Abstract
Telomere maintenance is an important genetic mechanism controlling cellular proliferation. Normally, telomeres are maintained by telomerase which is downregulated upon cellular differentiation in most somatic cell lineages. Telomerase activity is upregulated in immortalized cells and cancers to support an infinite lifespan and uncontrolled cell growth; however, some immortalized and transformed cells lack telomerase activity. Telomerase-negative tumors and immortalized cells utilize an alternative mechanism for maintaining telomeres termed alternative lengthening of telomeres (ALT). This research explored evidence for the ALT pathway in chicken cell lines by studying nontransformed immortalized cell lines (DF-1 and OU2) and comparing them to a normal (mortal) cell line and a transformed cell line (DT40). The research consisted of molecular and cellular analyses including profiling of telomeric DNA (array sizing and total content), telomerase activity, and expression of genes involved in the telomerase, recombination, and ALT pathways. In addition, an immunofluorescence analysis for an ALT marker, i.e. ALT-associated promyelocytic leukemia bodies (APBs), was conducted. Evidence for ALT was observed in the telomerase-negative immortalized cell lines. Additionally, the APB marker was also found in the other cell systems. The attributes of the chicken provide an additional vertebrate model for investigation of the ALT pathway.
Key Words: Alternative lengthening of telomeres, Chicken, DF-1, DT40, Immortalization, OU2, Promyelocytic leukemia, Recombination, Telomerase, Telomere
Introduction
The telomeres of the chicken genome are maintained by telomerase similar to other vertebrates [Delany et al., 2003; Swanberg and Delany, 2006; Swanberg et al., 2010]. Telomere maintenance, i.e. length and structure, is critical for control of genome stability and serves as a predominant genetic mechanism controlling lifespan of cell lineages. In both human and chicken, telomerase activity is downregulated when somatic cells differentiate and as a result telomeres shorten with each cell cycle, eventually resulting in cellular senescence. Additionally, telomerase is reactivated in most immortalized and transformed cells and thus the telomeres are maintained. However, there exist a number of human immortalized cell lines and cancers wherein telomerase activity is lacking; these cells utilize an alternative mechanism to telomerase to maintain their telomeres which is termed alternative lengthening of telomeres (ALT). This alternative telomere lengthening mechanism has been reported for 10–15% of human tumors and 30–40% of immortalized cell lines [Bryan and Reddel, 1997; Bryan et al., 1997; Cerone et al., 2005]. In addition, an alternative telomere maintenance mechanism was identified in yeast [Lundblad and Blackburn, 1993] and telomerase RNA null (Terc–/–) mouse cells [Niida et al., 2000; Chang et al., 2003], but has not been reported in other vertebrates.
A combination of markers provides evidence that the ALT pathway is operating to maintain telomeres [Pickett and Reddel, 2009]. The markers of ALT include the absence of telomerase activity in immortalized (or transformed) cells (i.e. cell types with unlimited proliferation potential), a heterogeneous terminal telomeric DNA profile (i.e. an enhanced variable size range of telomere array lengths), and presence of nuclei which exhibit ALT-associated promyelocytic leukemia (PML) bodies known as APBs. In particular, the APBs are considered a definitive marker for ALT [Yeager et al., 1999]. These nuclear bodies contain the PML protein with telomere-associated proteins (TRF1, TRF2) plus DNA repair and recombination proteins (RAD51, RAD52, MRE11, RAD50, NBS1). The current model suggests that the ALT mechanism utilizes telomere homologous recombination to maintain and even lengthen the telomeres [reviewed in Cesare and Reddel, 2008]. However, a single definitive assay for the ALT pathway does not exist and as mentioned, detection is dependent on markers shown experimentally to be associated with the pathway [Cesare and Reddel, 2010].
A characteristic feature of the chicken genome is that it possesses a highly heterogeneous telomeric DNA profile [Delany et al., 2000; Rodrigue et al., 2005; O'Hare and Delany, 2009] and at least in meiotic cells, evidence exists for high rates of telomeric DNA recombination as shown by the generation of novel telomere arrays in progeny not seen in parental genomes [Rodrigue et al., 2005]. Interestingly, the immortalized chicken cell line DF-1 maintains an unusually large amount of heterogeneously sized telomeric DNA and greater than 3-fold more total telomeric sequence content than normal chicken cells [O'Hare and Delany, 2009]. Further, it was reported by Christman et al. [2005] that telomerase activity was not detectable in the DF-1 cell line. Based on these combined results it seems plausible that the chicken which shares many telomere biology features with human [Swanberg and Delany, 2006; Swanberg et al., 2010] may also possess the capacity to employ ALT as a mechanism to maintain telomeres.
This research investigates the hypothesis that chicken, similar to human, possesses an alternative mechanism for maintaining telomeres, specifically ALT. Four cell lines with differing proliferation phenotypes were studied including 2 immortalized chicken embryo fibroblast cell lines (DF-1 and OU2), a normal (mortal) chicken embryo fibroblast cell line, and a transformed cell line (DT40). Telomerase activity, expression of genes associated with the telomerase and ALT pathways including telomere-associated, DNA repair and recombination genes, and the presence of an ALT marker (APBs) were investigated. Evidence for ALT was found in the immortalized lines as these were negative for telomerase activity, had normal or larger amounts of telomeric DNA with a heterogeneous profile, and exhibited APBs. Interestingly, albeit to a lesser extent, APBs were also observed in the telomerase-negative mortal cells as well as the telomerase-positive transformed cells. Overall, these results suggest the interesting possibility that the 2 telomere-lengthening pathways, i.e. telomerase and recombination-based ALT, coexist as redundant pathways with differential utilization depending on the cell system.
Materials and Methods
Cells
Chicken embryo fibroblast (CEF) cell lines, DF-1 (spontaneously immortalized; Himly et al. [1998]; ATCC CRL-12203), OU2 (chemically immortalized; Ogura and Fujiwara [1987]; ATCC CRL-12302), and CEF001 (a normal mortal primary cell line derived from a pool of 3 embryos from the UCD 001 red jungle fowl inbred line) were cultured in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen 11995-065), 10% fetal bovine serum (FBS, Invitrogen 10437-010), and 2% penicillin/streptomycin (Invitrogen 15140-122), at 38°C and 5% CO2. DT40 cells (transformed B-cell bursal lymphoma; Baba et al. [1985]) were cultured in RPMI 1640 with glutamine (Invitrogen 11875-093), 10% FBS (Invitrogen 10437-010), 2% penicillin/streptomycin (Invitrogen 15140-122), 2% chicken serum (Invitrogen 16110-082), and 91.7 μM 2-mercaptoethanol, at 41°C and 5% CO2.
Telomerase Activity – Telomeric Repeat Amplification Protocol
Telomerase activity was determined using the TRAPeze® Telomerase Detection Kit (Millipore S7700). Positive controls for activity were DT40 and gastrula embryos (pool of 3 chicken embryos incubated for 24 h, from the commercial stock Hy-Line CD20). Negative controls (lack of activity) were created by heat-treating (95°C for 30 min) an aliquot of the positive control samples. CEF001 and CHAPS lysis buffer only were also used as negative controls. For each sample, 1 μg of protein was used for the TRAPeze PCR. The PCR amplicons were separated by size on a 10% PAGE (20 ml 30% acrylamide, 40 ml 0.5× TBE, 125 μl 10% APS, 75 μl TEMED) at 40 W for 60 min. The resulting gel was stained with SYBR Safe (Invitrogen) and then imaged using a Fujifilm imager (FLA-5100).
Gene Expression – RNA Isolation, cDNA Synthesis, Quantitative PCR
RNA was extracted from the cells for gene expression analysis using the RNeasy® Mini Kit (Qiagen) including DNase treatment. The concentration of each RNA sample was determined using a Nanodrop 1000 Spectrophotometer (Thermo Scientific). The RNA was stored at −80°C until cDNA synthesis. Two micrograms of RNA were used in 40-μl cDNA reactions using iScript™ cDNA synthesis (Bio-Rad). The resulting cDNA was diluted with 60 μl of DEPC-treated water. The cDNA was stored at −20°C until Quantitative (q)PCR.
Eighteen genes were studied for their expression by qPCR using SsoFast™ EvaGreen® Supermix (Bio-Rad) and gene-specific primers (Eurofins MWG Operon); gene and primer details are provided in table 1. The qPCR protocol was as follows: 95°C for 30 s, 40 cycles of 95°C for 5 s, 57°C for 10 s, plate read, followed by a melt curve of 55–95°C with 10 s hold and plate read every 0.5°C. The reaction tube included 5 μl of the diluted cDNA in a total reaction volume of 20 μl with 10 μl of supermix, 500 nM forward primer, 500 nM reverse primer, and 3 μl of water. The PCRs were conducted using a Chromo4™ (Bio-Rad) and Opticon Monitor™ 3.1 with a cycle range of 3–12 and a threshold of 1.100. LinRegPCR version 11.5 [Ruijter et al., 2009] was used to determine amplification efficiencies. The overall average efficiency was 1.85. The comparative Ct method was used to analyze the samples [O'Hare and Delany, 2005]. All cell culture samples (DF-1, OU2, CEF001, DT40) were assessed using 2 different culture samples and each sample consisted of a pool of 3 T75 flasks. For the gastrula embryos, there were 2 samples which consisted of a pool of 3 embryos each. Every sample was run in triplicate. For each gene-primer set, qPCR amplicons were purified using QIAquick® PCR purification kit (Qiagen) and sequence verified (Davis Sequencing, Davis, Calif., USA). Statistical evaluation of the results was conducted for the normalized Ct values using Minitab 15 for ANOVA and Tukey's pairwise comparisons (α = 0.05).
Table 1.
Chicken genes and primers for quantitative gene expression analysis
| Gene | GGA | Accession number | Gene ID | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|---|---|---|
| TR | 9 | AY312571 | 379037 | GAAGCTCATGCTCTGTGCTG | ACCCTTCAAAATTCCCCATC |
| TERT | 2 | AY502592 | 420972 | TGCTTCCAGCAGTTATCCAG | CTGTTCTCATCCAGCAAGGAG |
| DKC1 | 4 | CR353445 | 422196 | CACCAGAGGAGCTGAGCAA | TGCTTTCACTGATCACTTCTATTTG |
| MYC | 2 | X68073 | 420332 | CAGCGACTCGGAAGAAGAAC | TTCTGTGCTGGACTCTGTGC |
| TRF1 | 2 | AY237359 | 378896 | CCGTCGCTTGATTCAAATTC | GGCTTATGTGATTCTGAGTCTGTG |
| TRF2 | 11 | AJ133783 | 395598 | GGCGAAAACCTAGATTGCAC | CAGGCACGCTGAATTCTTTT |
| RAP1 | 11 | AY083608 | 395122 | GCTCCGAGTCAGGAAGTGAC | AGCAGGGAGAGCACAGTCAT |
| KU70 | 1 | AB016529 | 395767 | TAGCTCCTCCAGGTTTCCAC | TGTCCACCTGTTCTCGATTG |
| KU80 | 7 | XM_422072 | 424222 | ACTGCACAGCAAAAGGAAGG | CGTTCAAGGCTCTCTCTGAAA |
| RAD51 | 5 | S59426 | 396086 | GGCTAGAGCAATGTGGTATAAATG | AGCTCCTTCTTGGGTGCAT |
| RAD52 | 1 | U01047 | 418152 | TTCTTTGCAGATGATCTTGAACTC | TGCTGTGCTGCTGGTGAG |
| MRE11 | 1 | AF166094 | 395555 | AGCGTCACATTGATGCAGAG | CATTGCCTCACGAACTTCTTC |
| RAD50 | 13 | BX929640 | 416329 | GAACAAAGCATGGTGAGAAGG | CATTGTTAAGCACTGACTTGGAA |
| NBS1 | 2 | AF230342 | 374246 | GCTGTCTTGGAAAGCAACAA | TTGTTATCAGGCTGCCTCTG |
| SMC5 | Z | AJ720299 | 431608 | GACCTTGGGTGGAATATGAAAC | TGGAGATTGCATTTCCTTCA |
| SMC6 | 3 | XM_419962 | 421950 | GGACAGAGTGAGGTTTAAATGGA | CAGATCAGCAAAGGGTTACTTG |
| PML-L | 10 | XM_413692 | 415304 | CAAACTGCTTGCCCCAAC | AGAAGGTCACTATCTGAGTCATGC |
| PML-S | 10 | XM_413690 | 415302 | CCACGTCACCAGCATCTTC | GCGGATGTCGCAGTAGAAAG |
| GAPDH | 1 | CBO18343 | 374193 | GGAAAGTCATCCCTGAGCTG | CTTGGCTGGTTTCTCCAGAC |
Accession number and Gene ID are from NCBI. At least one of the primers for each gene (except TR, a direct transcript) was created across an exon-exon boundary. GGA = Gallus gallus chromosome.
OU2 Cytogenetic Profile – Fluorescence in situ Hybridization
Mitotic chromosome harvest, slide preparation, and fluorescence in situ hybridization (FISH) procedures were followed as described in O'Hare and Delany [2009]. Telomeric PNA probe hybridization was performed to establish the cytogenetic telomeric profile of OU2 cells, as had been previously studied for the other cell types [O'Hare and Delany, 2009]. DF-1 and CEF001 samples were utilized as a point of comparison. To assess the OU2 karyotype status, 3 chromosome-specific probes were used and indirectly labeled with anti-digoxigenin-rhodamine (Roche Applied Science), including 5S rDNA (GGA9), ETS rDNA (GGA16), and TAM32-55E18 BAC (GGAW).
Telomeric Sizes – Southern Blot
Telomeric lengths were analyzed according to O'Hare and Delany [2009] by the terminal restriction fragment (TRF) method. Briefly, restriction-digested DNA samples (150 ng) of DF-1, OU2, CEF001, and DT40 were separated by size on an agarose gel (0.7% agarose, 1× TAE, 55 V for 4 h), transferred to a membrane, and hybridized with a telomeric probe.
Telomeric Sequence Content – Slot Blot
DNA was isolated from DF-1, OU2, CEF001, CEF003 (CEFs created from a pool of 5 UCD 003 embryos), and DT40 cells using DNeasy Blood and Tissue Kit (Qiagen). The slot blot procedures were conducted as described in O'Hare and Delany [2009]. Three different slot blot experiments were conducted and the results averaged for each cell system. The telomeric sequence content was determined as a percentage of total genomic DNA.
Chicken PML Genes – Bioinformatics
In order to study a marker of ALT in chicken, nuclear APBs which contain the PML protein, the available knowledge regarding PML was assessed, i.e. gene sequence, RNA and protein sequence. The chicken genome sequence (build 2.1) indicates the presence of 2 PML genes (NCBI Gene IDs 415304 and 415302) whereas other vertebrate genomes contain 1 PML gene as found in the NCBI database. Each of the 2 chicken PML genes has associated ESTs according to NCBI UniGene (http://www.ncbi.nlm.nih.gov/UniGene). The sizes of the predicted proteins of the 2 chicken genes differ. One PML gene (415304) predicts a protein (XP_413692.2) of 1,021 amino acids (aa) and herein is referred to as PML-L (L for long), while the other gene (415302) predicts a protein (XP_413690.2) of 530 aa and is referred to as PML-S (S for short). This descriptive nomenclature provides easy differentiation of the 2 PML proteins for the purpose of this study whereas in the NCBI database both of these genes are designated as PML (http://www.ncbi.nlm.nih.gov/gene/415304, http://www.ncbi.nlm.nih.gov/gene/415302). The 2 predicted chicken PML proteins are 77% identical to each other. The chicken PML similarity with other species including Homo sapiens, Mus musculus, Bos taurus, Rattus norvegicus, and Pan troglodytes ranges from 36–41% (considering both chicken PML proteins). Since other vertebrates have only 1 gene it was unexpected to find 2 in chicken. Both chicken PML genes were studied in these experiments.
PML and TRF2 Antibodies
Primary antibodies were custom-made for 2 chicken PML proteins, PML-L and PML-S. Polyclonal antibodies were raised in guinea pigs against the chicken PML-L peptide C-SHALVHDELLEAMKTKQD and the chicken PML-S peptide GRIPPSKASRSRTRDAE (Open Biosystems, Thermo Fisher Scientific). The 2 peptides were used as immunogens (separately) in 2 animals each; the resulting anti-sera (from both animals for each peptide) were pooled and affinity purified (Open Biosystems). The primary antibody used to detect chicken TRF2 was created using a chicken TRF2 peptide as an immunogen in rabbit [Cooley et al., 2009].
PML and TRF2 Cellular Immunofluorescence
Cells (DF-1, OU2, CEF001, DT40) were grown on chamber slides (Nunc Lab-Tek II CC2). These slides contained CC2 (mimics polylysine) which allowed the suspension cells (DT40) to adhere to the slides. The slides were fixed with HistoChoice® MB (Amresco) for 15 min, washed in 1× PBS/0.02% Tween-20 for 4 min, and placed in 70% EtOH. The slides were stored in 70% EtOH at −20°C until used. Briefly, slides were treated following immunofluorescence (IF) [Peddada et al., 2006; Swanberg et al., 2009] procedures as follows: incubated with primary antibody at a 1:100 dilution with IF stain buffer (1% FBS, 1% sodium azide, 0.5% Tween-20, in 1× PBS) at 37°C for 2 h; washed shaking 3 times in 1× PBS/0.5% Tween-20 for 5 min each at room temperature, followed by secondary antibody at a 1:100 or 1:200 dilution with IF stain buffer including 1 μg/μl RNase A at 37°C for 2 h followed by 3 washes with 1× PBS/0.5% Tween-20 for 5 min each. Finally, DAPI was applied and the slides were coverslipped, then stored at 4°C until image capture within 1–24 h. Secondary antibodies were goat anti-guinea pig IgG FITC (Millipore, AP108F) at a 1:100 dilution (for the PML-detecting primary antibodies) and donkey anti-rabbit IgG Texas Red (Jackson ImmunoResearch, 711-075-152) at a 1:200 dilution (for the TRF2-detecting primary). Slides were also incubated with secondary antibody only and normal IgG (rabbit or guinea pig) to test for background and nonspecific binding, respectively.
PML Antibody Analysis – Western Blot
Protein was extracted from cell pellets using CHAPS lysis buffer (Millipore). The protein samples were mixed 1:1 with Laemmli sample buffer (with 5% β-mercaptoethanol) and heated at 95°C for 5 min. The proteins were separated using Mini-PROTEAN® TGX™ Precast 4–15% gels (Bio-Rad) at 200 V for 30 min. Proteins were transferred using a Mini-Trans Blot (Bio-Rad) at 90 V for 1 h to polyvinylidene fluoride membranes (Bio-Rad). The membrane was placed in blocking buffer (5% nonfat dry milk in 1× Tris-buffered saline, TBS) overnight at 4°C. The immunoblotting protocol was performed following the Immun-Star™ HRP Chemiluminescent Kit Instruction Manual (Bio-Rad) with 1:300 dilution of the PML-L or PML-S primary antibodies in TTBS (1× TBS with 0.05% Tween 20) and a 1:15,000 dilution of goat anti-guinea pig IgG HRP (Millipore, AP108P) secondary antibody in 1× TTBS. The chemiluminescence signal was detected using Kodak X-Omat Blue XB-1 Film. The membrane was stained with Ponceau S solution (Sigma) to confirm that protein was present in all sample lanes. As a control, a blot was incubated with secondary antibody alone.
Precision Plus Protein™ WesternC Standards (Bio-Rad), which utilize Strep-Tactin®-HRP for chemiluminescent visualization, was used during western blotting. However, it was determined that Strep-Tactin (a modified form of streptavidin) was reacting to the chicken protein extract producing many nonspecific bands in all sample lanes for all cell lines. In order to eliminate the bands resulting from the Strep-Tactin, the standard lane was separated from the sample lanes for detection steps and then reassembled for exposure to film.
Results
Four chicken cell lines, including mortal (CEF001), immortalized (DF-1 and OU2), and transformed (DT40) cell lines, were studied to investigate evidence for the ALT pathway. The scope of the analysis included telomeric profiling including terminal array size distribution and total content/genome using both cytogenetic and molecular techniques, determination of telomerase activity, gene expression profiling (qPCR) of genes involved in the telomerase, recombination, and/or ALT pathways, and determination of the status of nuclear APBs (a commonly used marker for ALT) by IF using antibodies created against chicken PML proteins and TRF2.
Telomeric Arrays and Karyotype of OU2
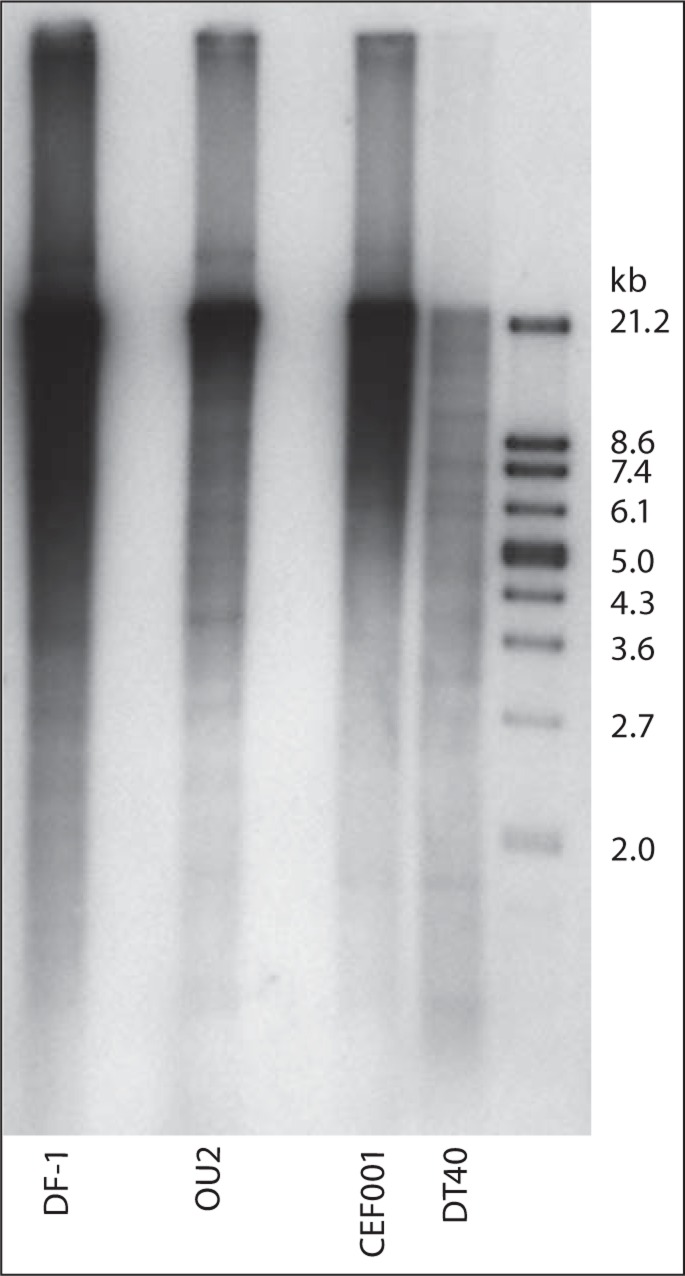
The OU2 cell line was investigated for its telomeric array profile to allow for comparisons with the other cell lines which were studied previously [O'Hare and Delany, 2009]. As per the cytogenetic analysis, the OU2 cell line lacks the size class of mega-telomere arrays found in other chicken genotypes [Delany et al., 2000; O'Hare and Delany, 2009], but shows evidence of moderately sized terminal telomeres and also interstitial arrays (fig. 1). Similar to other chicken cell types, OU2 displays a heterogeneous telomeric array profile, i.e. a wide range of terminal telomere lengths among homologous and non-homologous chromosomes. Also, the OU2 cell line indicated a heterogeneous telomeric array length profile by TRF analysis (fig. 2), similar to DF-1, CEF001, and DT40 (see fig. 2 and O'Hare and Delany [2009]). DF-1 exhibits the greatest telomeric content by both the percentage analysis (slot blot) as well as molecular array length analysis (Southern blot), while DT40 exhibits the least amount of content but still exhibits a heterogeneous profile. Total telomeric DNA content (as a percentage of the genome) ranged from 0.7 to 12.3%, specifically, 0.7% for DT40, 5% for CEF001 and OU2, 6% for CEF003, and 12.3% for DF-1. The OU2 telomere profile maintains heterogeneous telomeric lengths and a percentage of telomeric DNA similar to normal chicken cells, but lacks mega-telomeres by cytogenetic analysis.
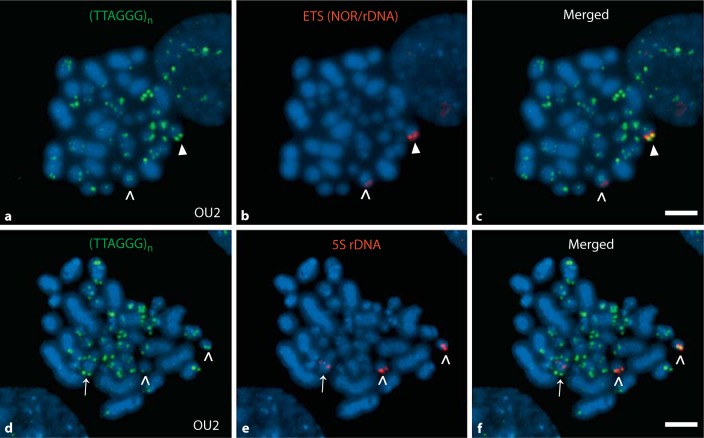
Fig. 1.
Cytogenetic profile of the immortalized OU2 cell line: heterogeneous telomeres and karyotype alterations. The immortalized CEF cell line OU2 exhibits a heterogeneous telomeric profile with telomeres of variable length between homologous and among nonhomologous chromosomes. Mapping of the rDNA-encoding chromosomes by FISH provides evidence for derivative chromosomes and aneuploidy. Telomeric profile of OU2 using a telomeric PNA probe for detecting telomeric array sequences (TTAGGG)n (a), an ETS (red) probe (ETS identifies the 18S-5.8S-28S rDNA, also known as the nucleolus organizer region, NOR) detected 2 chromosomes (>, ▸) both larger than the normal GGA16 (b) with the colors merged in image c. Although the ETS-detected chromosomes are similar in size, 1 homolog exhibited both a brighter/more intense ETS signal and a brighter/more intense telomere signal localized to the same end (b, c, ▸), than the other homolog (b, c, >). Telomeric profile of OU2 chromosomes at a less-condensed stage (d) which provides a generally enhanced signal profile compared to the signal in a. The 5S rDNA probe (red) hybridized to 3 chromosomes indicating a trisomic condition e. Two of the chromosomes were of a size expected for GGA9 (>) whereas the locus indicated by the third 5S rDNA signal was observed in the middle of a larger chromosome (→) suggesting an additional chromosome rearrangement. The telomere on one of the GGA9 chromosomes was typically brighter than the other one as seen in the merged-probes image (f, right >). Both GGA9 and 16 display mega-telomeres in other genotypes [Delany et al., 2007; O'Hare and Delany, 2009]. In general, OU2 contains a reduced number of microchromosomes for the diploid chicken genome, indicates more intermediate-sized chromosomes and as shown by FISH results possesses derivative chromosomes and aneuploidy. Scale bar = 5 μm.
Fig. 2.
Molecular analysis of telomeric array lengths of normal, immortalized, and transformed chicken cell lines highlights the heterogeneity of array sizes and genome-related concentration differences. Telomeric repeat sizes were determined using the TeloTAGGG Telomere Length Assay (Roche Applied Science) following O'Hare and Delany [2009]. The same amount of DNA (150 ng) was analyzed for each cell line. The immortalized cell line OU2 was not previously characterized for telomere length and was compared to the immortalized DF-1 line, normal CEF001, and transformed DT40. OU2 exhibits a heterogeneous telomeric profile with arrays across a broad range of molecular-weight size classes and not unlike other chicken genomes. As reported previously [O'Hare and Delany, 2009], DF-1 has the greatest telomeric content and DT40 the least telomeric content (and supported by total telomeric DNA contents, see Results), although both genomes maintain heterogeneously sized telomeric arrays.
The OU2 karyotype is abnormal in that it contains a number of derivative chromosomes, which is not unexpected for a chemically immortalized cell line cultured long term in vitro. In addition, the number of microchromosomes is reduced relative to the normal chicken karyotype. But there appears to be more intermediate-sized chromosomes, which could be a result of microchromosome fusions. Chromosome-specific probes for GGA9, 16, and W were utilized to gain insight regarding the nature of autosome and sex chromosome alterations within the OU2 karyotype. The karyotypes were described previously for DF-1 (derivative chromosomes and aneuploidies, and exhibiting diploid, haploid and tetraploid lineages; O'Hare and Delany [2009]) and DT40 (diploid with trisomy for GGA2; Chang and Delany [2004]), which both differ from the normal chicken karyotype [Ladjali-Mohammedi et al., 1999; Masabanda et al., 2004].
A 5S rDNA probe was utilized to examine the status of GGA9. This probe hybridized to 2 chromosomes of similar size and architecture, and a third larger chromosome with different architecture indicating a full or partial trisomy for GGA9 (fig. 1e). The larger chromosome exhibited a centrally located signal while the other 2 chromosomes indicated a signal near the terminus and as expected for a normal GGA9.
Chromosome 16 encodes the 18S-5.8S-28S rDNA array and is identified by the ETS probe. In OU2, GGA16 indicated unusual features, in that the ETS probe mapped to a larger chromosome than expected (analysis of normal GGA16 was described in Delany et al. [2009]), suggesting that the OU2 GGA16 is part of a derivative chromosome or that the 18S-5.8S-28S rDNA region alone is translocated (fig. 1b). The 2 chromosomes identified by the ETS probe differ in size, probe location, and probe intensity (i.e. one of the signals is stronger than the other). In addition, the chromosome with the stronger ETS rDNA signal exhibits a brighter terminal telomeric DNA signal than the other.
Interestingly, the GGAW chromosome probe did not hybridize to any of the OU2 chromosomes, nor a chromosome with the appropriate morphology, size, or DAPI-staining pattern was observed. The lack of a GGAW chromosome could be by happenstance, i.e. that the embryos used to create the original cell culture were male (ZZ) or that the W was lost during in vitroculture. In addition, although GGAZ is easily distinguishable by size (5th largest chromosome) and architecture (metacentric), a distinct Z was not visible in these cells; however, it is possible that the GGAZ of OU2 is part of a derivative chromosome (fused to another chromosome). Previously, it was discovered that DF-1 lacks a morphologically distinct GGAZ and the GGAW was present in 2 copies, 1 with normal appearance and 1 derivative [O'Hare and Delany, 2009].
DF-1 and OU2 Cell Growth and Telomerase Activity Profiles
DF-1 and OU2 cells were cultured over a period of several weeks to accommodate assay sampling which allowed a comparison of growth characteristics. In general, the DF-1 cells had a faster growth rate requiring more frequent passaging (faster time to confluency) than the OU2 cells, given the same culture conditions. Population doubling (PD) time (calculated as per Swanberg and Delany [2003]) was 0.23 PD/day for OU2 versus 0.72 for DF-1 based on representative flasks at passage (P) 5 and P6 for DF-1, and P12 and P13 for OU2. The P and days in culture (DIC) values reported for these cells are experimental (within this study) and do not reflect the overall values of these cell lines (i.e. DF-1 was obtained from ATCC at P149; overall passage number is unknown for OU2).
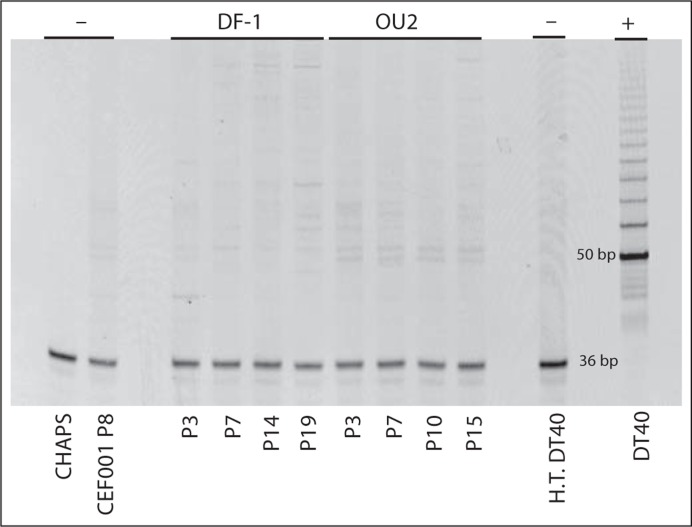
Telomerase activity was analyzed at multiple passages to establish the stability of the profile over time. DF-1 was analyzed at P3 (DIC 7), P7 (DIC 14), P14 (DIC 28), and P19 (DIC 41). OU2 was analyzed at P3 (DIC 13), P7 (DIC 28), P10 (DIC 41), and P15 (DIC 60). Both DF-1 and OU2 were telomerase negative at all sampling points. Figure 3 shows the telomerase activity assay results.
Fig. 3.
Absence of telomerase activity in the immortalized CEF lines DF-1 and OU2. Telomerase activity was determined using the TRAPeze Telomerase Detection Kit (S7700; Millipore). The immortalized cell lines, DF-1 and OU2, were collected at several passages (P) to detect telomerase activity, and both lines were negative for telomerase activity. Normal, i.e. mortal CEFs (CEF001) known to lack telomerase activity [Swanberg and Delany, 2003] were used as a negative control (–), along with CHAPS buffer-only and heat-treated (H.T.) DT40 cells. DT40 cells which maintain high telomerase activity [Swanberg and Delany, 2003] served as a positive control (+). The PCR control band (36 bp) shows that the lack of activity in DF-1 and OU2 was not due to a failed PCR reaction. Samples which are positive for telomerase activity show a band at 50 bp followed by bands at 6-bp increments.
Expression of Genes in the Telomerase and ALT Pathways
Genes were selected for expression analysis that are telomerase related, telomere binding, and/or required for ALT [Pickett and Reddel, 2009]. Notably, many genes in the ALT pathway are involved in the homologous recombination and DNA repair pathways and in some cases, genes of the telomerase, telomere-binding, and ALT pathways overlap. Table 1 lists gene and primer details. All qPCR amplicons were sequence verified to affirm identity of the expected sequence. Table 2 shows the gene expression results as fold differences and statistical significance among the values. The mortal telomerase-negative cell line CEF001 was used as the calibrator and all gene expression fold differences are expressed relative to CEF001 with the exception of TERT wherein gastrula was the calibrator since the CEF001 cells had no detectable TERT expression. The gastrula was included as a reference point and to evaluate results with regard to previous research [Swanberg et al., 2004; Swanberg and Delany, 2005; O'Hare and Delany, 2005]. As shown in table 2, TERT was not expressed in the telomerase-negative samples (CEF001, DF-1 and OU2) while TR and DKC1 (dyskeratosis congenita 1, also known as dyskerin) were expressed by all cell types. Interestingly, with regard to the recombination and ALT-associated genes including RAD51, RAD52, MRE11, RAD50, NBS1, SMC5, and SMC6, all show upregulation of transcripts in DF-1 and OU2. The greatest upregulation was exhibited by RAD51 and NBS1 for both DF-1 and OU2, with DF-1 being 5.2-fold and 11-fold upregulated and OU2 being 28.9-fold and 83.1-fold upregulated, respectively. Both PML genes were expressed in all of the cell lines; both PML-L and PML-S were upregulated in OU2, whereas in DF-1, PML-L was downregulated and PML-S was similar to CEF001. It is interesting that for DT40, which has a high level of telomerase activity (Swanberg and Delany [2003]; and this study, see fig. 3) and a high rate of homologous recombination [Takata et al., 1998], all of the genes in the telomerase, telomere-associated, recombination, and ALT pathways were upregulated, except PML-L and PML-S which were downregulated. MYC is dysregulated in DT40 due to the insertion of the avian leukosis virus (causative for the B-cell lymphoma from which the DT40 cell line was derived; Baba et al. [1985]) and, as expected and shown previously [Swanberg and Delany, 2005], MYC shows elevated expression (32-fold increase) in DT40.
Table 2.
. Quantitative PCR expression analysis of genes involved with telomerase and ALT pathways of telomere maintenance
| Genes | CEF001 | DF-1 | OU2 | DT40 | Gastrula | ANOVA p value |
|---|---|---|---|---|---|---|
| TR | 1a | 4.3 ↑a, b | 3.6 ↑a, b | 26.9 ↑b | 3.4 ↑a, b | 0.037 |
| TERT* | – | – | – | 1.4 ↑a | 1a | 0.849 |
| DKC1 | 1a | 3.8 ↓b | 1.5 ↓a, b | 2.1 ↑a | 1.5 ↑a | 0.008 |
| MYC | 1a, b | 1.9 ↑a | 4.2 ↑b | 32.1 ↑c | 2.3 ↓d | <0.001 |
| KU70 | 1a | 3.2 ↓a | 1.3 ↓a | 1.3 ↑a | 1.5 ↑a | 0.229 |
| KU80 | 1a, b | 1.5 ↑a, b | 2.1 ↑b | 1.2 ↑a, b | 1.2 ↓a | 0.037 |
| TRF1 | 1a | 5.0 ↓b | 1.8 ↑a | 1.5 ↑a | 3.0 ↓b | 0.003 |
| TRF2 | 1a | 1.2 ↓a | 1.3 ↓a | 1.5 ↑a | 1.1 ↓a | 0.137 |
| RAP1 | 1a | 3.1 ↓a | 1.4 ↓a | 1.4 ↑a | 2.6 ↓a | 0.074 |
| RAD51 | 1a | 5.2 ↑a, c | 28.9 ↑b | 8.0 ↑c | 3.1 ↑a, c | 0.005 |
| RAD52 | 1a | 1.9 ↑a | 4.4 ↑a | 2.1 ↑a | 1.4 ↓a | 0.178 |
| MRE11 | 1a | 2.0 ↑a, b | 5.4 ↑b | 1.7 ↑a, b | 1.1 ↓a | 0.027 |
| RAD50 | 1a | 1.3 ↑b, c | 2.8 ↑c | 1.7 ↑a, b | 1.4 ↓a | 0.002 |
| NBS1 | 1a | 11.0 ↑a | 83.1 ↑b | 5.7 ↑a | 2.3 ↑a | 0.003 |
| SMC5 | 1a | 2.5 ↑a, b | 5.8 ↑b | 2.1 ↑a | 1.0a | 0.005 |
| SMC6 | 1a | 1.1 ↑a | 2.9 ↑a | 1.7 ↑a | 1.4 ↓a | 0.181 |
| PML-L | 1a | 2.9 ↓b | 3.6 ↑c | 15.4 ↓d | 1.1 ↑a | <0.001 |
| PML-S | 1a, b | 1.2 ↑a, b | 7.1 ↑b | 6.0 ↓a | 4.5 ↓a | 0.032 |
The comparative Ct method was used to establish fold-difference expression changes using a calculated average efficiency of 1.85 for a final equation of 1.85−ΔΔCt [O'Hare and Delany, 2005]; CEF001 was used as the calibrator and is set to 1. Statistical evaluation of the normalized Ct values was conducted using ANOVA and Tukey's pairwise comparison. For each gene, the values with different superscript letters were significantly different (p < 0.05). The cells utilized for the expression analysis were at the following experimental passage and days in culture: CEF001 – P8 (DIC 24), DF-1 – P14 (DIC 28), OU2 – P7 (DIC 28).
↑ = Fold difference, increase; ↓ = fold difference, decrease.
TERT transcripts were not detectable (–) following 40 qPCR cycles for CEF001, DF-1, and OU2 whereas the gastrula embryo (calibrator) and DT40 had expression.
Nuclear Expression of PML and TRF2 Proteins
Both PML proteins were detected in the nuclei of the 4 cell lines by IF utilizing the chicken-specific antibodies (see Materials and Methods for details on antibodies). Table 3 indicates the percentage of cells positive for the PML and TRF2 proteins. DF-1 had the smallest percentage of cells positive for PML proteins (e.g. 48% for PML-L in DF-1 vs. 94% in CEF001). Interestingly, DF-1 is heterozygous for a derivative GGA10 (which encodes the PML genes) and has a high number of haploid cells [O'Hare and Delany, 2009], which could be one possible explanation for a lack of expression in some cells. Table 3 also shows the number of PML signals per cell. On average, DF-1 exhibited the lowest number of PML signals (2.6 for both PML proteins) and OU2 had the greatest number of signals (28.7 and 16.1 for PML-L and PML-S, respectively) relative to the other cell systems.
Table 3.
. Immunofluorescence analysis of PML and TRF2 proteins in nuclei of chicken cells
| Proteins | Immunofluorescence analysis | CEF001 | DF-1 | OU2 | DT40 |
|---|---|---|---|---|---|
| PML-L | positive cells (%) | 94 | 48 | 99 | 92 |
| signals/cell average ± S.D. | 12.8 ± 5.6 | 2.6 ± 1.9 | 28.7 ± 7.7 | 4.4 ± 2.5 | |
| signals/cell range (low–high) | 1–29 | 1–10 | 3–101 | 1–15 | |
| PML-S | positive cells (%) | 94 | 42 | 96 | 47 |
| signals/cell average ± S.D. | 7.6 ± 4.0 | 2.6 ± 1.5 | 16.1 ± 6.1 | 2.5 ± 1.3 | |
| signals/cell range (low–high) | 1–27 | 1–9 | 2–45 | 1–7 | |
| TRF2 | positive cells (%) | 100 | 100 | 98 | 99 |
| signals/cell average ± S.D. | 29.8 ± 7.0 | 29.8 ± 6.5 | 32.8 ± 6.7 | 16.7 ± 4.8 | |
| signals/cell range (low–high) | 14–83 | 8–64 | 8–64 | 4–34 | |
In addition to the percentage of positive cells for the protein, the number of protein signals (dots) was determined. Values are the average signals/cell ± standard deviation (S.D.) and the range (low–high) of signals/cell. At least 120 cells were counted for each cell line and protein assayed.
DF-1, OU2, and CEF001 shared a similar number of signals for TRF2 (averages of 29.8–32.8 per cell) and about twice as many TRF2 signals compared to DT40 (16.7 per cell) which aligns with expectations since TRF2 binds to the telomere [Smogorzewska et al., 2000] and DT40 has shorter telomeres with a reduced amount of telomeric DNA. Figure 4 shows a representative example of nuclei assayed by multi-color IF for the PML and TRF2 proteins individually, as well as PML/TRF2 colocalization.
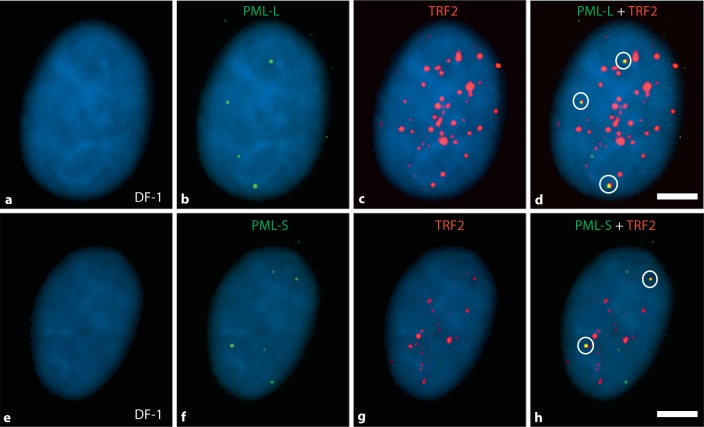
Fig. 4.
PML and TRF2 proteins in chicken nuclei: evidence for APB foci. Immunofluorescence detection of PML-L, PML-S, and TRF2 proteins was conducted using chicken-specific antibodies to detect colocalization of PML/TRF2 as an assay for the presence of ALT. Representative results are shown here for the DF-1 cell line. Images a–d show 1 cell from an experiment employing PML-L (green) and TRF2 (red); images e–h show a cell from an experiment employing PML-S (green) and TRF2 (red). Images d and h show merged images of all colors illustrating colocalization of PML-L or PML-S with TRF2, i.e. APB foci (yellow signals, 3 in image d and 2 in image h). Signal counts were conducted for PML-L, PML-S, and TRF2, and the data are shown in table 3. Colocalization of the PML and TRF2 (APB foci) was determined for each of the cell lines, and the results are shown in figure 5. Scale bar = 5 μm.
Since colocalization of PML and TRF2 proteins serves as a marker for presence of the ALT pathway [Jiang et al., 2009], each cell line was analyzed for the degree of colocalization, i.e. the percentage of cells with colocalized signals (APB foci) and the number of APB foci per cell. A cell was counted as positive for APB foci if there was at least 1 PML signal that colocalized with a TRF2 signal. Only cells positive for both PML and TRF2 were analyzed for the presence of APB foci. Each of the cell lines exhibited APB foci, however, there was a hierarchical order observed. The immortalized cell lines DF-1 and OU2 exhibited the greatest percentage of cells exhibiting APB foci followed by CEF001, and then DT40 with the lowest level. The total percentage of cells positive for APB foci per cell line was as follows: DF-1 − 80% and 75%, OU2 − 70% and 63%, CEF001 − 54% and 49%, and DT40 − 32% and 32%, for PML-L/TRF2 and PML-S/TRF2, respectively. The frequency of APB foci signals (range of 0–7 for PML-L/TRF2 signals; range of 0–5 for PML-S/TRF2 signals) are shown in figure 5. Interestingly, although DF-1 had the lowest average number of PML signals per cell (2.6 on average, see table 3) and the lowest percentage of cells positive for the PML-proteins (48% for PML-L and 42% for PML-S), DF-1 exhibited the greatest percentage of cells positive for APB foci.
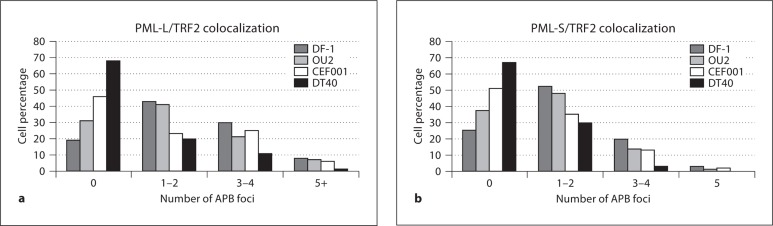
Fig. 5.
APB foci frequency in chicken cells having different proliferation phenotypes including mortal (CEF), immortalized (DF-1 and OU2) and transformed (DT40). For each cell line, a minimum of 100 cells were analyzed and the number of APB foci (colocalized PML/TRF2 signals in cells positive for these proteins). Shown in these graphs are results for PML-L/TRF2 (a) and PML-S/TRF2 (b). The percentage of cells (y-axis) with 0–7 (PML-L/TRF2) or 0–5 (PML-S/TRF2) APB foci (x-axis). The percentages of cells positive for APB foci were 80% and 75% in DF-1, 70% and 63% in OU2, 54% and 49% in CEF001, and 32% and 32% in DT40 for the PML-L/TRF2 and PML-S/TRF2 assays, respectively.
PML-L and PML-S Western Blot Analysis
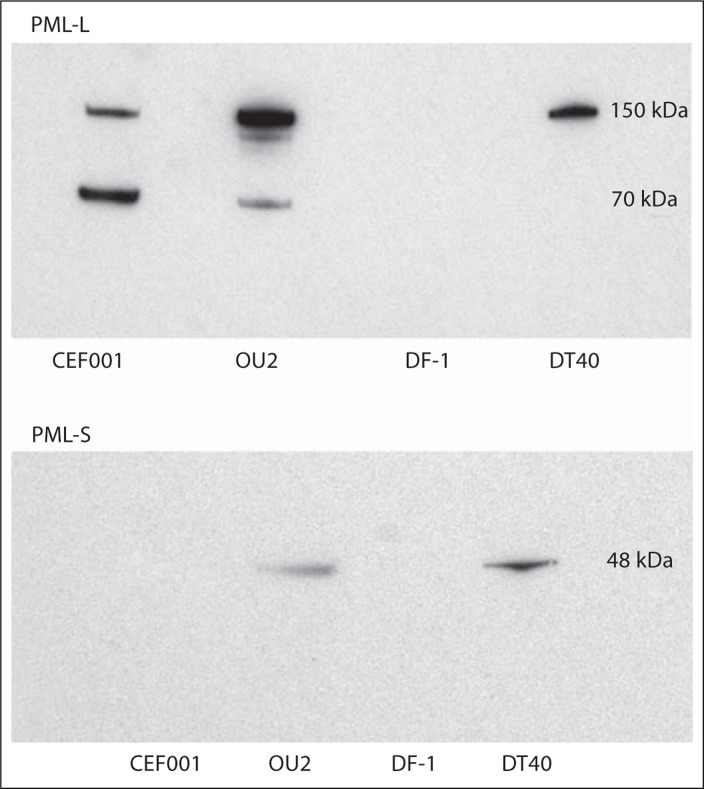
The PML chicken-specific antibodies were utilized for immunoblot analysis (Western blots) to investigate the size of the PML proteins (fig. 6). With regard to PML-L, DT40 exhibited a protein band at 150 kDa, while OU2 and CEF001 indicated protein bands at 150 kDa and 70 kDa. These results suggest the possibility of cell-specific PML-L isoforms. For PML-S, DT40 and OU2 indicated a protein band at 48 kDa. The 150-kDa size for PML-L and the 48-kDa size for PML-S generally fit the predicted sizes of 113 kDa (PML-L) and 59 kDa (PML-S) for the aa sequences as calculated utilizing the Science Gateway molecular-weight calculator (http://www.sciencegateway.org/tools/proteinmw.htm). Interestingly, CEF001 did not exhibit a protein band for PML-S and DF-1 did not exhibit a band for either PML protein even when the concentration of protein was increased by more than 50% (to 160 μg from 100 μg of protein for the standard assay). The reason for this variability in immunoblot results is not clear; the most likely cause for cell-type-specific variability is a concentration issue especially since the antibodies worked by cellular IF analysis. That is, the PML protein concentration of CEF001 and DF-1 may be below the limits of detection by immunoblotting; noteworthy in this regard is the overall low percentage of PML-positive cells in DF-1 (42–48%) and the low signal number (2.6 signals per cell on average) relative to the other cells. Additionally, DF-1 being heterozygous for a derivative GGA10 and maintaining a large proportion (14%) of haploid cells [O'Hare and Delany, 2009] could account for the detection issues, and this cell line may require larger amounts of total protein for immunoblot detection of the PML proteins. The issue with the CEF001 cells, given the high level of PML-S-positive cells (94%) by IF analysis, is very likely a technical issue related to protein concentration, which was not explored in these experiments.
Fig. 6.
Western blots of chicken PML-L and PML-S proteins detected in different cell types. Each of the 4 cell lines was analyzed using the 2 anti-chicken PML antibodies designed for this study (see Materials and Methods). One hundred micrograms of total protein were analyzed for each cell line. For PML-L, DT40 exhibited a protein at 150 kDa, while OU2 and CEF001 exhibited proteins at both 150 kDa and 70 kDa. For PML-S, DT40 and OU2 showed a protein at 48 kDa. Interestingly, no PML-S proteins were detected in CEF001 and neither PML-L nor PML-S proteins were detected in DF-1 even when the concentration of protein was increased from 100 μg to 160 μg.
Discussion
Human immortalized cells which lack telomerase maintain telomeres by employing a pathway called ALT. Hallmark characteristics of cells utilizing the ALT pathway include being telomerase negative, possessing unusually heterogeneous telomere lengths among chromosomes (relative to normal cells), presence of APBs, and displaying extrachromosomal telomeric DNA [Cesare and Reddel, 2010]. The APBs are PML bodies that also contain telomeric DNA, telomere-associated proteins (TRF1, TRF2, and RAP1), and DNA repair and recombination proteins (RAD51, RAD52, MRE11, RAD50, and NBS1). The PML protein forms nuclear bodies in most cells [reviewed in Lamond and Earnshaw, 1998; Lallemand-Breitenbach and de Thé, 2010] with 5–30 PML nuclear bodies per nucleus ranging in size from 0.1–1 μm [Zhong et al., 2000; Lallemand-Breitenbach and de Thé, 2010]. Many of the protein components within PML nuclear bodies are involved in transcriptional regulation [Zhong et al., 2000]. The PML-containing APBs are a marker of the ALT pathway [Cesare and Reddel, 2008].
The chicken shares many telomere biology features with human and has the advantage that a number of well-studied versatile in vitro cell systems with phenotype including mortal, immortalized, and transformed exist [Delany et al., 2003; Swanberg and Delany, 2006; Swanberg et al., 2010]. Our study explored the hypothesis that ALT operates in immortalized chicken cells. The results provide evidence that immortalized telomerase-negative chicken cells are positive for markers of ALT including the presence of APBs. In addition, the APBs were found in telomerase-positive cells (DT40) and in telomerase-negative mortal cells (CEF001). Clearly, there was a hierarchical order to the extent of the APB marker, with the telomerase-negative immortalized cells having the greatest number of cells positive for APB foci relative to the other cell types.
Numerous studies support the hypothesis that telomere recombination is involved in yeast and human ALT pathways [Lundblad and Blackburn, 1993; Bryan et al., 1995; Teng and Zakian, 1999; Dunham et al., 2000; Cesare and Reddel, 2008, 2010; Morrish and Greider, 2009]. Recently, Draskovic et al. [2009] showed the involvement of APBs in telomere recombination in ALT human cell lines via anaphase bridges between nonhomologous chromosomes proximal to APBs. Therein, APBs beyond being a marker for ALT appear to be a mechanistic component fostering telomere-telomere recombination.
Knowledge and understanding of the ALT pathway for telomere maintenance continues to emerge. Fasching et al. [2005] reported a lack of APBs in telomerase-negative immortalized cells having heterogeneous telomere lengths which displayed rapid changes suggesting a mechanism similar to other ALT cells. The existence of ALT-like APB-negative cells suggests the possibility that more than one ALT pathway exists. Interestingly, when ALT cells and telomerase-positive cells were fused, telomeric loss was observed along with repression of the ALT pathway [Perrem et al., 1999], suggesting that telomerase could repress the ALT pathway. However, telomerase activity produced by transfecting TERT and TR into ALT cells led to the creation of a cell system with both ALT and telomerase [Cerone et al., 2001], affirming the possibility that ALT and telomerase pathways could coexist. Additionally, HeLa and HT1080 (telomerase-positive transformed human cell lines) transfected with TR were observed with APB foci in about 3% of the cells [Pickett et al., 2009], indicating that APBs could be present in telomerase-positive cells without functional ALT. Combined, these results illustrate that there is variability in the ALT pathway and possibly redundant telomere maintenance pathways operating simultaneously. Herein, results of this study suggest that chicken may offer a model having inherent redundancy for telomere maintenance, wherein both ALT and telomerase pathways may be present, a hypothesis for which further study is warranted.
Beside the observations of colocalization of PML and protein components which form APB foci [Yeager et al., 1999; Jiang et al., 2009], individual components have been disrupted or altered in order to investigate their role in APB formation and/or ALT function. A conditional, truncated NBS1 protein expression system established that MRE11 and RAD50 recruitment to APBs and assembly of APBs require NBS1 [Wu et al., 2003]. Using RNA interference, it was determined that PML, TRF1, TRF2, TIN2, RAP1, MRE11, and RAD50 are required for APB formation [Jiang et al., 2007]. The MRN complex (MRE11, RAD50, NBS1) proteins were examined using shRNA-mediated knockdown; loss of each of the proteins was shown to decrease the number of APBs and decrease telomere length [Zhong et al., 2007], providing evidence that the MRN complex is necessary for the ALT pathway.
The chicken model has several unique features useful for research on the molecular and cellular aspects of ALT. The chicken genome has a high basal rate of recombination [International Chicken Genome Sequencing Consortium (ICGSC), 2004; Groenen et al., 2009; Elferink et al., 2010], additionally, telomere recombination is a feature of meiosis [Rodrigue et al., 2005]. The chicken genome normally has an extensive heterogeneous telomeric DNA profile [Delany et al., 2000; Rodrigue et al., 2005; O'Hare and Delany, 2009] relative to other well-studied vertebrates. In addition to normally heterogeneous telomeric lengths, it was observed that telomerase-negative mortal (CEF001) and telomerase-positive (DT40) cells exhibit APBs, albeit at lower levels than the telomerase-negative immortalized cells. Thus, the presence of 2 predominant markers of ALT, APBs and telomeric heterogeneity in a variety of chicken cell types, suggests that both pathways may be operational in chicken cells. The DT40 cell system could be particularly valuable for future research of the interplay between ALT and telomerase pathways given its high rate of homologous recombination [Takata et al., 1998], versatility in knock-out/knock-in experiments [Buerstedde and Takeda, 1991; Barr et al., 2009], high telomerase activity [Swanberg and Delany, 2003] and that it was herein shown to exhibit APB foci.
Conclusion
The analysis of ALT in chicken in vitro cell systems indicates that ALT is present within immortalized cells as per the key markers: absence of telomerase activity, heterogeneous telomeric DNA profiles, and presence of APB foci. Gene expression profiles show that many of the genes involved in the ALT and recombination pathways are upregulated in immortalized chicken cells, some in a striking amount (e.g. RAD51, NBS1) relative to levels of nonimmortalized cells. In addition, these results suggest that the telomerase and telomere-recombination (ALT) pathways may coexist in chicken cells; however, depending on the cell type, one or the other mechanism may dominate.
Acknowledgements
The authors appreciate the generous gift of anti-chicken TRF2 antibody from Dr. Christine Farr (University of Cambridge). Discussions with Drs. Jerry Dodgson (Michigan State University) and Janet Weber (National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health) were helpful for the genomic analysis of the chicken (Gallus gallus) PML sequences. This project was supported by National Research Initiative Grant no. 2005-35205-16679 from the USDA National Institute of Food and Agriculture (NIFA) Animal Genomics program and USDA-NIFA Multistate Research Projects NE-1016 (CA-D*-ASC-7281-RR) and NRSP-8 (CA-D*-ASC-7233-RR). Graduate student financial support was provided in part by the University of California Austin Eugene Lyons fellowship. We gratefully acknowledge the UC Davis Department of Animal Science and College of Agricultural and Environmental Sciences, and the California Agricultural Experiment Station for infrastructure support.
References
- Baba TW, Giroir BP, Humphries EH. Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology. 1985;144:139–151. doi: 10.1016/0042-6822(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Barr AR, Zyss D, Gergely F. Knock-in and knock-out: The use of reverse genetics in somatic cells to dissect mitotic pathways. Methods Mol Biol. 2009;545:1–19. doi: 10.1007/978-1-60327-993-2_1. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Reddel RR. Telomere dynamics and telomerase activity in in vitro immortalised human cells. Eur J Cancer. 1997;33:767–773. doi: 10.1016/S0959-8049(97)00065-8. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- Buerstedde JM, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- Cerone MA, Londono-Vallejo JA, Bacchetti S. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum Mol Genet. 2001;10:1945–1952. doi: 10.1093/hmg/10.18.1945. [DOI] [PubMed] [Google Scholar]
- Cerone MA, Autexier C, Londono-Vallejo JA, Bacchetti S. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT. Oncogene. 2005;24:7893–7901. doi: 10.1038/sj.onc.1208934. [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR. Alternative lengthening of telomeres in mammalian cells. In: Nosek J, Tomáška Ľ, editors. Origin and Evolution of Telomeres. Austin: Landes Bioscience; 2008. pp. 45–57. [Google Scholar]
- Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- Chang H, Delany ME. Karyotype stability of the DT40 chicken B cell line: Macrochromosome variation and cytogenetic mosaicism. Chromosome Res. 2004;12:299–307. doi: 10.1023/b:chro.0000021947.17128.96. [DOI] [PubMed] [Google Scholar]
- Chang S, Khoo CM, Naylor ML, Maser RS, DePinho RA. Telomere-based crisis: Functional differences between telomerase activation and ALT in tumor progression. Genes Dev. 2003;17:88–100. doi: 10.1101/gad.1029903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman SA, Kong BW, Landry MM, Kim H, Foster DN. Modulation of p53 expression and its role in the conversion to a fully immortalized chicken embryo fibroblast line. FEBS Lett. 2005;579:6705–6715. doi: 10.1016/j.febslet.2005.10.066. [DOI] [PubMed] [Google Scholar]
- Cooley C, Baird KM, Faure V, Wenner T, Stewart JL, et al. Trf1 is not required for proliferation or functional telomere maintenance in chicken DT40 cells. Mol Biol Cell. 2009;20:2563–2571. doi: 10.1091/mbc.E08-10-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany ME, Krupkin AB, Miller MM. Organization of telomere sequences in birds: Evidence for arrays of extreme length and for in vivo shortening. Cytogenet Cell Genet. 2000;90:139–145. doi: 10.1159/000015649. [DOI] [PubMed] [Google Scholar]
- Delany ME, Daniels LM, Swanberg SE, Taylor HA. Telomeres in the chicken: genome stability and chromosome ends. Poult Sci. 2003;82:917–926. doi: 10.1093/ps/82.6.917. [DOI] [PubMed] [Google Scholar]
- Delany ME, Gessaro TM, Rodrigue KL, Daniels LM. Chromosomal mapping of chicken mega-telomere arrays to GGA9, 16, 28, and W using a cytogenomic approach. Cytogenet Genome Res. 2007;117:54–63. doi: 10.1159/000103165. [DOI] [PubMed] [Google Scholar]
- Delany ME, Robinson CM, Goto RM, Miller MM. Architecture and organization of chicken microchromosome 16: Order of the NOR, MHC-Y, and MHC-B subregions. J Hered. 2009;100:507–514. doi: 10.1093/jhered/esp044. [DOI] [PubMed] [Google Scholar]
- Draskovic I, Arnoult N, Steiner V, Bacchetti S, Lomonte P, Londono-Vallejo A. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc Natl Acad Sci USA. 2009;106:15726–15731. doi: 10.1073/pnas.0907689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Elferink MG, van As P, Veenendaal T, Crooijmans RP, Groenen MA. Regional differences in recombination hotspots between two chicken populations. BMC Genet. 2010;11:11. doi: 10.1186/1471-2156-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching CL, Bower K, Reddel RR. Telomerase-independent telomere length maintenance in the absence of alternative lengthening of telomeres-associated promyelocytic leukemia bodies. Cancer Res. 2005;65:2722–2729. doi: 10.1158/0008-5472.CAN-04-2881. [DOI] [PubMed] [Google Scholar]
- Groenen MA, Wahlberg P, Foglio M, Cheng HH, Megens HJ, et al. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 2009;19:510–519. doi: 10.1101/gr.086538.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himly M, Foster DN, Bottoli I, Iacovoni JS, Vogt PK. The DF-1 chicken fibroblast cell line: Transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology. 1998;248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium (ICGSC) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Jiang WQ, Zhong ZH, Henson JD, Reddel RR. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene. 2007;26:4635–4647. doi: 10.1038/sj.onc.1210260. [DOI] [PubMed] [Google Scholar]
- Jiang WQ, Henson JD, Axel AN, Roger RR. Protocol III: detection of alternative lengthening of telomeres. In: Hiyama K, editor. Telomeres and Telomerase in Cancer. New York City: Humana Press; 2009. pp. 351–364. [Google Scholar]
- Ladjali-Mohammedi K, Bitgood JJ, Tixier-Boichard M, Ponce de Leon FA. International system for standardized avian karyotypes (ISSAK): standardized banded karyotypes of the domestic fowl (Gallus domesticus) Cytogenet Cell Genet. 1999;86:271–276. doi: 10.1159/000015318. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de Thé H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Masabanda JS, Burt DW, O'Brien PC, Vignal A, Fillon V, et al. Molecular cytogenetic definition of the chicken genome: The first complete avian karyotype. Genetics. 2004;166:1367–1373. doi: 10.1534/genetics.166.3.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish TA, Greider CW. Short telomeres initiate telomere recombination in primary and tumor cells. PLoS Genet. 2009;5:e1000357. doi: 10.1371/journal.pgen.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Shinkai Y, Hande MP, Matsumoto T, Takehara S, et al. Telomere maintenance in telomerase-deficient mouse embryonic stem cells: Characterization of an amplified telomeric DNA. Mol Cell Biol. 2000;20:4115–4127. doi: 10.1128/mcb.20.11.4115-4127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura H, Fujiwara T. Establishment and characterization of a virus-free chick cell line. Acta Med Okayama. 1987;41:141–143. doi: 10.18926/AMO/31758. [DOI] [PubMed] [Google Scholar]
- O'Hare TH, Delany ME. Telomerase gene expression in the chicken: telomerase RNA (TR) and reverse transcriptase (TERT) transcript profiles are tissue-specific and correlate with telomerase activity. AGE. 2005;27:257–266. doi: 10.1007/s11357-005-4558-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare TH, Delany ME. Genetic variation exists for telomeric array organization within and among the genomes of normal, immortalized, and transformed chicken systems. Chromosome Res. 2009;17:947–964. doi: 10.1007/s10577-009-9082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddada S, Yasui DH, LaSalle JM. Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome. Hum Mol Genet. 2006;15:2003–2014. doi: 10.1093/hmg/ddl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrem K, Bryan TM, Englezou A, Hackl T, Moy EL, Reddel RR. Repression of an alternative mechanism for lengthening of telomeres in somatic cell hybrids. Oncogene. 1999;18:3383–3390. doi: 10.1038/sj.onc.1202752. [DOI] [PubMed] [Google Scholar]
- Pickett HA, Reddel RR. Alternative lengthening of telomeres in human cells. In: Hiyama K, editor. Telomeres and Telomerase in Cancer. New York City: Humana Press; 2009. pp. 127–148. [Google Scholar]
- Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue KL, May BP, Famula TR, Delany ME. Meiotic instability of chicken ultra-long telomeres and mapping of a 2.8 megabase array to the W-sex chromosome. Chromosome Res. 2005;13:581–591. doi: 10.1007/s10577-005-0984-7. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, et al. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, et al. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanberg SE, Delany ME. Dynamics of telomere erosion in transformed and non-transformed avian cells in vitro. Cytogenet Genome Res. 2003;102:318–325. doi: 10.1159/000075769. [DOI] [PubMed] [Google Scholar]
- Swanberg SE, Delany ME. Differential expression of genes associated with telomere length homeostasis and oncogenesis in an avian model. Mech Ageing Dev. 2005;126:1060–1070. doi: 10.1016/j.mad.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Swanberg SE, Delany ME. Telomeres in aging: birds. In: Conn PM, editor. Handbook of Models for Human Aging. Burlington: Elsevier Academic Press; 2006. pp. 339–349. [Google Scholar]
- Swanberg SE, Payne WS, Hunt HD, Dodgson JB, Delany ME. Telomerase activity and differential expression of telomerase genes and c-myc in chicken cells in vitro. Dev Dyn. 2004;231:14–21. doi: 10.1002/dvdy.20149. [DOI] [PubMed] [Google Scholar]
- Swanberg SE, Nagarajan RP, Peddada S, Yasui DH, LaSalle JM. Reciprocal co-regulation of EGR2 and MECP2 is disrupted in Rett syndrome and autism. Hum Mol Genet. 2009;18:525–534. doi: 10.1093/hmg/ddn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanberg SE, O'Hare TH, Robb EA, Robinson CM, Chang H, Delany ME. Telomere biology of the chicken: A model for aging research. Exp Gerontol. 2010;45:647–654. doi: 10.1016/j.exger.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Jiang X, Lee WH, Chen PL. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res. 2003;63:2589–2595. [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- Zhong ZH, Jiang WQ, Cesare AJ, Neumann AA, Wadhwa R, Reddel RR. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]