Abstract
We isolated a cDNA encoding a 568-amino acid, heat-stress-induced peptidyl prolyl isomerase belonging to the FK506-binding-protein (FKBP) family. The open reading frame encodes for a peptidyl prolyl isomerase that possesses three FKBP-12-like domains, a putative tetratricopeptide motif, and a calmodulin-binding domain. Specific antibodies showed that the open reading frame encodes a heat-induced 77-kD protein, the wheat FKBP77 (wFKBP77), which exhibits 84% identity with the wFKBP73 and 42% identity with the human FKBP59. Because of the high similarity in sequence to wFKBP73, wFKBP77 was designated as the heat-induced isoform. The wFKBP77 mRNA steady-state level was 14-fold higher at 37°C than at 25°C. The wFKBP77 transcript abundance was the highest in mature embryos that had imbibed and 2-d-old green shoots exposed to 37°C, and decreased to 6% in 6-d-old green shoots. The transcript level returned to the level detected at 25°C after recovery of the embryos for 90 min at 25°C. We compared wFKBP73 and wFKBP77 with the heat-shock proteins having cognate and heat-stress-induced counterparts.
Protein folding in vivo is mediated by an array of proteins that act as molecular chaperones, as foldases, or both. The molecular chaperones were originally defined as unrelated classes of proteins that mediate the correct assembly of other proteins, but are not themselves components of the final functional structures (Ruddon and Bedows, 1997). They occur ubiquitously and many of them are classified as stress proteins, although they have essential functions under normal growth conditions (Boston et al., 1996; Buchner, 1996; Hartl, 1996). When cells are exposed to elevated temperatures or other environmental stresses, heat-shock proteins belonging to several gene families are induced. Many heat-shock proteins function as chaperones and play important roles in normal growth and stress tolerance. There is increasing evidence that the function of chaperones in preventing or reversing protein aggregation is linked to their role in presenting misfolded intermediates to the cellular machinery for proteolytic degradation (Hayes and Dice, 1996).
The foldases are catalysts accelerating slow steps in the folding of some proteins by rearrangements of disulfide bonds by the protein disulfide isomerase or isomerization of peptide bonds by PPIases (Galat and Metcalfe, 1995; Schmid, 1998; Walker and Gilbert, 1998). PPIases are ubiquitous proteins found in the cytosol of both prokaryotic and eukaryotic cells (Fruman et al., 1994; Galat and Metcalfe, 1995; Chou and Gasser, 1997) and in organelles such as mitochondria and chloroplasts (Breiman et al., 1992; Luan et al., 1994a; Matouschek et al., 1995; Fulgosi et al., 1998).
There are three structurally distinct classes of PPIases: cyclophilins, which bind the immunosuppressive drug cyclosporin A (Handschumacher et al., 1984); FKBPs, which bind the macrolide drugs FK506 and rapamycin (Harding et al., 1989; Siekierka et al., 1989); and the parvulin family (Dolinski and Heitman, 1997). Because of their drug-binding activities, PPIases have also been called immunophilins. The binding of the drugs inhibits PPIase activity (Schreiber, 1991), but the drug can still serve as a molecular glue, affecting the formation of novel protein complexes involved in signal transduction in the immune-response pathway (Crabtree and Schreiber, 1996).
The FKBP family comprises several members, the names of which are suffixed with their molecular masses. The main FKBP is the cytosolic FKBP12 (Standaert et al., 1990), the structure of which has been established by high-resolution radiographic crystallography and NMR spectroscopy (Michnick et al., 1991; van Duyne et al., 1991, 1993). In mammalian cells a large cytosolic PPIase, FKBP59 (also called FKBP52, Hsp56 [heat-shock protein 56], p59, or HBI [heat-shock-protein binding immunophilin]), and CyP40 (cyclophilin 40) are components of the steroid hormone complex that includes the protein HSP90 (Sanchez, 1990; Callebaut et al., 1992; Peattie et al., 1992; Ratajczak et al., 1993). FKBP59 and CyP40 bind to HSP90 via the TPR domain and form stable complexes (Sanchez, 1990; Radanyi et al., 1994; Owens-Grillo et al., 1996a). A novel function was recently demonstrated for CyP40, the inhibition of c-myb DNA activity, which implicates the prolyl isomerase in the regulation of transcription, transformation, and differentiation (Leverson and Ness, 1998).
The mammalian FKBP59 has two FKBP12-like domains, of which only the first possesses PPIase activity that is inhibited by the drugs FK506 and rapamycin (Callebaut et al., 1992; Chambraud et al., 1993). The protein contains a TPR domain (Radanyi et al., 1994; Owens-Grillo et al., 1995) and a calmodulin-binding domain (Massol et al., 1992) at the C terminus.
Plant FKBPs have been identified by their capacity to bind to an FK506 column (Luan et al., 1994a). A fava bean FKBP, VfFKBP15 (Vicia fava FKBP15), which is homologous to the mammalian FKBP13, and two cDNAs from Arabidopsis showing homology to the VfFKBP15 have been cloned (Luan et al., 1996).
Plant homologs of FKBP59 have been cloned and characterized: the Arabidopsis 62-kD ROF1 (Vucich and Gasser, 1996) and PAS1FKBP70 (pasticcino; Vittorioso et al., 1998), the wheat wFKBP73 (Blecher et al., 1996), and the maize mzFKBP-66 (Hueros et al., 1998).
The large FKBPs from plants possess a similar structure, with three or four FKBP12-like domains in the N-terminal portion of the protein, the TPR, which is thought to be involved in protein-to-protein interaction, and a calmodulin-binding domain. However, the mRNA expression patterns of these FKBPs are different. For example, ROF1 is expressed at low levels in all organs and is induced by wounding and NaCl (Vucich and Gasser, 1996), whereas wFKBP73 is highly expressed in young tissues and is not induced by stress (Blecher et al., 1996). Characterization of the pas1 mutants of Arabidopsis revealed for the first time the involvement of a member of the FKBP family in plant development (Vittorioso et al., 1998). The inactivation of PAS1 causes ectopic cell proliferation in cotyledons, extra layers of cells in the hypocotyl, and an abnormal apical meristem, a phenotype correlated with cell division and the cell-elongation effect. It was also demonstrated that the mRNA steady-state level of pas1 was increased in the presence of cytokinin. This was the first example of involvement of FKBP in the control of cell proliferation possibly connected to the cytokinin signal (Vittorioso et al., 1998).
The ability of plant FKBPs to interact with other proteins has recently been documented for wFKBP73 (Owens-Grillo et al., 1996b; Breiman et al., 1997) and mzFKBP66, which was demonstrated to be associated with an unknown 36-kD polypeptide (Hueros et al., 1998). The biochemical evidence for the participation of plant FKBPs in multicomponent protein complexes, as has been shown for the mammalian immunophilins, and the involvement of PAS1 in cell proliferation, suggest an important role of this class of proteins in the control of plant development.
In this study we report the characterization of the heat-stress-induced wFKBP. This protein shows identity of 84% to wFKBP73, but its mRNA steady-state level increased 14-fold after exposure to 37°C, whereas the wFKBP73 was expressed at a similar level at 25°C and 37°C. The heat-stress-induced FKBP is developmentally regulated: it was detected only in young tissues, was more abundant in dark-grown seedlings, and accumulated during caryopsis maturation.
MATERIALS AND METHODS
Plant Material: Growth and Stress Conditions
Wheat (Triticum aestivum cv Atir) seeds were germinated on wet paper overnight or for 2 d in the dark at 25°C. Seedlings were transferred to vermiculite and grown for up to 6 d in the dark or in 16-h light/8-h dark cycles at 25°C. Anthers and pistils were taken from mature wheat plants grown in the nethouse. The most basal 1 cm of the shoots, the most distal 1 cm of the roots, and the most basal 1 cm of the leaves were dissected out and frozen immediately. The anthers and pistils were removed by dissection from plants 6 d before anthesis.
Heat-stress treatments were performed by exposing the tissue to 37°C for 2 h, unless indicated otherwise. The embryos (which were allowed to imbibe for 24 h), seedlings, and other dissected tissues were placed in a Petri dish with wet filter paper under conditions of humidity to avoid drought stress. Recovery from heat stress was performed by transfer of the tissues to Petri dishes at 25°C.
The illumination effect after heat stress was determined in 3-d-old etiolated seedlings by exposure to 8 h of light or darkness. As a control, seedlings were grown in light conditions (16-h light/8-h dark cycles) for 3 d and then kept for an additional 8 h in the light. All seedlings were kept at 25°C and exposed for 2 h at 37°C at the end of the treatment. The basal first 1 cm of each of the shoots was dissected and frozen immediately.
Stresses were then applied to 2-d-old etiolated seedlings. Cold stress was accomplished by placing the seedlings on ice in the cold room (4°C) for 4 d. The ABA treatment consisted of exposure of seedlings to 100 μm solutions for 1 or 2 d according to the method of Hong et al. (1992). The NaCl treatment consisted of incubation of the seedlings in 100, 200, or 300 mm for 4 or 8 h. The salicylic acid treatment consisted of watering the seedlings with a 10 mm solution, as described previously (Marivet et al., 1995). The CdCl2 treatment consisted of incubation of the seedlings in 0.05, 0.5, and 5 mm solution according to the method of Winter et al. (1988).
Isolation of the wFKBP77 cDNA
To isolate wFKBP77, a root-tip cDNA λ-ZAPII library (Clontech, Palo Alto, CA) was screened with the following two probes (Blecher et al., 1996): 524 bp of wFKBP73 NcoI, BamHI DNA fragment that included the first FKBP12-like domain, and 12 amino acids of the second FKBP12-like domain (common probe); a 260 bp of HindIII, EcoRI fragment from the 3′ noncoding sequence (starting from the stop codon) of the wFKBP73 (specific probe). Probes were labeled by random-primed DNA labeling (Boehringer Mannheim).
Plaques (2 × 105) were screened using the common probe under moderate-stringency conditions (hybridization in 5× SSPE, 2.5× Denhardt's solution, 0.25% [w/v] SDS, and 100 μg mL−1 sheared, single-stranded DNA) at 55°C. Final washes were done in 0.2× SSPE and 0.1% (w/v) SDS at 50°C. To eliminate the wFKBP73 clones, the 150 positive clones obtained were hybridized with the specific FKBP73 probe at 65°C. The 149 clones that hybridized with the wFKBP73 specific probe were discarded, and the single clone that was left was named wFKBP77. The wFKBP77 clone was isolated, rescued from the λ-ZAPII phage, cloned into Bluescript plasmid (according to the instructions supplied by Clontech), and analyzed by restriction enzymes. The sequence was obtained by Sequenase version 2.0 DNA polymerase (United States Biochemical). The sequence appears in the EMBL database (accession no. Y07636), and all of the nucleotide sequences refer to the submitted sequence. The sequence revealed an open reading frame of 1707 bp. The cDNA clone includes 81 bp at the 5′ noncoding region, 211 bp at the 3′ noncoding region, and a poly(A+) tail of 21 bp.
Computer Analysis of the Deduced wFKBP77 Sequence
Hydrophobic cluster analysis, a signature motif search (using the PROSITE dictionary), and a phosphorylation site search (using FIND PATTERNS) were performed as described previously (Blecher et al., 1996). The amino acid sequences of the human hFKBP12 (accession no. M80199), hFKBP59 (accession no. M88279), ROF1 (accession no. U49453), and wFKBP73 (accession no. X86903) were aligned by FASTA analysis.
RNA Isolation and Analysis
Total RNA was extracted from frozen tissue samples using TRIzol reagent (GIBCO-BRL). Ten-microgram samples of total RNA were separated on denaturing Glyoxal and DMSO 1.4% agarose gels and blotted onto nylon membranes (Hybond-N, Amersham) (Blecher et al., 1996).
Poly(A+) RNA was extracted from wheat embryos that had been allowed to imbibe for 24 h exposed for 2 h at 37°C or 25°C. The oligo(dT) cellulose was used according to the instructions supplied by Pharmacia. Samples of 4 μg (for detection of wFKBP77 at 25°C) and 1 μg were separated as described above.
For quantitative determination of transcript abundance, 10 μg of total RNA was loaded on a nylon membrane according to the instructions supplied by Amersham for manual slot-blot analysis. The radiospecific activity of each probe was diluted to a constant value (1 × 109 cpm μg−1 DNA), and the membrane was hybridized with 1 × 106 cpm mL−1 of the probe of interest. In anthers and pistils the blot was hybridized with 3 × 105 cpm mL−1 of the wFKBP77 probe. All blots were exposed for 6 h and scanned in a phosphor imager (Fujix BAS1000, Fuji, Tokyo, Japan). The results were normalized by hybridization of 100 ng of total RNA of each sample with an rRNA probe, which served as a control for an equal amount of total RNA. The ratio of mRNA abundance was determined by the average of three independent experiments.
Northern-blot and manual slot-blot analyses were detected by the following five probes: (a) FKBP77, a 130-bp probe derived from the N terminus of wFKBP77 60 bp upstream of the first ATG and 70 bp after the first ATG amplified by PCR (Promega) using a T7 primer and a specific primer (5′ GGGAGTCGGCCTCCTCCT 3′); (b) FKBP73, a 178-bp probe derived from the N terminus of the wFKBP73 100 bp upstream of the first ATG and 78 bp after the first ATG amplified by PCR using a T3 primer and a specific primer (5′ CGGCGCCGCCGAAGTC- AC 3′) (these two probes were checked for specificity by hybridization with the full cDNA sequence of wFKBP73 and wFKBP77, and cross-hybridization could not be detected); (c) FKBP73/77, a 620-bp probe (EcoRI, BamHI fragment) derived from the N terminus of the wFKBP73 100 bp upstream of the first ATG and 520 bp after the first ATG (this probe possesses the common region of the first FKBP12-like domain and 36 nucleotides of the second FKBP12-like domain; FKBP73/77 and the first 620 bp of the wFKBP77 share 80.5% identity); (d) HSP82, a 2.4-kb DNA fragment possessing the full-length coding sequence of the wHSP80 (Bessudo, 1996); and (e) 26SrRNA, a 2.8-kb wheat rDNA probe derived from pTA71 (Gerlach and Bedbrook, 1979).
Preparation of Polyclonal Antibodies (Anti-HS24C)
A 24-mer peptide encoding the deduced C-terminal sequence of the wFKBP77 amino acids A-545 to A-568 (AKWRKTENAAGKQEAQPMASDSTA) was synthesized with an automated synthesizer (Applied Biosystems). This sequence is unique to wFKBP77. An N-terminal Cys was added to the synthesized peptide and used to couple the peptide to activated hemocyanin (Pierce). Antiserum was elicited in a rabbit using standard injection protocols. Monospecific IgG was prepared by coupling the peptide to a thiol-reactive column (Pierce) and eluting the bound antibody with 0.1 m Gly, pH 3.0, and dialyzing the purified antibody against a large volume of TBS (25 mm Tris-HCl, pH 7.4, 0.15 m NaCl).
Preparation of Plant Extract for Electrophoresis of Proteins
Plant tissue (2–3 g fresh weight) was ground to a powder in liquid nitrogen with a mortar and pestle, and 1 mL of extraction buffer (50 mm Tris-HCl, pH 7.8, 10 mm EDTA, pH 8.0, 300 mm Suc, 100 mm NaCl, 4 mm PMSF, 5 mm benzamidine, and 1 mm DTT) was added. The crude extract was cleared by successive centrifugation for 6 min at 700g, 6 min at 3,000g, and 20 min at 12,000g. The supernatant after the 12,000g centrifugation step was used for western-blot analysis.
SDS-PAGE and Western-Blot Analysis
Twenty micrograms of total protein extract, unless stated otherwise, was separated on 7.5% SDS-polyacrylamide gels and transferred to a nitrocellulose membrane (Blecher et al., 1996). For detection of wFKBP77 in immature embryos exposed to heat stress, 80 μg of total protein was loaded in each lane. To obtain optimal resolution of the two FKBPs and to determine the molecular mass of the wFKBPs, SDS-PAGE minigels were run at 100 V for 2 h, whereas all other gels were run for 75 min at 100 V. The western blots were immunodecorated with the polyclonal antibodies that had been raised against the recombinant wFKBP73 (anti-FKBP73) (Blecher et al., 1996) at 1:10,000 dilution for 1 h at room temperature. For wFKBP77 detection, western blots were immunodecorated with the polyclonal antibodies raised against the synthetic peptide starting 24 amino acids from the C terminus of the wFKBP77 (anti-HS24C) at 1:500 dilution for 4 h at room temperature. For the detection of barley HVA1 (Hordeum vulgare 1) the blots were immunodecorated with the polyclonal antibodies raised against the recombinant HVA1 (anti-HVA1) (Hong et al., 1992) at 1:10,000 dilution for 1 h at room temperature. The secondary antibody used was goat anti-rabbit IgG conjugated with horseradish peroxidase at a 1:15,000 dilution for 1 h at room temperature. Proteins were detected using enhanced chemiluminescence reagents (Amersham) according to the supplier's instructions.
DNA Isolation and Restriction Fragment-Length Polymorphism Analysis
The genome location of the wFKBP77 gene in wheat was determined using a set of 21 nullisomic-tetrasomic lines, with each line lacking one chromosome pair (Sears, 1954). Mapping in wheat and rye was done in F2 populations of the crosses Chinese Spring × Synthetic and Ds2 × RxL10, respectively. All methods for DNA isolation, restriction enzyme digestion, gel electrophoresis, Southern blotting, probe labeling, and hybridization were carried out as described previously (Devos et al., 1992). The blots were hybridized with the wFKBP77 probe and exposed to either radiographic film or scanned in the phosphor imager.
RESULTS
Isolation and Sequence Analysis of a Wheat cDNA Coding for an FKBP
In our search for members of the wFKBP family we isolated a 2.1-kb cDNA sequence with an open reading frame of 1707 bp with two ATG codons (nucleotide positions 1 and 21). Examination of the nucleotide sequences surrounding the two ATG codons revealed that with respect to the consensus translation sequence (GCCGCCpurCCATGpur) (Kozak, 1989), the first Met codon (GTGAAATGA) is located in a more suitable translation start in that it possesses a conserved purine at position −3, whereas for the Met-7, although the sequence has 5 of 9 conserved nucleotides, it is lacking the conserved purine (AGCCGATGA). Therefore, we assume that the translation starts from the first Met.
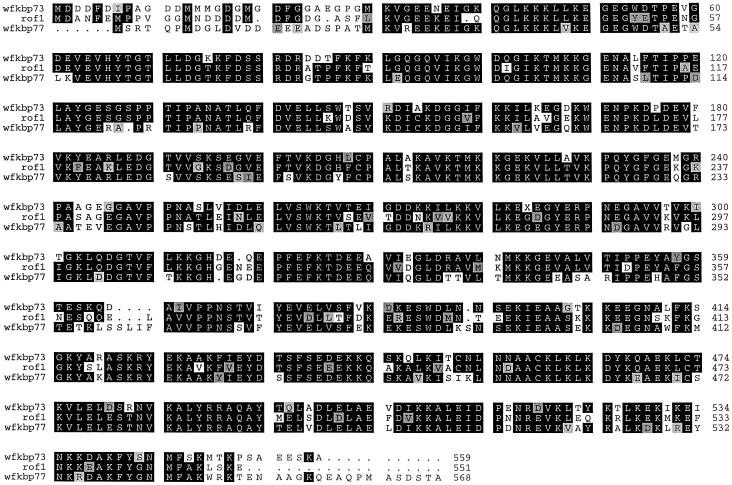
Comparison of the sequence with other FKBPs revealed similarity between the 1707-bp open reading frame and wFKBP73, ROF1, and hFKBP59; therefore, similarity spans the entire deduced amino acid sequence, being the highest to wFKBP73 (84%) and the lowest to hFKBP59 (42%) (Table I). The cDNA possesses three FKBP12-like domains, a putative TPR domain, and a putative calmodulin-binding domain (Fig. 1).
Table I.
Comparison of the deduced amino acid identity between hFKBP59, ROF1, wFKBP73, and wFKBP77
| Protein | hFKBP59 | ROF1 | wFKBP73 | wFKBP77 |
|---|---|---|---|---|
| % | ||||
| wFKBP77 | 42 | 64 | 84 | 100 |
| wFKBP73 | 50 | 68 | 100 | |
| ROF1 | 44 | 100 | ||
| hFKBP59 | 100 | |||
Figure 1.
Alignment of the deduced amino acid sequences of wFKBP73 (Blecher et al., 1996) and ROF1 (Vucich and Gasser, 1996) with that of the heat-induced wFKBP77 according to FASTA analysis. Single-letter codons are used for amino acid residues, black or gray boxes indicate identical or similar residues, respectively, and dots indicate gaps introduced to allow optimal alignment of the sequence.
The first FKBP12-like domain spans amino acids G-36 to A-141 and possesses the 10 amino acids reported to be essential for binding and maintaining the hydrophobic core of FK506 (van Duyne et al., 1993): Y-61, G-63, F-71, D-72, V-90, I-91, W-94, Y-117, I-126, and F-134.
The second and third FKBP12-like domains span amino acids G-151 to K-258 and R-268 to K-382, respectively. The second domain possesses only one of the critical amino acids (Y-176). The third domain possesses 4 of 10 conserved amino acids (G-295, F-303, V-323, and I-324). The putative TPR domain spans from K-401 to F-435, K-451 to A-484, and L-485 to K-519, and the putative calmodulin-binding domain (K-528 to F-545) has five positively charged residues (K-528, R-530, K-534, R-535, and K-538) and eight hydrophobic residues (L-529, Y-532, A-537, F-539, Y-540, G-541, M-544, and F-545).
Because the signature of the FKBP family resides in the first FKBP12-like domain that possesses PPIase activity, we compared the sequence identity between the first, second, and third domains of hFKBP59, wFKBP73, the newly isolated wFKBP77, and ROF1 with hFKBP12, the structure of which has been determined by high-resolution radiographic crystallography and NMR spectroscopy (Michnick et al., 1991; van Duyne et al., 1991, 1993). The first FKBP-like domain of each of the four proteins showed the highest identity to FKBP12 (Table II), conserving all of the amino acids defined as directly involved in maintaining the hydrophobic core and interacting with the drugs FK506 and rapamycin (van Duyne et al., 1993). Analysis of conserved amino acids of the binding pocket reveals an additional five amino acids involved in drug binding of the first FKBP12-like domain of FKBP59 (Futer et al., 1995; Craescu et al., 1996). These amino acids, also conserved in the wFKBP77 first domain, are S-73, R-77, F-81, L-132, and the fifth amino acid, Q-189, is polar and similar to the E-54 present in hFKBP12.
Table II.
Comparison of the deduced amino acid identity between hFKBP12 and the FKBP12-like domains of hFKBP59, ROF1, wFKBP73, and wFKBP77
| Protein | Domain I | Domain II | Domain III |
|---|---|---|---|
| % | |||
| hFKBP12a | 100 | ||
| hFKBP59 | 49.5 | 29.7 | |
| ROF1 | 51 | 30 | 30 |
| wFKBP73 | 50.5 | 29.1 | 34.6 |
| wFKBP77 | 50.1 | 25.7 | 28.7 |
The sequences compared are as published: hFKBP12 (Staendart et al., 1990), hFKBP59 (Peattie et al., 1992), ROF1 (Vucich and Gasser, 1996), and wFKBP73 (Blecher et al., 1996).
wFKBP77 possesses 15 putative phosphorylation sites belonging to four groups: myosin I heavy chain kinase (KXXS*X, i.e. KSE-S-91-I), cGMP-dependent protein kinase (XS*RX, i.e. SS-72-RD), casein kinase I (XSXXS*X, i.e. DSSFS-436-E), and casein kinase II (XS*XXEX, i.e. VS-187-KSE). These phosphorylation sites reflect possible sites for posttranslational modification of the protein.
The wFKBP77 was mapped on the wheat genome and found to be located on the distal region of the long arms of the group 2 chromosomes (data not shown) according to the mapping system developed for wheat (Devos et al., 1992). Based on the known relationships between the genomes of wheat and rye (Devos et al., 1993), F2 individuals of the wheat and rye mapping populations were tested to reveal polymorphism, but none was detected (data not shown).
Effect of Temperature on the Steady-State Level of the wFKBP77 Transcripts
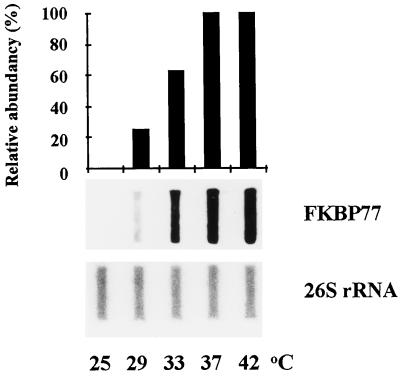
The level of the wFKBP77 transcripts in mature embryos revealed a maximal level after exposure to 37°C and 42°C; 60% of the maximum level of the transcripts was detected at 33°C, and 22% of the transcript accumulated at 29°C (Fig. 2).
Figure 2.
Temperature-dependent accumulation of wFKBP77 RNA in embryos. Slot-blot analysis of RNA extracted from embryos that were exposed for 2 h to temperatures from 25°C to 42°C. The blots were hybridized with the FKBP77 and the 26S rRNA probes. Scale represents 100% for the wFKBP77 mRNA extracted from embryos that were exposed for 2 h at 37°C.
For accurate estimation of wFKBP77 abundance, poly(A+) RNA was extracted and the northern blot was hybridized with the specific probes FKBP77 and FKBP73 (Fig. 3). wFKBP77 was detected at 25°C after loading of 4-fold more RNA than the RNA loaded in the other samples, and the calculated ratio of its level at 37°C indicated a 14-fold increase in its abundance. The wFKBP73 transcript level did not change after exposure to 37°C, and we calculated that it was 12-fold more abundant at 25°C than wFKBP77. Because wFKBP77 is highly induced by the heat stress, the transcript ratio of wFKBP73 to wFKBP77 became 1:1.2 at 37°C.
Figure 3.
Northern-blot analysis of poly(A+) RNA extracted from embryos that were exposed to 25°C or 37°C for 2 h. One microgram of mRNA from FKBP77 (25°C) was hybridized with the FKBP77 probe, and 4 μg with the FKBP73 probe.
Kinetics of Accumulation and Decay of the wFKBP77 Transcripts at 37°C and at 25°C in Embryos That Had Been Allowed to Imbibe
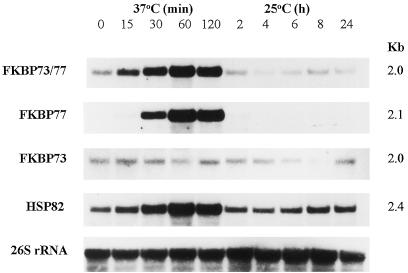
Like HSP82, wFKBP77 transcript was detected after exposure to 37°C for 15 min, reaching its maximal level after 60 min of exposure (Fig. 4). This accumulation was revealed by the FKBP73/77 and the FKBP77 probes, whereas the wFKBP73 transcript level did not change during heat stress.
Figure 4.
Accumulation of wFKBP77 in mature embryos exposed to 37°C. Northern blot of RNA extracted from embryos that were exposed to 37°C or 25°C after 2 h at 37°C and hybridized with the cDNA probes from the common region of the wFKBP73 (FKBP73/77) and the specific probes FKBP77, FKBP73, wHSP82 (HSP82), and the wheat 26S rRNA as a control for the amount of RNA that was loaded.
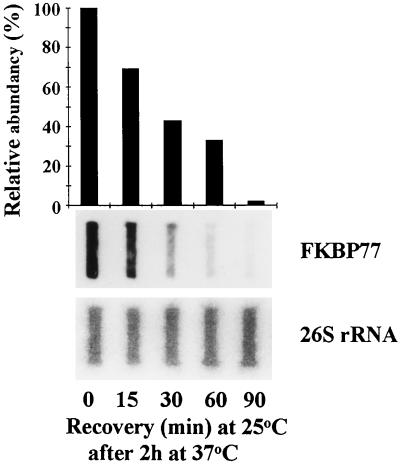
The decay of the wFKBP77 signal occurred after 2 h of recovery at 25°C. The HSP82 mRNA transcript returned to its basal level after 2 h at 25°C, and the wFKBP73 transcript level did not change during the treatment. Because no wFKBP77 transcript was detected after 2 h at 25°C, shorter time intervals were examined and it was found that 60% of the transcripts disappeared after 30 min at 25°C, and after 90 min only very low levels of the transcripts were detected (Fig. 5).
Figure 5.
Disappearance of wFKBP77 after heat stress in embryos that had imbibed. Slot-blot analysis of RNA extracted from embryos that were exposed to 25°C after 2 h at 37°C. The blots were hybridized with the FKBP77 and the 26S rRNA probes. Scale represents 100% for the wFKBP77 mRNA extracted from embryos that were exposed to 37°C for 2 h.
Tissue Specificity of wFKBP77 and wFKBP73 Gene Expression
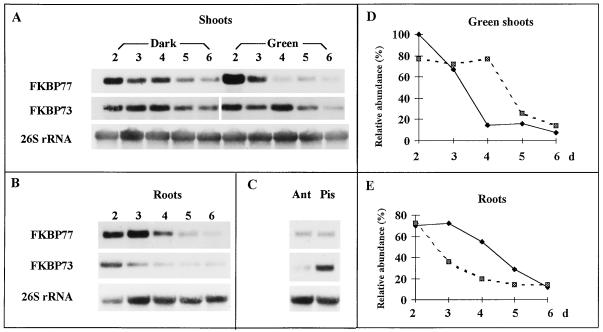
wFKBP77 and wFKBP73 were highly expressed in young and reproductive tissues (Fig. 6). A decrease in the expression of wFKBP77 was detected during seedling maturation mainly in green shoots, being 15% and 8% in 4- and 6-d-old shoots, respectively, compared with 2-d-old shoots (Fig. 6, A and D). The expression of wFKBP73 was reduced in the green shoots but to a lesser extent: 77% in the 4-d-old shoots and reaching a minimal expression of 14% in 6-d-old shoots. The transcript level of both wFKBPs in etiolated seedlings was the highest in the 2-d-old seedlings and decreased to 43% and 46% in the 6-d-old shoots. Because a difference in transcript level between green and etiolated shoots was found, 3-d-old dark-grown seedlings were illuminated for 10 h, exposed for 2 h to heat stress, and compared with light-grown and etiolated seedlings, both of which had been exposed to heat stress. Illumination treatment did not affect the wFKBP77 transcript level compared with the etiolated seedlings (data not shown).
Figure 6.
Expression of wFKBP77 and wFKBP73 in tissues exposed to heat stress. Northern-blot analysis of RNA extracted from 2- to 6-d-old etiolated and green shoots (A), 2- to 6-d-old roots (B), anthers (Ant), and pistils (Pis) (results of an independent experiment; see Methods) (C). RNA was extracted only from the basal first 1 cm of the shoots and the distal first 1 cm of the roots. Blots were hybridized with the FKBP77, FKBP73, and 26S rRNA probes. Scale of green shoots (D) and roots (E) represents 100% for the transcripts abundance of 2-d-old green shoots exposed for 2 h at 37°C. Solid lines, wFKBP77; dashed lines, wFKBP73.
In roots the wFKBP77 transcript level decreased gradually during the 4-d period, whereas wFKBP73 transcripts were reduced drastically, reaching 50% in the 3-d-old seedlings compared with the 2-d-old seedlings (Fig. 6, B and E). wFKBP77 was expressed in anthers and pistils at similar levels, whereas wFKBP73 was 8-fold higher in pistils than in anthers (Fig. 6C). For wFKBP73 the same transcript pattern was obtained at 25°C. In mature leaves, leaf sheaths, and culms neither wFKBP77 nor wFKBP73 transcript could be detected (data not shown).
wFKBP Is a Heat-Stress-Induced 77-kD Protein
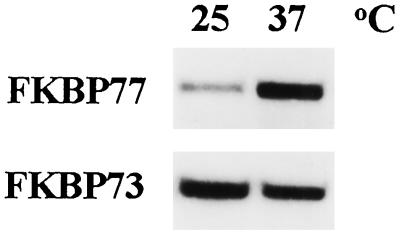
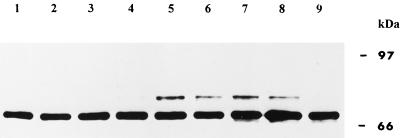
When wheat tissues were exposed to 37°C and the extracted proteins were visualized with antibodies raised against wFKBP73 (Blecher et al., 1996), an additional cross-reacting protein of an estimated molecular mass of 77 kD was observed (Fig. 7). The protein was detected after incubation of the embryos that had been allowed to imbibe at 37°C for 2 h, and disappeared after the exposure of the tissue to 25°C for more than 8 h.
Figure 7.
Western blot of proteins (20 μg/lane) extracted from embryos that were exposed at 25°C for 2 h (lane 1) or at 37°C for 15 min (lane 2), 30 min (lane 3), 60 min (lane 4), or 120 min (lane 5). For recovery the embryos that were exposed for 2 h at 37°C were incubated at 25°C for an additional 4 h (lane 6), 6 h (lane 7), 8 h (lane 8), or 24 h (lane 9). The blot was immunodecorated with the polyclonal antibodies raised against recombinant wFKBP73 (anti 73).
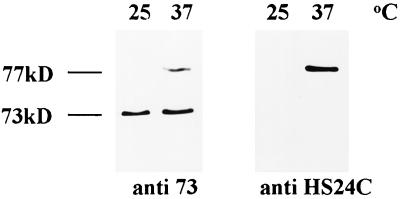
Specific antibodies raised against a synthetic peptide derived from the 24 deduced amino acids of the C terminus (HS24C) (see Methods) detected a single protein with an estimated molecular mass of 77 kD (Fig. 8). However, although the antibodies against the recombinant FKBP73 recognized the 73- and 77-kD cross-reacting proteins, anti-HS24C immunodecorated only the heat-stress protein we named wFKBP77 (Fig. 8).
Figure 8.
Western blot of proteins (20 μg/lane) extracted from embryos exposed to 37°C for 2 h and immunodecorated with the polyclonal anti 73 or with polyclonal antibodies produced against a synthetic peptide of 24 amino acids from the C terminus of wFKBP77 (anti HS24C).
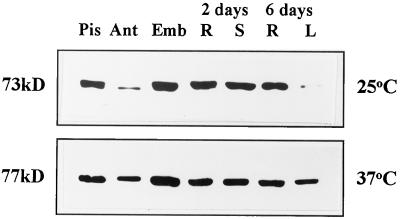
wFKBP77 was expressed in all tissues after exposure to 37°C but could not be detected at 25°C, whereas wFKBP73 was expressed in the same tissues at 25°C (Fig. 9) and its level did not change after exposure to 37°C (data not shown). The wFKBP77 protein was most abundant in embryos and least abundant in anthers and 6-d-old leaves (Fig. 9).
Figure 9.
Expression of wFKBP77 and wFKBP73 in various tissues. Western blot of proteins (20 μg/lane) extracted from pistils (Pis), anthers (Ant), embryos that had been allowed to imbibe for 24 h (Emb), shoots (S), or roots (R) that had been allowed to imbibe for 2 d, and leaves (L) or roots (R) that had been allowed to imbibe for 6 d exposed to 25°C or 37°C. The antibodies anti 73 and anti HS24C were used to immunodecorate the blots.
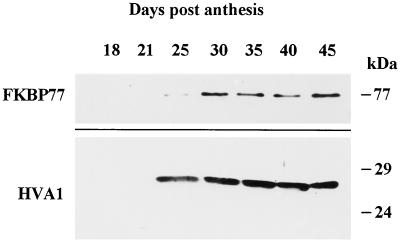
To determine whether the wFKBP77 is developmentally regulated during seed maturation, immature embryos dissected at various intervals during seed maturation were heat stressed and the extracted proteins analyzed by western blots. The amount of heat-stress wFKBP77 was very low in immature embryos, so to detect its presence 4-fold more protein was loaded (Fig. 10). wFKBP77 could be detected as soon as 25 d after anthesis, reaching a maximal level after 45 d. Its accumulation coincided with the accumulation of HVA1, a barley protein belonging to the LEA (late-embryogenesis abundant) proteins reported to accumulate during seed maturation and desiccation (Hughes and Galau, 1989; Hong et al., 1992).
Figure 10.
Expression of wFKBP77 in wheat immature embryos exposed to heat stress. Western blot of proteins (80 μg/lane) extracted from immature embryos (18–45 d after anthesis) exposed to 37°C for 2 h and immunodecorated with anti HS24C or anti HVA1 antibodies.
The effect of several abiotic stresses on the expression of wFKBP77 was assessed by RNA blots in 2-d-old roots and shoots exposed to the following treatments: 0.1 to 0.3 m NaCl, 0.05 to 5 mm CdCl2, 10 mm salicylic acid, or 100 μm ABA. The level of RNA transcripts was not induced by these stresses or by cold stress. When the chemical stresses were applied during exposure to heat stress, the induction of wFKBP77 mRNA was found to be identical to the induction obtained by the control heat-stress treatment (data not shown).
DISCUSSION
PPIases belong to several gene families, one of which is the FKBP family, defined by the conservation of the FKBP-type PPIase domain (for reviews, see Trandinh et al., 1992; Fruman et al., 1994; Galat and Metcalfe, 1995). The number of FKBP-12-like domains may vary among family members: one in vfFKBP15 and Arabidopsis PAS1 (Luan et al., 1996; Vittorioso et al., 1998), three in wFKBP and ROF1 (Fig. 1) (Blecher et al., 1996; Vucich and Gasser, 1996), and four in mzFKBP-66 (Hueros et al., 1998).
wFKBP77, like hFKBP59, wFKBP73, and ROF1 (Radanyi et al., 1994; Blecher et al., 1996; Vucich and Gasser, 1996), possesses a TPR motif (Sikorski et al., 1990; Goebel and Yanagida, 1991). The TPRs of the hFKBP59 have been found to be responsible for the interaction with HSP90 in the untransformed glucocorticoid receptor (Owens-Grillo et al., 1995, 1996a, 1996b). Recently, we and others have demonstrated that both wFKBP73 and wFKBP77 bind to the wHSP90 via the TPR motif (Owens-Grillo et al., 1996b; Breiman et al., 1997; Reddy et al., 1998).
wFKBP77 also has a putative calmodulin-binding domain with five positively charged residues and eight hydrophobic residues. The calmodulin-binding domain shows extreme variability in sequence, but its common feature is the ability to form an amphipathic basic α-helix with a large number of positively charged residues (O'Neil and DeGrado, 1990). The estimated size difference between wFKBP73 and wFKBP77 may be the result of posttranslational modifications (in addition to the calculated molecular mass) occurring on wFKBP77, which possesses more putative phosphorylation sites than wFKBP73 (Blecher et al., 1996).
In view of the postulated role of PPIases in protein folding, the response of the novel wFKBP to stress was studied. The expression of wFKBP77, which is very low at 25°C, was induced 14-fold after heat stress, unlike that of VfFKBP15, which was induced only 4- to 5-fold (Luan et al., 1996). This observation defines wFKBP77 as a heat-stress-dependent FKBP isoform, whereas wFKBP73 can be defined as a cognate isoform, because it was expressed under physiological conditions and was not induced by heat stress (Fig. 3).
The higher abundance of wFKBP77 and wFKBP73 transcripts in young tissues and reproductive tissues can be compared with the expression of hFKBP51, which shows the highest levels in testis (Nair et al., 1997), and Arabidopsis cyclophilins, which are highly expressed in floral tissues (Gasser et al., 1990), but is different from that of ROF1 and VfFKBP15, which show similar levels of expression (Luan et al., 1996; Vucich and Gasser, 1996), or Vf FKBP13, which is expressed preferentially in green tissues (Luan et al., 1994b). A decrease from the highest expression (100%) in 2-d-old seedlings to the lowest expression (8%) in 6-d-old seedlings reflects a temporal regulation of expression. wFKBP77 expression was abundant in young tissues and the transcript steady-state level decreased during differentiation in light, being lower than in etiolated shoots (Fig. 6).
Because the N-terminal amino acid sequence of wFKBP77 did not reveal any targeting presequence, we refer to this isoform as cytosolic. Members of the FKBP family have been found to reside in several subcellular compartments, such as the cytosolic FKBP59, the chloroplastic VfFKBP13, and the ER-located VfFKBP15 and pea mitochondrial FKBP18 (Breiman et al., 1992; Massol et al., 1992; Luan et al., 1994a, 1994b, 1996). The fact that FKBPs are present in most of the cell compartments is consistent with their role in protein folding, similar to the cyclophilins and the heat-stress-induced molecular chaperones.
The parallels between the physiology of PPIase and the HSP70 family is striking. Both families are highly conserved in all cellular compartments and contain several family members. Both families contain proteins that are stress inducible and others that are expressed constitutively, and members of the two families facilitate the survival of cells exposed to high temperatures (Sykes et al., 1993). In addition, many stress proteins function as chaperones during protein folding and assembly in vitro (Bose et al., 1996; Freeman and Morimoto, 1996; Freeman et al., 1996; Ruddon and Bedows, 1997). In plants several members of the FKBP and cyclophilin family were found to be affected by heat and other stresses (Marivet et al., 1994, 1995; Luan et al., 1996; Vucich and Gasser, 1996). The presence of two wheat FKBPs is comparable with the heat-shock proteins. For example, developmental and heat-stress-dependent expression was reported for HSP81 and HSP82 in maize, where the highest expression was detected in young tassels. HSP81 is only mildly heat inducible, whereas HSP82 shows strong inducibility (Marrs et al., 1993). wFKBP expression revealed tissue specificity, with wFKBP73 being the cognate isoform (Blecher et al., 1996) recently found to function as a molecular chaperone in vitro (Breiman et al., 1997), and wFKBP77 being the heat-stress-induced isoform (Fig. 4).
Although the normal function of the large PPIases is not fully defined in vivo, there is evidence of their role as molecular chaperones (Bose et al., 1996; Freeman et al., 1996), and PPIase activity is apparently only one of their activities, in that inhibition of the enzymatic activity does not abolish chaperone activity (Bose et al., 1996).
The characterization of the novel heat-stress-induced plant foldase wFKBP77 provides additional evidence for the presence of many plant molecular chaperones that are regulated by external signals and exhibit tissue specificity. We hope to understand more about the function of wFKBP77 in vivo and the contribution of the individual domains to the biological activity by analyzing transgenic plants overexpressing wFKBPs.
In mammals the HSP56 (also known as FKBP52) has been characterized as a heat-stress-inducible protein (Sanchez, 1990; Tai et al., 1993). In wheat the presence of two isoforms, one expressed under normal growth and the other after heat stress, indicates a more complex regulatory situation. The immunophilins emerge as an important multifunctional gene family probably involved in the basic regulatory system by acting as molecular chaperones, performing housekeeping and regulatory functions, and providing protection against stresses. Further studies on their biological roles in various systems will contribute to an understanding of their specific roles.
ACKNOWLEDGMENTS
We thank Ms. Moshit Lindzen for help with the library screening and the gene isolation and Dr. Katrien Devos (John Innes Centre, Norwich Research Park, Colney, Norwich, UK) for mapping the gene on the wheat genome. We also thank Prof. David Ho (Washington University, St. Louis, MO) for providing the anti-HVA1 antibodies and Prof. Moshe Feldman (Weizmann Institute of Science, Rehovot, Israel) for providing the HSP80 cDNA clone. I.K. thanks Mr. and Mrs. Marejn for financial support from the Don and Sara Marejn Scholarship Fund and the Joan and Chaime Constantiner Institute for Molecular Genetics at Tel Aviv University.
Abbreviations:
- FKBP
FK506-binding protein
- PPIase
peptidyl prolyl cis-trans isomerase
- TPR
tetratricopeptide
Footnotes
This work was supported by a grant from the Israeli Academy of Science to A.B.
LITERATURE CITED
- Bessudo A (1996) Molecular characterization of HSP80: a microtubule-associated protein in common wheat. Its gene copy number, location and expression. PhD thesis. Weizmann Institute of Science, Rehovot, Israel
- Blecher O, Erel N, Callebaut I, Aviezer K, Breiman A. A novel wheat peptidyl-prolyl cis trans isomerase: cDNA isolation, structure, enzymatic activity and expression. Plant Mol Biol. 1996;32:493–504. doi: 10.1007/BF00019101. [DOI] [PubMed] [Google Scholar]
- Bose S, Weikl T, Bugl H, Buchner J. Chaperone function of HSP90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- Boston RS, Viitaner PV, Vierling E. Molecular chaperones and protein folding in plants. Plant Mol Biol. 1996;32:191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- Breiman A, Fawcett TW, Ghirardi ML, Mattoo AK. Plant organelles contain distinct peptidyl-prolyl-cis-trans-isomerases. J Biol Chem. 1992;267:21293–21296. [PubMed] [Google Scholar]
- Breiman A, Kurek I, Herman E, Erel N, Aviezer K, Blecher O. Wheat novel molecular chaperones exhibiting peptidyl prolyl cis-trans isomerase activity, tissue specificity and abiotic stress induction (abstract no. 131) Plant Physiol. 1997;114:S-44. [Google Scholar]
- Buchner J. Supervising the fold: functional principles of molecular chaperones. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- Callebaut I, Renoir JM, Lebeau MC, Massol N, Burny A, Baulieu EE, Mornon JP. An immunophilin that binds Mr 90,000 heat shock protein: main structural features of a mammalian p59 protein. Proc Natl Acad Sci USA. 1992;89:6270–6274. doi: 10.1073/pnas.89.14.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambraud B, Rouvierefourmy N, Radanyi C, Hsiao K, Peattie DA, Livingston DJ, Baulieu EE. Overexpression of p59 (FKBP59), full length and domains, and characterization of PPIase activity. Biochem Biophys Res Commun. 1993;196:160–166. doi: 10.1006/bbrc.1993.2229. [DOI] [PubMed] [Google Scholar]
- Chou IT, Gasser CS. Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol Biol. 1997;35:873–892. doi: 10.1023/a:1005930024796. [DOI] [PubMed] [Google Scholar]
- Connern CP, Halestrap AP. Purification and N-terminal sequencing of peptidyl-prolyl cis-trans isomerase from rat liver mitochondrial matrix reveals the existence of a distinct mitochondrial cyclophilin. Biochem J. 1992;284:381–385. doi: 10.1042/bj2840381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GR, Schreiber SL. Three part inventions: intracellular signaling and induced proximity. Trends Biochem Sci. 1996;21:418–422. doi: 10.1016/s0968-0004(96)20027-1. [DOI] [PubMed] [Google Scholar]
- Craescu CT, Rouviere N, Popescu A, Cerpolini E, Lebeau MC, Baulieu EE, Mispelter J. Three dimensional structure of the immunophilin-like domain of FKBP59 in solution. Biochemistry. 1996;35:11045–11052. doi: 10.1021/bi960975p. [DOI] [PubMed] [Google Scholar]
- Devos KM, Atkinson MD, Chinoy CN, Harcourt RL, Koebner RMD, Liu CJ, Masojc P, Xie DX, Gale MD. Chromosome rearrangements in rye genome relative to that of wheat. Theor Appl Genet. 1993;85:673–680. doi: 10.1007/BF00225004. [DOI] [PubMed] [Google Scholar]
- Devos KM, Atkinson MD, Chinoy CN, Liu CJ, Gale MD. RFLP based genetic map of the homoeologous group 3 chromosomes of wheat and rye. Theor Appl Genet. 1992;83:931–939. doi: 10.1007/BF00232953. [DOI] [PubMed] [Google Scholar]
- Dolinski K, Heitman J. Peptidyl-prolyl isomerases: an overview of the cyclophilin, FKBP and parvulin families. In: Gething M-J, editor. Guidebook to Molecular Chaperones and Protein Folding Catalysis. Oxford, UK: Oxford University Press; 1997. pp. 359–369. [Google Scholar]
- Freeman BC, Morimoto RI. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70), and hdj1 have distinct roles in recognition of nonnative protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Toft DO, Morimoto RI. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Burakoff SJ, Bierer BE. Immunophilins in protein folding and immunosuppression. FASEB J. 1994;8:391–400. doi: 10.1096/fasebj.8.6.7513288. [DOI] [PubMed] [Google Scholar]
- Fulgosi H, Vener AV, Altschmied L, Herrmann RG, Andersson B. A novel multi-functional chloroplast protein: identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J. 1998;17:1577–1587. doi: 10.1093/emboj/17.6.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futer O, DeCenzo MT, Aldape RA, Livingston DJ. FK506 binding protein mutational analysis: defining the surface residue's contributions to stability of the calcineurin co-complex. J Biol Chem. 1995;270:18935–18940. doi: 10.1074/jbc.270.32.18935. [DOI] [PubMed] [Google Scholar]
- Galat A, Metcalfe S. Peptidyl prolyl cis-trans isomerases. Prog Biophys Mol Biol. 1995;63:67–118. doi: 10.1016/0079-6107(94)00009-x. [DOI] [PubMed] [Google Scholar]
- Gasser CS, Gunning DA, Budelier KA, Brown SM. Structure and expression of cytosolic cyclophilin/peptidyl-prolyl cis-trans isomerase of higher plants and production of active tomato cyclophilin in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:9519–9523. doi: 10.1073/pnas.87.24.9519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilins: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–546. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- Harding MW, Galat A, Uehling DE, Schreiber SL. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature. 1989;341:758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hayes SA, Dice FJ. Roles of molecular chaperones in protein degradation. J Cell Biol. 1996;132:255–258. doi: 10.1083/jcb.132.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Barg R, Ho TD. Developmental and organ-specific expression of an ABA- and stress-induced protein in barley. Plant Mol Biol. 1992;18:663–674. doi: 10.1007/BF00020009. [DOI] [PubMed] [Google Scholar]
- Hueros G, Rahfeld J, Salamini F, Thompson R. A maize FK506-sensitive immunophilin, mzFKBP-66, is a peptidylproline cis-trans-isomerase that interacts with calmodulin and a 36-kDa cytoplasmic protein. Planta. 1998;205:121–131. doi: 10.1007/s004250050303. [DOI] [PubMed] [Google Scholar]
- Hughes DW, Galau GU. Temporally modular gene expression during cotyledon development. Genes Dev. 1989;3:3358–3369. doi: 10.1101/gad.3.3.358. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson JD, Ness SA. Point mutations in v-Myb disrupt a cyclophilin-catalyzed negative regulatory mechanism. Mol Cell. 1988;1:203–211. doi: 10.1016/s1097-2765(00)80021-0. [DOI] [PubMed] [Google Scholar]
- Luan S, Albers MW, Schreiber SL. Light-regulated, tissue-specific immunophilins in higher plants. Proc Natl Acad Sci USA. 1994a;91:984–988. doi: 10.1073/pnas.91.3.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Kudla J, Gruissem W, Schreiber SL. Molecular characterization of a FKBP-type immunophilin from higher plants. Proc Natl Acad Sci USA. 1996;93:6964–6969. doi: 10.1073/pnas.93.14.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Lane WS, Schreiber SL. pCyP: a chloroplast-localized, heat shock-responsive cyclophilin from fava bean. Plant Cell. 1994b;6:885–892. doi: 10.1105/tpc.6.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marivet J, Frendo P, Burkard G. DNA sequence analysis of a cyclophilin gene from maize: developmental expression and regulation by salicylic acid. Mol Gen Genet. 1995;247:222–228. doi: 10.1007/BF00705653. [DOI] [PubMed] [Google Scholar]
- Marivet J, Margis-Pinhero M, Frendo P, Burkard G. Bean cyclophilin gene expression during plant development and stress condition. Plant Mol Biol. 1994;26:1181–1189. doi: 10.1007/BF00040698. [DOI] [PubMed] [Google Scholar]
- Marrs KA, Casey ES, Capitant SA, Bouchard RA, Dietrich PS, Mettler IJ, Sinibaldi RM. Characterization of two maize HSP90 heat shock protein genes: expression during heat shock, embryogenesis, and pollen development. Dev Genet. 1993;14:27–41. doi: 10.1002/dvg.1020140105. [DOI] [PubMed] [Google Scholar]
- Massol N, Lebeau MC, Renoir JM, Faber LE, Baulieu EE. Rabbit FKBP59-heat shock protein binding immunophilin (HBI) is a calmodulin binding protein. Biochem Biophys Res Commun. 1992;187:1330–1335. doi: 10.1016/0006-291x(92)90448-t. [DOI] [PubMed] [Google Scholar]
- Matouschek A, Rospert S, Schmid K, Glick BS, Schatz G. Cyclophilin catalyzes protein folding in yeast mitochondria. Proc Natl Acad Sci USA. 1995;92:6319–6323. doi: 10.1073/pnas.92.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michnick SW, Rosen MK, Wandless TJ, Karplus M, Schreiber SL. Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin. Science. 1991;252:836–839. doi: 10.1126/science.1709301. [DOI] [PubMed] [Google Scholar]
- Nair SC, Rimerman RA, Toran EJ, Chen S, Prapapanich V, Butts RN, Smith DF. Molecular cloning of human FKBP51 and comparisons of immunophilin interactions with HSP90 and progesterone receptor. Mol Cell Biol. 1997;17:594–603. doi: 10.1128/mcb.17.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil KT, Degrado WF. How calmodulin binds its targets: sequence independent recognition of amphiphilic a-helices. Trends Biochem Sci. 1990;15:59–64. doi: 10.1016/0968-0004(90)90177-d. [DOI] [PubMed] [Google Scholar]
- Owens-Grillo JK, Czar MJ, Hutchison KA, Hoffmann K, Perdew GH, Pratt WB. A model of protein targeting mediated by immunophilins and other proteins that bind to the hsp90 via tetratricopeptide repeat domains. J Biol Chem. 1996a;271:13468–13475. doi: 10.1074/jbc.271.23.13468. [DOI] [PubMed] [Google Scholar]
- Owens-Grillo JK, Hofman K, Hutchison KA, Yem AW, Deibel MR, Handschumacher JRE, Pratt WB. The cyclosporin A-binding immunophilin Cyp-40 and the FK506-binding immunophilin HSP56 bind to a common site on HSP90 and exist in independent cytosolic heterocomplexes with the untransformed glucocorticoid receptor. J Biol Chem. 1995;270:20479–20484. doi: 10.1074/jbc.270.35.20479. [DOI] [PubMed] [Google Scholar]
- Owens-Grillo JK, Stancato LF, Hoffmann K, Pratt WB, Krishna P. Binding of immunophilins to the 90 kDa heat shock protein (hsp90) via a tetratricopeptide repeat domain is a conserved protein interaction in plants. Biochemistry. 1996b;35:15249–15255. doi: 10.1021/bi9615349. [DOI] [PubMed] [Google Scholar]
- Peattie DA, Harding MW, Fleming MA, DeCenzo MT, Lippke JA, Livingston DJ, Benasutti MT. Expression and characterization of human FKBP52, an immunophilin that associates with 90 kDa heat shock protein and is a component of steroid receptor complexes. Proc Natl Acad Sci USA. 1992;89:10974–10978. doi: 10.1073/pnas.89.22.10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radanyi C, Chambraud B, Baulieu EE. The ability of the immunophilin FKBP59-HBI to interact with the 90 kDa heat shock protein is encoded by its tetratricopeptide repeat domain. Proc Natl Acad Sci USA. 1994;91:11197–11201. doi: 10.1073/pnas.91.23.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J, Carrello A, Mark PJ, Warner BJ, Simpson RJ, Moritz RL, House AK. The cyclophilin component of the unactivated estrogen receptor contains a tetratricopeptide repeat domain and shares identity with p59 (FKBP59) J Biol Chem. 1993;268:13187–13192. [PubMed] [Google Scholar]
- Reddy RK, Kurek I, Silverstein AM, Clinkers M, Breiman A., Krishna P (1998) High-molecular-weight FKBPs are components of HSP90 heterocomplexes in wheat germ lysate. Plant Physiol 118: 1395–1401 [DOI] [PMC free article] [PubMed]
- Ruddon RW, Bedows E. Assisted protein folding. J Biol Chem. 1997;272:3125–3128. doi: 10.1074/jbc.272.6.3125. [DOI] [PubMed] [Google Scholar]
- Sanchez ER. Hsp56: a novel heat shock protein associated with untransformed steroid receptor complexes. J Biol Chem. 1990;265:22067–22070. [PubMed] [Google Scholar]
- Schmid FX (1998) Catalysis of protein folding by prolyl isomerases. In AL Fink, Y Goto, eds, Molecular Chaperones in the Life Cycle of Proteins: Structure, Function and Mode of Action. Marcel Dekker, New York, pp 361–390
- Schreiber SL. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science. 1991;251:283–287. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- Sears ER. The aneuploids of common wheat. Mo Agric Exp Stn Res Bull. 1954;572:1–59. [Google Scholar]
- Siekierka JJ, Wiederrecht G, Greulich H, Boulton D, Hung SHY, Cryan J, Hodges PJ, Sigal NH. The cytosolic-binding protein for the immunosuppressant FK506 is both a ubiquitous and highly conserved peptidyl-prolyl cis-trans isomerase. J Biol Chem. 1989;265:21011–21015. [PubMed] [Google Scholar]
- Sikorski RS, Boguski MS, Gobl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- Standaert RF, Galat A, Verdine GL, Schreiber SL. Molecular cloning and overexpression of the human FK506-binding protein FKBP. Nature. 1990;346:671–674. doi: 10.1038/346671a0. [DOI] [PubMed] [Google Scholar]
- Sykes K, Gething MJ, Sambrook J. Proline isomerases function during heat shock. Proc Natl Acad Sci USA. 1993;90:5853–5857. doi: 10.1073/pnas.90.12.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai PK, Chang H, Albers MW, Schreiber SL, Toft DO, Faber LE. p59 (FK506 binding protein 59) interaction with heat shock protein is highly conserved and may involve proteins other than steroid receptors. Biochemistry. 1993;32:8842–8847. doi: 10.1021/bi00085a015. [DOI] [PubMed] [Google Scholar]
- Trandinh CC, Pao GM, Saier MH. Structural and evolutionary relationships among the immunophilins: two ubiquitous families of peptidyl prolyl cis-trans isomerases. FASEB J. 1992;6:3410–3420. doi: 10.1096/fasebj.6.15.1464374. [DOI] [PubMed] [Google Scholar]
- van Duyne GD, Standaert RF, Karplus M, Schreiber SL, Clardy J. Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science. 1991;252:839–842. doi: 10.1126/science.1709302. [DOI] [PubMed] [Google Scholar]
- van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structure of the human immunophilin FKBP12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- Vittorioso P, Cowling R, Faure JD, Caboche M, Bellini C. Mutation in the Arabidopsis pasticcino 1 gene, a new FKBP-like protein, has a dramatic effect on plant development. Mol Cell Biol. 1998;18:3034–3043. doi: 10.1128/mcb.18.5.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucich VA, Gasser CS. Novel structure of a high molecular weight FK506 binding protein from Arabidopsis thaliana. Mol Gen Genet. 1996;252:510–517. doi: 10.1007/BF02172397. [DOI] [PubMed] [Google Scholar]
- Walker KW, Gilbert HF (1998) Protein disulfide isomerase. In AL Fink, Y Goto, eds, Molecular Chaperones in the Life Cycle of Proteins: Structure, Function and Mode of Action. Marcel Dekker, New York, pp 331–360
- Winter J, Wright R, Duck N, Gasser C, Fraley R, Shan D. The inhibition of petunia hsp70 mRNA processing during CdCl2 stress. Mol Gen Genet. 1988;211:315–319. [Google Scholar]