et al., 2007). Since the pKa of Lys is at pH 10, a high pH (optimal for 2'-O methylation; Fig. 2A) would facilitate the deprotonation of K182. Such deprotonation would in turn favor the deprotonation of the ribose 2'-OH, leading to formation of the SN2-like transition state to accomplish the methyl transfer (Hodel et al., 1998).
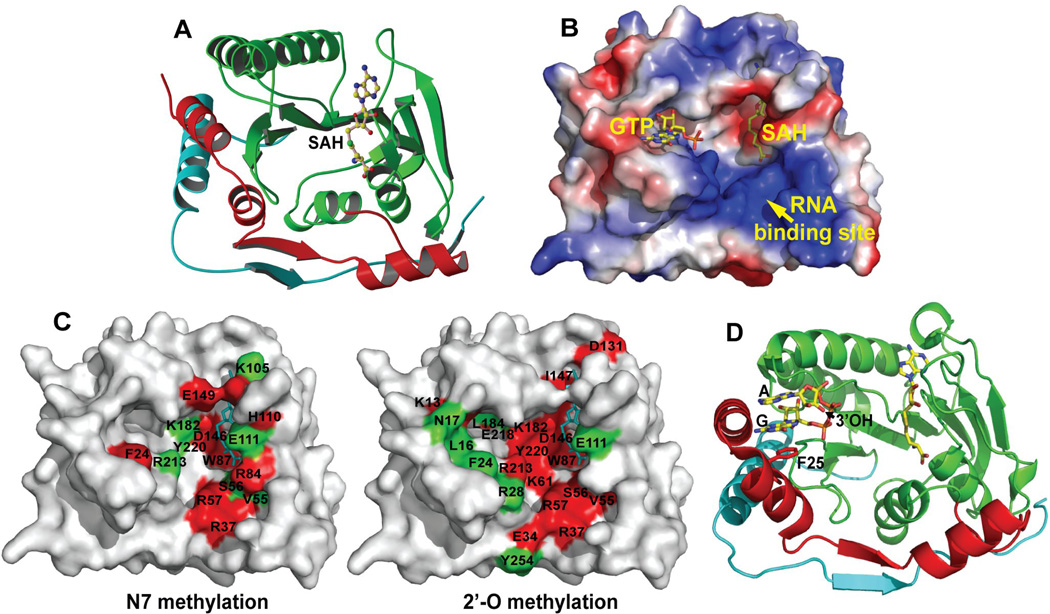
FIG. 3. Structure of the flavivirus MTase.
(A) Ribbon representation of the crystal structure of the WNV MTase. Three domains of the MTase structure are colored: N-terminal domain, red; MTase core, green; and C-terminal domain, cyan. The bound SAH molecule is shown in a ball-and-stick representation, with atom colors as follow: carbon, yellow; oxygen, red; nitrogen, blue; and sulfur, green. (B) Three flavivirus-conserved binding sites for SAH/SAM, GTP, and RNA. The binding sites are shown on the surface of WNV MTase, which in this panel is depicted as an electrostatic potential map: positively charged residues in blue, negatively charged in red, and neutral in white. The SAH/SAM-binding pocket is identified based on the co-crystal structure of the WNV MTase with SAH. The GTP-binding site of the WNV MTase was modeled through structural alignment with the DENV-2 GTP-SAH-MTase tertiary complex using the PyMOL program. The RNA-binding site is indicated by the positively charged residues. (C) Amino acids important for N-7 methylation (left panel) and 2'-O methylation (right panel) of the WNV MTase. Residues whose mutations reduce activity to a level <20% of wild-type are labeled in red; residues whose mutations reduce activity to a level 20%–60% of wild-type are indicated in green. SAH is shown in a stick presentation, and colored in cyan. (D) Crystal structure of DENV-2 MTase-SAH-GpppA tertiary complex. The guanine cap is sandwiched between the aromatic ring of F25 and the adenosine of GpppA in an S2 conformation, so that the ribose 3' hydroxyl of the adenosine points toward the RNA-binding site. For details, see text and also (Egloff et al., 2007).
In contrast to the residues involved in 2'-O methylation, the residues of the N-7 active site remain to be determined. Although several amino acids have been identified as critical for N-7 methylation (including D146 of the K-D-K-E motif, and others residues described below), the N-7 methylation could use a different mechanism from that of the 2'-O methylation. The structure of the Encephalitozoon cuniculi (Ecm1) N-7 MTase-substrate complex suggests that catalysis of N-7 methylation is achieved via close proximity of the methyl donor and acceptor, without the need for direct contact between the enzyme and either the attacking nucleophile N-7 atom of guanine, the methyl carbon of SAM, or the leaving group sulfur of SAH (Fabrega et al., 2004). More biochemical and structural studies are required before we can fully understand the mechanism of flavivirus N-7 cap methylation.
4. Structure and function of flavivirus MTase
Crystal structures of MTases from a number of flaviviruses have been solved, including DENV-2 [PDB code 1L9K (Egloff et al., 2002); PDB code 2P3L (Egloff et al., 2007)], WNV [PDB code 2OY0 (Zhou et al., 2007)], Murray Valley encephalitis virus (MVEV) [PDB code 2PXC (Assenberg et al., 2007)], and Meaban virus [PDB code 2OXT (Mastrangelo et al., 2007)]. All known flavivirus MTases exhibit a conserved structure consisting of three motifs (Fig. 3A): an N-terminal domain (red), a core domain that shares a structural fold with nearly all SAM-dependent MTases (green), and a C-terminal domain (cyan). Three distinct binding sites have been identified on the enzyme surface (Fig. 3B).
4.1. SAH/SAM-binding site
Each flavivirus MTase contains a single SAH molecule. Crystals of most flavivirus MTases contained the SAH molecule that co-purifies with the enzyme. The SAH-binding site is assumed to serve as the SAM-binding pocket during methylation reactions. If we are to reconcile the single SAM-binding site with the two methylation activities that are observed in biochemical assays, we must posit that the 5' terminal cap becomes repositioned between one methylation reaction and the next. Indeed, mutagenesis of WNV MTase showed that mutations within the SAH-binding site impair both the N- and the 2'-O methylations. Furthermore, sinefungin (Fig. 1D), a SAM analogue in which the methyl group of SAM is replaced by an amino group and the sulphur atom was substituted with a carbon atom, inhibits both N-7 and 2'-O methylations (IC50 = 14 µM) through a mechanism of competitive binding to the SAM-binding site of the MTase (Dong et al., 2008b). Consistent with the biochemical results, viral titer reduction assays showed that sinefungin could suppress WNV replication in cell culture (EC50 = 27 µM; CC50 = 4.5 mM; TI = 167) (Dong et al., 2008b). However, selection of sinefungin-resistant virus is required to confirm whether the reduction in viral titer is truly due to the sinefungin-mediated suppression of cap methylation.
Virtual screening, using the DENV-2 MTase structure, recently revealed a compound that inhibited DENV 2'-O cap methylation (IC50 = 60 µM) (Luzhkov et al., 2007). The study used SAM as a starting structure, in a search for analogues that could specifically dock into the SAM-binding pocket of the DENV-2 MTase. Since the SAM molecule (bound in the same SAH-binding pocket) donates methyl groups to both N-7 and 2'-O positions during cap methylations, the compound identified in the search is predicted to inhibit N-7 methylation. Given that the flavivirus MTases are highly conserved in structure and sequence, this compound is further expected to inhibit other flaviviruses MTases. However, experiments are needed to verify these speculations and to demonstrate the potency of the compound in a viral infection assay. Finally, it should be pointed out that, because the SAH can compete with SAM in binding to enzymes, the co-purifying SAH serves to lower the methylation activity of flavivirus MTases, making it difficult to perform enzyme kinetics.
4.2. GTP-binding site
A GTP-binding pocket was first identified when crystals of the DENV-2 MTase were soaked with GTP (Fig. 3B). The GTP-binding site is conserved among flavivirus MTases. Functional analysis of WNV MTase showed that, except for F24 (equivalent to F25 in DENV-2 MTase; the aromatic ring of Phe forms a stacking interaction with the guanine moiety of GTP), residues within the GTP-binding pocket are selectively important for 2'-O methylation, but not for N-7 methylation. The GTP molecule preferentially inhibits 2'-O methylation rather than N-7 methylation (Dong et al., 2008b). GTP was also shown to inhibit binding of small RNAs with methylated caps, which function as substrates of 2'-O methylation (Egloff et al., 2007). Ribavirin 5'-triphosphate (a GTP analogue) was found to inhibit 2'-O methylation of DENV-2 MTase; furthermore, analysis of the co-crystal structure demonstrated that ribavirin 5'-triphosphate binds to the MTase through the GTP-binding site (Benarroch et al., 2004). The latter results suggest that, besides acting to deplete intracellular GTP pools (Leyssen et al., 2005) and as a mutagen to induce error catastrophe of a viral population (Crotty et al., 2000, Day et al., 2005), ribavirin also exerts its antiviral activity through suppression of cap formation. However, based on the report by Leyssen and colleagues (2005), it seems unlikely that inhibition of cap formation plays a significant role in suppression of viral replication by ribavirin. Collectively, the results support the hypothesis that the GTP-binding site is critical in positioning the m7Gppp moiety of RNA in such a way that the 2'-OH of the ribose of the first transcribed adenosine is presented to SAM during 2'-O methylation (Egloff et al., 2007).
4.3. RNA-binding site
Electrostatic potential analysis of flavivirus MTase structures identifies a putative RNA-binding site which is mainly formed by a number of conserved, positively charged residues (Fig. 3B). Mutagenesis of WNV MTase showed that distinct amino acids within the putative RNA-binding site are required for N-7 and 2'-O methylation (Fig. 3C). The mutational effects of individual residues on the two methylation events can be categorized into four groups: (i) amino acids that are critical for both methylation activities (i.e., R37 and R57); (ii) residues that are important only for N-7 methylation activity (i.e., R84); and (iii) amino acids that are important only for 2'-O methylation activity (i.e., E34 and Y254); and (iv) residues not important for either methylation activity (Dong et al., 2008b).
Direct analysis of RNA-MTase binding has been challenging, because the RNA is positioned on the enzyme through multiple contact sites, including the cap structure and the downstream nucleotides, as suggested by footprinting results (Dong et al., 2007). Since cap analogues are not active substrates for methylation, it remains to be determined whether the binding position of a cap analogue alone would represent the cap conformation in the context of viral RNA. In a gel-shift assay, MTase was found to bind tightly to viral RNA; mutations within the RNA-binding site (which abolished both methylation activities) did not produce detectable differences in RNA-binding activity (Dong et al., 2008b). However, addition of viral sequence to a short RNA (GpppA followed by 2 to 4 nucleotides) was found to enhance the short RNA's binding to DENV-2 MTase (Egloff et al., 2007). The latter result again suggests that viral RNA elements specifically contribute to the RNA-MTase interaction.
5. A molecular repositioning model for flavivirus RNA cap methylation
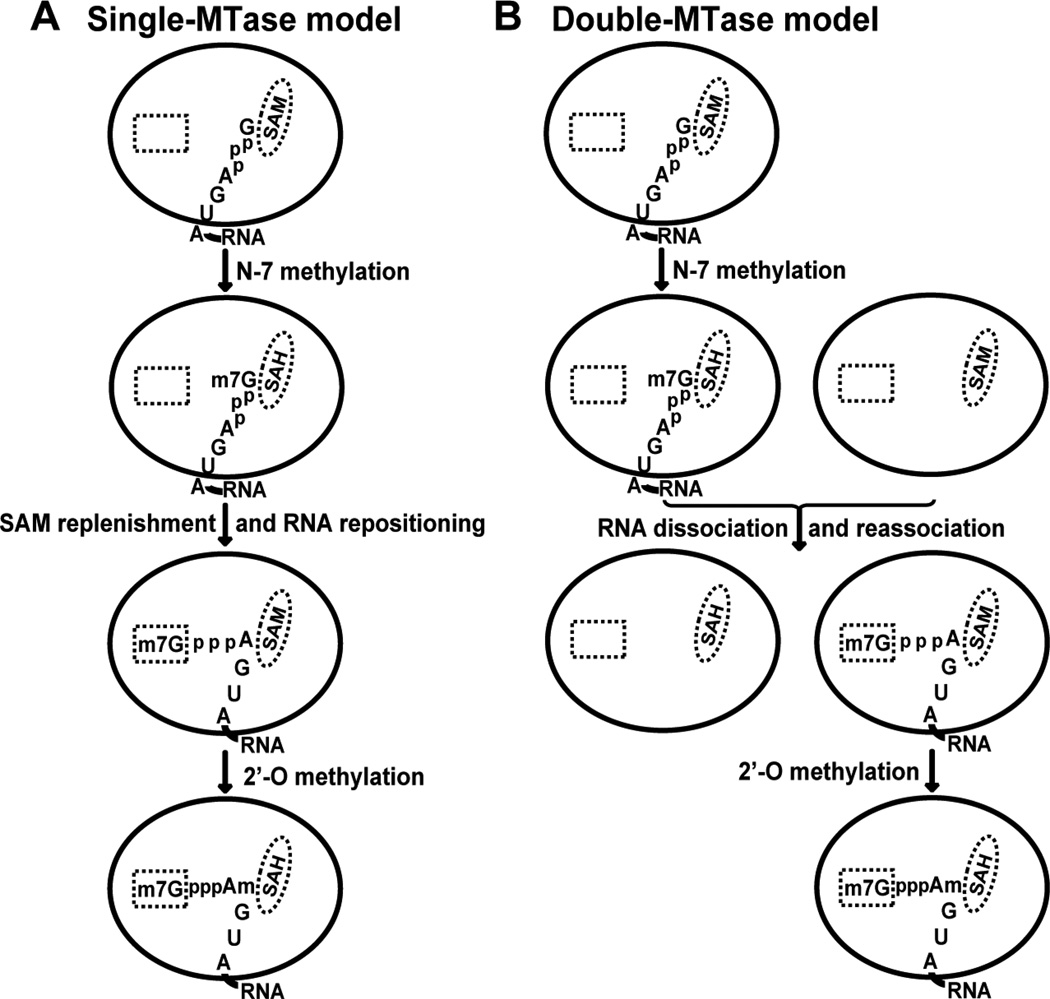
Figure 3C summarizes the mutagenesis results for WNV MTase, including the K-D-K-E motif and SAM-, GTP-, and RNA-binding sites. Two distinct sets of amino acids on the enzyme's surface are required for N-7 and 2'-O cap methylation. In general, residues critical for 2'-O methylation are more dispersed across the MTase surface than are those important for N-7 methylation. Furthermore, the number of residues critically involved in 2'-O methylation is larger than those involved in the N-7 methylation. These results suggest that the flavivirus MTase catalyzes two cap methylations through a substrate-repositioning mechanism. In this mechanism, guanine N-7 of substrate GpppA-RNA is first positioned to SAM to generate m7GpppA-RNA, after which the m7G moiety is repositioned to the GTP-binding pocket, so as to register the 2'-OH of the adenosine with SAM, generating m7GpppAm-RNA (Fig. 4). It is currently not known how the 5' cap of flavivirus RNA is translocated. Experiments are needed to determine whether one or two MTase molecules are involved in the two methylation reactions. If a single MTase catalyzes both methylations, the SAH should be replenished with a fresh SAM molecule after the N-7 methylation. It is possible that, upon N-7 methylation, repositioning of the m7G moiety of the m7GpppA-RNA to the GTP-binding site is coordinated with the SAM replenishment step (single-MTase model; Fig. 4A). Alternatively, upon N-7 methylation, the m7GpppA-RNA could dissociate from the SAH-bound MTase and reassociate with a new SAM-bound MTase for 2'-O methylation (double-MTase model; Fig. 4B). The double-MTase model is similar to the process of reovirus RNA cap formation, in which the N-7 and 2'-O methylations are carried out sequentially by two separate MTase domains of the λ2 protein (Reinisch et al., 2000, Sutton et al., 2007).
FIG. 4. Molecular repositioning between sequential flavivirus cap methylations.
(A) Single-MTase model. The guanine N-7 is first positioned in proximity to the methyl group of SAM, to generate m7GpppA-RNA. Once the guanine N-7 has been methylated, SAH is replaced by SAM, and the 5' terminus of m7GpppA-RNA is repositioned into the GTP-binding pocket. As a result, the ribose 2'-OH of the first transcribed adenosine is precisely registered with SAM, to generate m7GpppAm-RNA. (B) Double-MTase model. Once the guanine N-7 has been methylated, the m7GpppA-RNA molecule is dissociated from the MTase and reassociates with a second MTase molecule, which positions the m7Gppp to the GTP-binding pocket and generates m7GpppAm-RNA. The SAM- and GTP-binding sites are represented by an oval and rectangle, respectively. The 5'-terminal nucleotides of the WNV genome, GpppAGUA-RNA, are shown.
Using distinct mutant MTases of WNV, Dong and colleagues recently showed that the two methylation events are independent, and can complement each other (Dong et al., 2008a). For N-7 methylation, the conversion of GpppA-RNA→m7GpppA-RNA and the conversion of GpppAm-RNA→m7GpppAm-RNA are equally efficient. In contrast, for 2'-O methylation, the conversion of GpppA-RNA→GpppAm-RNA is much less efficient than the conversion of m7GpppA-RNA→m7GpppAm-RNA. The latter results indicate that 2'-O methylation prefers the substrate m7GpppA-RNA to GpppA-RNA, thereby determining the dominant methylation pathway as GpppA-RNA→m7GpppA-RNA→m7GpppAm-RNA. In addition, the authors showed that mutant enzymes with different methylation defects can trans complement one another in vitro. Furthermore, sequential treatment of GpppA-RNA with distinct methyltransferase mutants generates fully methylated m7GpppAm-RNA, demonstrating that separate molecules of the enzyme can independently catalyze the two cap methylations in vitro.
6. Co-crystal structures of flavivirus MTases in complex with cap analogues
Besides structures of DENV-2 MTase-SAH-GTP (Egloff et al., 2002) and MTase-SAH-ribavirin 5'-triphosphate (Benarroch et al., 2004), structures of DENV-2 MTase-SAH-cap analogues have recently been reported (Egloff et al., 2007). Interestingly, the guanosine moiety of the cap analogues, including GpppA, m7GpppA, GpppG, m7GpppG, and m7GpppGm, is always bound at the GTP-binding site. However, the triphosphates and the following nucleotide can adopt one of three distinct conformations. The first conformation (termed the S1 conformation) was found in complexes with N-7 methylated cap analogues (i.e., m7GpppA, m7GpppG, and m7GpppGm). In these complexes, the m7G is sandwiched between aromatic residue F25 and the second base moiety of the cap analogue, and the triphosphate linker adopts a U shape. The 3'-hydroxyl group of the second nucleotide in S1 conformation points toward the protein and, therefore, may not represent a biologically functional conformation.
The second conformation of the cap analogue (termed the S2 conformation) is adopted by GpppA (Fig. 3D). The S1 and S2 conformations of cap analogues are similar except that the second base of the cap analogue is flipped, so that the 3'-hydroxyl of the adenosine ribose in S2 conformation points down to the RNA-binding site. It was hypothesized that the S2 conformation of GpppA mimics the reaction product of a putative flavivirus guanylyltransferase activity, i.e., pppG + ppA-RNA → GpppA-RNA (Egloff et al., 2007). Experiments are needed to demonstrate such activity. The third conformation is found for the GpppG analogue. The GpppG molecule is in an extended conformation, in which the triphosphates point toward the SAH-binding pocket (similar to the GTP conformation observed in the MTase-GTP-SAH complex).
Besides the tertiary structures of DENV-2 MTase-SAH-cap analogues discussed above, crystal structures of MTase-SAH-cap analogues of MVEV were also reported (Assenberg et al., 2007). However, the GpppA dimer does not seem to be a biologically functional conformation in the MVEV MTase-SAH-cap complex.
7. Functions of MTase in flavivirus replication
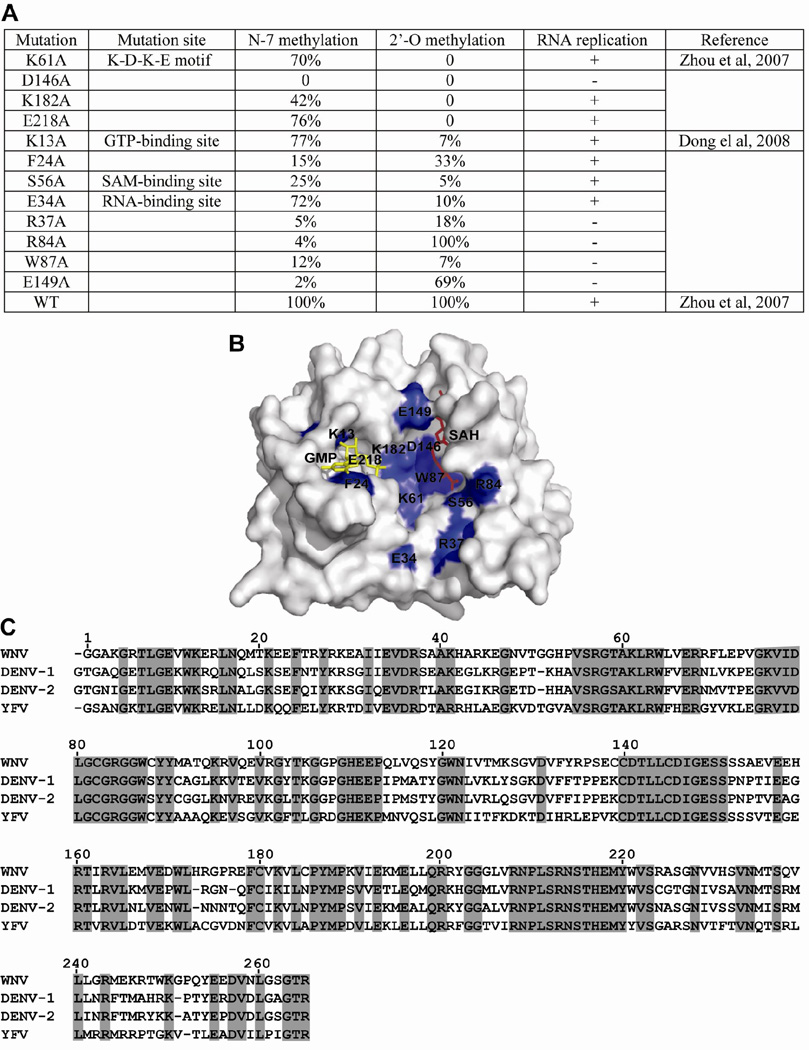
The function of cap methylation in viral replication was analyzed using both replicon RNA and genome-length RNA of WNV. In both systems, RNAs containing mutations that abolished N-7 methylation were non-replicative. In contrast, RNAs with mutations that knocked out 2'-O methylation were replicative, but at a lower level than wild-type RNA (Ray et al., 2006, Zhou et al., 2007). Figure 5 summarizes the effects of a panel of mutations on cap methylations and viral replication; the data clearly suggest that viral replication correlates with the competence of N-7 methylation, not with 2'-O methylation. The mutant MTase-mediated replication defect could not be complemented in trans by a wild-type replicase complex (Khromykh et al., 1999, Ray et al., 2006). Interestingly, WNVs defective in 2'-O methylation were attenuated both in vitro and in vivo, and could protect mice from later wild-type WNV challenge (Zhou et al., 2007). These results demonstrate that the N-7 methylation is essential for viral replication. Therefore, the N-7 MTase represents a valid target for WNV therapy.
FIG. 5. Correlation between N-7 methylation and WNV replication.
(A) Effects of a panel of mutations of WNV MTase on cap methylations and viral replication. Mutations of representative amino acids within the K-D-K-E motif, GTP-binding site, SAM-binding site, and RNA-binding site were assayed for their effects on cap methylation and viral replication. N-7 methylation was determined by conversion of GpppA-RNA→m7GpppA-RNA. 2'-O methylation was determined by conversion of m7GpppA-RNA→m7GpppAm-RNA. WT methylation activity was set as 100%. Viral RNA replication was indicated by plaque formation upon transfection of genome-length RNA containing the indicated mutation. Symbols “+” and “−“ indicate that genome-length RNA containing the MTase mutations are replicative or non-replicative, respectively. Note that the K-D-K-E motif forms the active site for 2'-O methylation; the D146 residue of the K-D-K-E motif is essential for N-7 methylation (Ray et al., 2006). (B) Crystal structure of WNV MTases displaying the mutated amino acids. Mutated residues (blue) from (A) are shown on the surface presentation of the WNV MTase [PDB code 2OY0 (Zhou et al., 2007)]. GTP and SAH molecules are colored in yellow and red, respectively. (C) Sequence alignment of flavivirus MTases. The MTase sequences of WNV, DENV-1, DENV-2, and YFV are derived from GenBank accession number AF404756, DVU88535, U87411, and YFU17-66, respectively. The alignment was performed using GCG software (Genetics Computer Group). Identical amino acids among all MTases are shaded.
Using a luciferase-reporting replicon of WNV, Ray and co-workers (2006) showed that, compared with a replicon without a 5' cap (i.e., pppA-RNA), RNA with a 5' GpppA, m7GpppA, or m7GpppAm cap increased the level of translation by approximately by 16-, 25-, and 25-fold. These results suggest that (i) the guanine cap is essential for translation, (ii) N-7 cap methylation enhances translation efficiency, and (iii) 2'-O methylation does not contribute to viral translation. To exclude the possibility that the difference in viral RNA translation was due to the effect of cap structures on RNA stability, the authors used real-time RT-PCR to quantify viral RNAs in cells at various time points post transfection. The RNA stability results showed that neither N-7 nor 2'-O methylation significantly contributed to RNA stability inside host cells. Therefore, the biological function of 2'-O cap methylation remains to be determined.
Taking a genetic selection approach, we recently showed that replication of WNV genome-length RNA containing an MTase-lethal mutation (D146S) can be restored by two types of compensatory mutations (Zhang et al., 2008). Type one mutations were located in the 5’-terminal stem-loop of the genomic RNA (G35U substitution or U38-insertion). Type two mutations resided in NS5 (K61Q in MTase and W751R in RdRp). Mutagenesis analysis, using a genome-length RNA and a replicon of WNV, demonstrated that the compensatory mutations can rescue viral replication. Biochemical analysis showed that the MTase K61Q mutation facilitates viral replication through an improved N-7 methylation activity, while the RdRp W751R mutation improves viral replication through an enhanced polymerase activity. These results may have implications for development of flavivirus MTase inhibitors. Escape viruses that are resistant to MTase inhibitors may emerge through mutations within the MTase and RdRp domains.
8. Methods for screening inhibitors of flavivirus MTase
Both enzyme-based and cell-based assays can potentially be developed for high-throughput screening (HTS) to identify inhibitors of flavivirus MTase. Each has its advantages and disadvantages. For enzyme-based assays, the advantage is that the identified compounds possess known targets. The major disadvantage is that cellular uptake and nonspecific binding of the compounds, two potential problems for many inhibitors, are not evaluated within the assay. For cell-based assays, the advantage is that the uptake of compounds into cells is assessed during screening; the identified inhibitors would have a higher success rate in subsequent animal experiments. However, since the cell-based assay usually employs viral genetic systems (i.e., replicon and complete virus), inhibitors of any of the targets involved in viral replication could be identified; therefore, the exact targets of the inhibitors need to be determined in subsequent studies.
8.1. MTase-based HTS assay
A scintillation proximity assay (SPA) could be developed for HTS, to search for MTase inhibitors. SPA is a radioactive homogeneous technology that relies upon the fact that the energy emitted from a radioisotope will only travel a limited distance in an aqueous environment. When a radioisotope-labeled molecule binds to the microsphere beads, the radioisotope is brought into close proximity to the scintillant, and effective energy transfer from the β-particle occurs, resulting in light emission. When the radioisotope remains free in solution, it is too distant from the scintillant; the β-particle dissipates its energy into the aqueous medium and remains undetected. Because the flavivirus MTase specifically requires viral RNA for cap methylation (Fig. 2B), the RNA substrate should contain the 5'-terminal stem-loop of genomic RNA. The 5' viral RNA should be biotin-labeled. Upon transfer of the 3H-labeled methyl group from SAM to RNA cap, the biotin-labeled RNA can be captured on the streptavidin SPA beads and measured with a scintillation counter.
8.2. Cell-based HTS assays
Three types of genetics-launched, cell-based HTS assays have been developed for flavivirus antiviral discovery. These assays have been optimized in 96- and 384-well formats and validated with known flavivirus inhibitors; they have proven useful in identifying new inhibitors through library screening (Puig-Basagoiti et al., 2005). Type I assays involve cell lines expressing flavivirus replicons (with a deletion of viral structural genes). Many of these cell lines harbor replicons contain a luciferase and a neomycin phosphotransferase gene (for selection and maintenance of cells harboring the replicon) (Khromykh and Westaway, 1997, Lo et al., 2003). The reporting cell lines enable screening for inhibitors at any step of viral translation and RNA replication, but they do not allow screening for inhibitors of assembly and entry.
Type II assays employ packaged virus-like particles (VLPs) containing a reporting-replicon RNA. The reporting-replicon can be packaged into VLPs by supplying structural proteins in trans (Khromykh et al., 1998). The structural proteins can be supplied by three different means: (i) transiently expressed by an alphavirus expression vector (Khromykh et al., 1998), (ii) conditionally provided in an inducible expression cell line (Harvey et al., 2004), or (iii) constitutively expressed in a cell line harboring a noncytopathic Venezuelan equine encephalitis virus (VEEV) replicon (Fayzulin et al., 2006). The VLP-infection system allows screening for inhibitors of viral entry, as well as for inhibitors of translation and RNA synthesis.
Type III assays employ a full-length reporting virus. A reporter gene (luciferase or green fluorescent protein) is engineered into the full-length viral genome to monitor viral replication (Deas et al., 2005, Shustov et al., 2007). The reporting virus-infection assay allows the screening for inhibitors of all steps of viral life cycle. Since N-7 methylation is essential for WNV replication, screening efforts using any of the assay types can identify potential MTase inhibitors.
Besides the above cell-based assays, a raw viral infection assay can also be used to screen compound libraries. Cytopathic effect (CPE), induced by viral infection, could be monitored to measure the antiviral effect of potential inhibitors. A number of methods have been developed to quantify the CPE. For example, the CPE can be quantified by ATP levels in infected cells, because the amounts of intracellular ATP correlate with cell viability. The CellTiter-Glo Luminescent assay (Progema) is a homogeneous method, based on ATP-dependent luciferase activity to determine cell viability. This assay can be readily adapted to HTS of compound libraries. However, as mentioned before, inhibitors identified in cell-based assays may not necessarily target the viral MTase. Target deconvolution is needed to pinpoint the mechanism of the inhibitors. In general, target de-convolution could be accomplished by testing compounds in individual enzyme assays (protease, helicase, NTPase, MTase, and RdRp) or through selection of compound-resistant viruses.
9. Concluding remarks
Although significant progress has been made in our understanding of flavivirus cap formation, many important questions remain. It is not known how cap formation is modulated during viral RNA synthesis. During cap formation, the 5' end of flavivirus RNA must interact with various components of the replication complex in a highly orchestrated manner. After synthesis from RdRp domain of NS5, nascent 5' pppA-RNA first interacts with NS3, to remove the γ–phosphate. The resulting ppA-RNA is capped via an unidentified guanylyltransferase. Next, the GpppA-RNA interacts with MTase domain for N-7 and 2'-O methylations. Capping of cellular mRNA is coupled to RNA synthesis through direct binding of the capping apparatus to the RNA polymerase II elongation complex (Cho et al., 1997, McCracken et al., 1997, Yue et al., 1997). It is likely that flavivirus RNA capping and methylation are coupled to RNA synthesis. Based on genetic selection results, a putative interface between the MTase and RdRP domains was recently proposed for WNV NS5 (Malet et al., 2007). Further experiments are needed to examine the interactions between the flavivirus capping and RNA replication.
Identification of features unique to viral targets will direct antiviral drug development. The flavivirus NS5 protein shares several characteristics with the L protein from VSV. Both possess MTase and RdRp activities. Recent studies showed that mutations of the predicted SAM-binding site of the VSV L protein are defective in both N-7 and 2'-O cap methylation, indicating that the two reactions share a SAM-binding site (Li et al., 2006). A recombinant L protein of Sendai virus catalyzes N-7 cap methylation in a viral sequence-dependent manner (Ogino et al., 2005). Cap formation of VSV mRNA was recently shown to require specific cis-acting signals in the RNA (Wang et al., 2007). However, major differences in cap formation exist between the two taxonomically distant virus groups. As noted earlier, the type 1 cap of VSV mRNA is formed through transfer of a GDP moiety from GTP to the 5' monophosphate of the acceptor RNA (Abraham et al., 1975), with subsequent methylations at 2'-O and guanine N-7 positions of the cap (Moyer et al., 1975, Testa and Banerjee, 1977). The mechanism of the L protein-mediated guanylyltransferase is also distinct from that of cellular and other known viral counterparts (Ogino and Banerjee, 2007).
The critical function of N-7 methylation in viral replication needs to be generalized in flaviviruses other than WNV. We must emphasize that the essential function of N-7 methylation in viral replication has only been demonstrated in WNV. It is critical that the WNV results be validated in other flaviviruses. Another important question pertinent to the use of MTase as an antiviral target is viral specificity. If the geometry of the MTases from virus and host is similar, an inhibitor is likely to suppress both host and viral MTases, resulting in toxicity. In the case of WNV, this concern is argued against by the fact that sinefungin inhibited WNV cap methylation and viral replication with a TI value of greater than 167 (Dong et al., 2008b). Furthermore, flavivirus cap methylation is viral RNA sequence-specific (Dong et al., 2007), raising the possibility that inhibitors can be developed to specifically block viral RNA-MTase interactions, without affecting host mRNA cap methylation. If so, the crystal structures of flavivirus MTases and the residues identified for N-7 methylation should aid in the rational design of antiviral agents.
ACKNOWLEDGEMENTS
The work was partially supported by grants AI061193 and U54-AI057158 (Northeast Biodefense Center), and contract NOI-AI25490 from NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abraham G, Rhodes DP, Banerjee AK. The 5' terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell. 1975;5:51–58. doi: 10.1016/0092-8674(75)90091-4. [DOI] [PubMed] [Google Scholar]
- Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Ahola T, Kaariainen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. U .S. A. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenberg R, Ren J, Verma A, Walter TS, David Alderton, Hurrelbrink RJ, Fuller SD, Bressanelli S, Owens RJ, Stuart DI, Grimes JM. Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues. J. Gen. Virol. 2007;88:2228–2236. doi: 10.1099/vir.0.82757-0. [DOI] [PubMed] [Google Scholar]
- Bartelma G, Padmanabhan R. Expression, purification, and characterization of the RNA 5'-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology. 2002;299:122–132. doi: 10.1006/viro.2002.1504. [DOI] [PubMed] [Google Scholar]
- Benarroch D, Egloff MP, Mulard L, Guerreiro C, Romette JL, Canard B. A structural basis for the inhibition of the NS5 dengue virus mRNA 2'-O-methyltransferase domain by ribavirin 5'-triphosphate. J. Biol. Chem. 2004;279:35638–35643. doi: 10.1074/jbc.M400460200. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaves GR, Dubin DT. Methylation status of intracellular Dengue type 2 40S RNA. Virology. 1979;96:159–165. doi: 10.1016/0042-6822(79)90181-8. [DOI] [PubMed] [Google Scholar]
- Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nature Medicine. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- Day CW, Smee DF, Julander JG, Yamshchikov VF, Sidwell RW, Morrey JD. Error-prone replication of West Nile virus caused by ribavirin. Antiviral Res. 2005;67:38–45. doi: 10.1016/j.antiviral.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Deas TS, Binduga-Gajewska I, Tilgner M, Ren P, Stein DA, Moulton HM, Iversen PL, Kauffman EB, Kramer LD, Shi P-Y. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 2005;79:4599–4609. doi: 10.1128/JVI.79.8.4599-4609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Ray D, Ren S, Zhang B, Puig-Basagoiti F, Takagi Y, Ho C, Li H, Shi PY. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J. Virol. 2007;8:4412–4421. doi: 10.1128/JVI.02455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Ren S, Li H, Shi PY. Separate molecules of West Nile virus methyltransferase can independently catalyze the N7 and 2'-O methylations of viral RNA cap. Virology. 2008a doi: 10.1016/j.virol.2008.04.026. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Ren S, Zhang B, Zhou Y, Puig-Basagoiti F, Li H, Shi PY. Flavivirus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate repositioning mechanism. J. Virol. 2008b;82:4295–4307. doi: 10.1128/JVI.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2'-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Decroly E, Malet H, Selisko B, Benarroch D, Ferron F, Canard B. Structural and Functional Analysis of Methylation and 5'-RNA Sequence Requirements of Short Capped RNAs by the Methyltransferase Domain of Dengue Virus NS5. J Mol Biol. 2007;372:723–736. doi: 10.1016/j.jmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- Fayzulin R, Scholle F, Petrakova O, Frolov I, Mason PW. Evaluation of replicative capacity and genetic stability of West Nile virus replicons using highly efficient packaging cell lines. Virology. 2006;351:196–209. doi: 10.1016/j.virol.2006.02.036. [DOI] [PubMed] [Google Scholar]
- Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould EA, Solomon T, Mackenzie JS. Does antiviral therapy have a role in the control of Japanese encephalitis? Antiviral Res. 2008;78:140–149. doi: 10.1016/j.antiviral.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Gubler D, Kuno G, Markoff L. Flaviviruses. In: Knipe DM, Howley PM, editors. Fields virology. 5th. vol. 1. Philadelphia, Pa: Lippincott William & Wilkins; 2007. pp. 1153–1253. [Google Scholar]
- Guyatt KJ, Westaway EG, Khromykh AA. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods. 2001;92:37–44. doi: 10.1016/s0166-0934(00)00270-6. [DOI] [PubMed] [Google Scholar]
- Hager J, Staker BL, Bugl H, Jakob U. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 2002;277:41978–41986. doi: 10.1074/jbc.M205423200. [DOI] [PubMed] [Google Scholar]
- Harvey T, Liu W, Wang X, Linedale R, Jacobs M, Davidson A, Le T, Anraku I, Suhrbier A, Shi P, Khromykh A. Tetracycline-inducible packaging cell line for production of flavivirus replicon particles. J. Virol. 2004;78:531–538. doi: 10.1128/JVI.78.1.531-538.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel AE, Gershon PD, Quiocho FA. Structural basis for sequence-nonspecific recognition of 5'-capped mRNA by a cap-modifying enzyme. Mol. Cell. 1998;1:443–447. doi: 10.1016/s1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Sedlak PL, Westaway EG. Trans-Complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J. Virol. 1999;73:9247–9255. doi: 10.1128/jvi.73.11.9247-9255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Varnavski AN, Westaway EG. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 1998;72:5967–5977. doi: 10.1128/jvi.72.7.5967-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Westaway EG. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 1993;74:733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- Kramer L, Li J, Shi PY. West Nile virus. The Lancet Neurology. 2007;6:171–182. doi: 10.1016/S1474-4422(07)70030-3. [DOI] [PubMed] [Google Scholar]
- Leyssen P, Balzarini J, De Clercq E, Neyts J. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J. Virol. 2005;79:1943–1947. doi: 10.1128/JVI.79.3.1943-1947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyssen P, De Clercq E, Neyts J. Molecular strategies to inhibit the replication of RNA viruses. Antiviral Res. 2008;78:9–25. doi: 10.1016/j.antiviral.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Clum S, You S, Ebner KE, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 1999;73:3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang J, Whelan S. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 2006;103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel H-J, Rice CM. Flaviviridae: The Virus and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Fourth ed. Lippincott William & Wilkins; 2007. [Google Scholar]
- Lo L, Tilgner M, Shi P-Y. A potential high-throughput assay for screening inhibitors of West Nile virus replication. J. Virol. 2003;77:12901–12906. doi: 10.1128/JVI.77.23.12901-12906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzhkov V, Selisko B, Nordqvist A, Peyrane F, Decroly E, Alvarez K, Karlen A, Canard B, Qvist J. Virtual screening and bioassay study of novel inhibitors for dengue virus mRNA cap (nucleoside-2'O)-methyltransferase. Bioorg. Med. Chem. 2007;15:7795–7802. doi: 10.1016/j.bmc.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Malet H, Egloff M, Selisko B, Butcher R, Wright P, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, Mackenzie J, Khromykh AD, Davidson A, Canard B. Crystal structure of the RNA polymerase domain of the West Nile virus nonstructural protein 5. J. Biol. Chem. 2007;282:10678–10689. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- Mastrangelo E, Bollati M, Milani M, Selisko B, Peyrane F, Canard B, Grard G, De Lamballerie X, Bolognesi M. Structural bases for substrate recognition and activity in Meaban virus nucleoside-2'-O-methyltransferase. Protein Sci. 2007;16:1133–1145. doi: 10.1110/ps.072758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5'-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath TP. Treatment of yellow fever. Antiviral Res. 2008;78:116–124. doi: 10.1016/j.antiviral.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Moyer SA, Abraham G, Adler R, Banerjee AK. Methylated and blocked 5' termini in vesicular stomatitis virus in vivo mRNAs. Cell. 1975;5:59–67. doi: 10.1016/0092-8674(75)90092-6. [DOI] [PubMed] [Google Scholar]
- Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Ogino T, Kobayashi M, Iwama M, Mizumoto K. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 2005;280:4429–4435. doi: 10.1074/jbc.M411167200. [DOI] [PubMed] [Google Scholar]
- Puig-Basagoiti F, Deas TS, Ren P, Tilgner M, Ferguson DM, Shi PY. High-throughput assays using luciferase-expressing replicon, virus-like particle, and full-length virus for West Nile virus drug discovery. Antimicrob. Agent. Chemother. 2005;49:4980–4988. doi: 10.1128/AAC.49.12.4980-4988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas T, Zhou Y, Li H, Shi PY. West nile virus 5'-cap structure is formed by sequential guanine N-7 and ribose 2'-O methylations by nonstructural protein 5. J. Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 A resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- Schnierle BS, Gershon PD, Moss B. Mutational analysis of a multifunctional protein, with mRNA 5' cap-specific (nucleoside-2'-O-)-methyltransferase and 3'-adenylyltransferase stimulatory activities, encoded by vaccinia virus. J. Biol. Chem. 1994;269:20700–20706. [PubMed] [Google Scholar]
- Shustov A, Mason P, Frolov I. Production of pseudoinfectious yellow fever virus with a two-component genome. J Virol. 2007;81:11737–11748. doi: 10.1128/JVI.01112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G, Grimes JM, Stuart DI, Roy P. Bluetongue virus VP4 is an RNA-capping assembly line. Nat. Struc. Mol. Biol. 2007;14:449–451. doi: 10.1038/nsmb1225. [DOI] [PubMed] [Google Scholar]
- Tan BH, Fu J, Sugrue RJ, Yap EH, Chan YC, Tan YH. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216:317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- Testa D, Banerjee AK. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J. Virol. 1977;24:786–793. doi: 10.1128/jvi.24.3.786-793.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, McElvain L, Whelan S. Vesicular stomatitis virus mRNA capping machinery requires specific cis-acting signals in the RNA. J. Virol. 2007;81:11499–11506. doi: 10.1128/JVI.01057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrener P, Tamura JK, Collett MS. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 1993;67:989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G. Terminal sequences of the genome and replicative-form RNA of the flavivirus West Nile virus: absence of poly(A) and possible role in RNA replication. Virology. 1981;113:544–555. doi: 10.1016/0042-6822(81)90182-3. [DOI] [PubMed] [Google Scholar]
- Wengler G, Wengler G. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184:707–715. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- Wengler G, Wengler G. The NS3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology. 1993;197:265–273. doi: 10.1006/viro.1993.1587. [DOI] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. U .S. A. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Dong H, Zhou Y, Shi PY. Genetic interactions among the West Nile virus methyltransferase, the RNA-dependent RNA polymerase, and the 5' stem-loop of genomic RNA. J. Virol. 2008 doi: 10.1128/JVI.00654-08. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard K, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]