Abstract
We describe here a new lactococcal abortive phage infection system, designated AbiP. AbiP is effective against some lactococcal phages of one prevalent group, 936, but not against phages from the other two groups (c6A and P335). It was identified in the Lactococcus lactis subsp. cremoris strain IL420, on the native plasmid pIL2614. AbiP is encoded by a single gene, expressed in an operon with a second gene. In this work, abiP is shown to affect both the replication and transcription of phage DNA. In AbiP+ cells, phage DNA replication is arrested approximately 10 min after infection. Levels of middle and late phage transcripts are lower in AbiP+ than in AbiP− cells, probably due to the smaller amount of phage DNA. By contrast, early phage transcripts are more abundant in AbiP+ than in AbiP− cells, suggesting that the switch-off, which occurs 15 min after infection in AbiP− cells, is prevented in AbiP+ cells.
Dairy fermentations using lactococci are highly susceptible to bacteriophage attacks. Therefore, special attention has been given to lactococcal phage defense mechanisms and in particular to abortive phage infection systems (Abi). These systems arrest phage multiplication and cause premature cell death upon infection. This decreases the number of progeny particles produced and limits their spread to other cells. As a consequence, the cell population survives (reviewed in references 32 and 39).
Twenty-two lactococcal Abi genes, designated abiA to abiT, have been described (7, 9, 43; for a review, see reference 20). Three of these (abiD, abiD1, and abiF) code for proteins sharing 28 to 46% identity throughout the protein sequence. Similarly, AbiA and AbiK share 23% identity. Putative products of the other abi genes show no homology. Lactococcal phages fall into three prevalent groups of DNA homology (29). Two of these groups, designated 936 and c6A, are composed of virulent phages responsible for industrial fermentation failures, and the third, designated P335, is composed mostly of temperate phages. Phages from one of these groups share essentially no DNA homology with members of the other groups (13). The activities of the Abi proteins have been checked against some phages representative of each of these three groups. Ten Abi's are effective on a single group of phages. Eight Abi's are effective on two groups of phages: either 936 and c6A or 936 and P335. Since DNA and protein homologies between phages of different groups are limited, we can speculate that these Abi's are directed against the few DNA sequences or proteins that are conserved in phage groups on which they are effective. This is indeed the case for AbiD1. This mechanism, effective on all 936 and c6A phages tested (references 5 and 19, respectively; M. C. Chopin, unpublished data), interferes with a gene coding for an essential endonuclease homologous to RuvC (4, 5), which is highly conserved in the two phage groups. Finally, AbiA (24) is the only lactococcal system that is effective on all three groups.
In a group of phage, not all individuals are sensitive to a given Abi system. In order to understand how some phages escape Abi, spontaneous variants of sensitive phages, able to overcome AbiA, AbiC, AbiD1, AbiF, or AbiK, have been selected. Among these, P335 phages have been shown to acquire resistance to Abi's either by recombination with host chromosomal sequences (8, 31) or by spontaneous mutation (18). By contrast, virulent phages of the 936 or c6A group, having no prophage counterpart on the host chromosome, have been shown to evolve resistance to Abi's by mutation only (4, 20).
Some Abi's have been studied in more detail, especially with regard to their effect on phage DNA replication and transcription, two functions crucial to phage multiplication. These studies revealed a variety of modes of action. AbiA (24), AbiF (21), AbiK (9), and AbiR (43) arrest phage DNA replication by an unknown mechanism. AbiG inhibits mRNA synthesis (33). AbiB allows normal onset of phage transcripts but promotes their degradation 10 to 15 min after infection. This degradation has been shown to result from endonucleolytic cleavage at specific sites. It has been proposed that AbiB either induces the synthesis of an RNase or stimulates its activity (34). AbiT, which has no effect on either DNA replication or transcription, has been proposed to be activated by one of the late phage mRNAs or proteins and to rapidly cause premature cell death (7). AbiD1 has been proposed to decrease an essential RuvC-like endonucleolytic activity (4, 5), resolving branched DNA structures, and thus conceivably to be involved in phage DNA replication and/or packaging.
The presence of numerous and diverse Abi mechanisms in Lactococcus spp. is probably a consequence of the abundance and diversity of phages in their dairy environment. This genus is therefore a good source of such systems, both for studies on phage-host interactions and for the development of phage defense strategies for improvement of industrial strains. In this work, we describe a new lactococcal abortive infection system designated AbiP. This mechanism is shown to arrest phage DNA replication 10 min after infection and to prevent the switch-off of early transcripts, normally observed 15 min after infection.
MATERIALS AND METHODS
Bacterial strains, plasmids, phages, and media.
Lactococcus lactis subsp. lactis IL1403 (12) and derivatives were grown at 30°C in M17 medium (42) in which lactose had been replaced by glucose (M17-G). Lactococcus lactis subsp. cremoris IL420 was grown at 30°C in M17. When needed, 5 μg of erythromycin/ml was added to the culture medium. Phages bIL32, bIL41, bIL66M1, bIL67, bIL170, bIL198 (from our laboratory collection [4]), and c2 (35) were enumerated, as described elsewhere (42), on IL1403 and derivatives. Phage sk1 (35), which was unable to grow on IL1403, was enumerated on L. lactis MG1363 (22) and MG1363(pIL2617).
Phage adsorption and one-step growth experiments were performed as described previously (14). Results are means of two experiments. To select phage-resistant transformants, cells were grown overnight in M17-G broth containing erythromycin. The pool of transformants was then checked for phage resistance as described previously (14).
Molecular cloning and DNA sequence analysis.
L. lactis was transformed by electroporation as described previously (25). DNA manipulation and cloning were performed essentially as described by Maniatis et al. (30). Sequences were determined on double-stranded DNA in a cycle extension reaction by using appropriate primers, Taq polymerase (Applied Biosystems), and fluorescent dye-coupled dideoxynucleotides on a 370A DNA sequencer (Applied Biosystems). Each sequence was determined twice on both strands. DNA and protein sequences were analyzed with the Genetics Computer Group software (16) and the Blast program (1).
DNA extraction and hybridization.
Phages were added, in the presence of 10 mM CaCl2, at a multiplicity of infection of 5, to strains grown as described above, at an optical density at 600 nm of 0.4. Samples were taken at different time points, mixed with glycerol to a final concentration of 10%, and immediately frozen in liquid nitrogen. Total intracellular DNA was extracted by the method of Hill et al. (24). Ten microliters of each DNA sample was digested with HincII, and the fragments were separated by agarose gel electrophoresis. DNA was transferred to a nylon N+ membrane (Amersham Pharmacia Biotech) and probed with total phage DNA labeled with [α-32P]dCTP by using the Ready To Go DNA Labeling kit (-dCTP) (Amersham Pharmacia Biotech). Hybridization and washing conditions were those described by Maniatis et al. (30). DNA was quantified with a PhosphorImager by using ImageQuant (version 5.2; Molecular Dynamics) software.
RNA extraction and Northern hybridization.
Cells were grown as described above to an optical density at 600 nm of 0.4, and phages were added at a multiplicity of infection of 5, in the presence of 10 mM CaCl2. Aliquots were taken at different time points and centrifuged, and the pellet was immediately frozen in liquid nitrogen. Total RNA was extracted according to the work of Glatron and Rapoport (23), with the modifications described previously (2). For Northern blot analysis, 20 μg of RNA was denatured for 1 h at 50°C in glyoxal-dimethyl sulfoxide, essentially as described by Williams and Mason (46), and was separated by electrophoresis, which was carried out in a 10 mM phosphate buffer (pH 7.0) on a 1% RNase-free agarose gel (FMC BioProducts) in the presence of 0.2% sodium iodoacetate. The 0.24- to 9.5-kb RNA ladder from Gibco-BRL was used as a molecular weight marker. The membranes were stained with ethylene blue (45) to assess the integrity of the rRNAs and to visualize the RNA markers. The RNA was transferred to a nylon Hybond N+ membrane (Amersham Pharmacia Biotech) according to the method of Maniatis et al. (30) and was hybridized either with Long Range PCR fragments, obtained as described below, labeled with [α-32P]dCTP, by using the Ready To Go DNA Labeling kit (-dCTP) (Amersham Pharmacia Biotech), or with oligonucleotide probes labeled with [γ-32P]ATP by using T4 polynucleotide kinase according to the supplier's instructions (New England Biolabs). Hybridization and washing were carried out under standard conditions (30) for long-range PCR probes. For oligonucleotide probes, the concentration of formamide in the hybridization solution was reduced from 50 to 5%. RNA was quantified with a PhosphorImager, as described above for DNA.
Long-range PCR DNA amplification.
DNA amplification was carried out using the GeneAmp PCR system 9700 (Applied Biosystems) and Ex Taq (TaKaRa Biomedicals), essentially as described by the supplier, with an elongation time of 5 min. Primers used throughout this study are described in Table 1. Long-range PCR fragments were purified with the Wizard PCR Preps DNA Purification system (Promega).
TABLE 1.
Oligonucleotides used in this study
| Primer
|
DNA | Accession no. | Coordinates (bp) | |
|---|---|---|---|---|
| No. | Sequence (5′-3′)a | |||
| 1 | ggaattccGTTGCGGATTGGATTAGTC | pIL2614 | U90222 | 9191-9209 |
| 2 | gctctagagcCTTGTGTTTGGGTGTATTG | pIL2614 | U90222 | 10054-10036 |
| 3 | GGGATTTGTGAGGGGTTATTATC | pIL2614 | U90222 | 8538-8553 |
| 4 | GCTGAGATAATAACAACAAGC | pIL2614 | U90222 | 8686-8706 |
| 5 | CGTACATTCTGAAACAAACCC | pIL2614 | U90222 | 9439-9459 |
| 6 | ATGGATATTTTATTTTTAGAAAAAGC | pIL2614 | U90222 | 9239-9264 |
| 7 | CTAGTTTCCTTTTATAAGTTCCTC | pIL2614 | U90222 | 9950-9973 |
| 8 | CGAACGCTGGCGGCGTGCCT | IL1403 16S rRNA | NC_002662 | |
| 9 | CACTCACGCGGCGTTGCTCG | IL1403 16S rRNA | NC_002662 | |
| 10 | TCACTCGAATCTAATTCCTC | pIL2614 | U90222 | 9480-9461 |
| 11 | CGTACATTCTGAAACAAACCC | pIL2614 | U90222 | 9459-9439 |
| 12 | TTCTTGTCTACTTCTGGCTG | bIL66M1 | AY249139 | 5779-5798 |
| 13 | GGGCTCGTATGAGCGTGTTT | bIL66 | L35175 | 2376-2395 |
| 14 | AGTATGGCTTGCAATTAAGC | bIL66 | L35175 | 2124-2143 |
| 15 | CCCAAAAAATCAAAAAGAAAAGTTTTAGCT | bIL170 | NC_001909 | 27-56 (late region) |
| 16 | GCTTCTTTGCAAGAGTTAATGTATTATGGA | bIL170 | NC_001909 | 5761-5790 (late region) |
| 17 | TCCATAATACATTAACTCTTGCAAAGAAGC | bIL170 | NC_001909 | 5790-5761 (late region) |
| 18 | TCTTTGGTAATGAAGCTCTAATCGTAGCTG | bIL170 | NC_001909 | 8860-8889 (late region) |
| 19 | CAGCTACGATTAGAGCTTCATTACCAAAGA | bIL170 | NC_001909 | 8889-8860 (late region) |
| 20 | GCCATACTAATAGCCATAGCAATACGAACA | bIL170 | NC_001909 | 31642-31613 (middle region) |
| 21 | GAAGAACAGCTACTATTTAAGCAAGAAACA | bIL170 | NC_001909 | 30158-30187 (middle region) |
| 22 | GTTGCGTTTGGTCCAAATTCAGCGTCTAGT | bIL170 | NC_001909 | 17500-17471 (late region) |
| 23 | TCAAAGGTTCTATGGTGGTAGGTTTACCAG | bIL170 | NC_001909 | 13151-13180 (late region) |
| 24 | CTGGTAAACCTACCACCATAGAACCTTTGA | bIL170 | NC_001909 | 13180-13151 (late region) |
| 25 | TTTTTGGGCTTTCTCTGCACGTTTAGCAAG | bIL170 | NC_001909 | 24021-24050 (early region) |
| 26 | CTTGCTAAACGTGCAGAGAAAGCCCAAAAA | bIL170 | NC_001909 | 24050-24021 (early region) |
| 27 | TTTTCTTCACTAATTCGTTGTTCTTCAAGT | bIL170 | NC_001909 | 18513-18542 (early region) |
| 28 | GCTAATGAAATCGAACGCAAACTTAAAGAA | bIL170 | NC_001909 | 28705-28734 (early region) |
| 29 | atcgaattcGATCTAAAACAAGTAAATATTT | pIL2614 | U90222 | 8360-8381 |
| 30 | taaggatccTTAACTTAATATATGACTAATC | pIL2614 | U90222 | 9223-9202 |
Extensions containing restriction sites are lowercase.
RT-PCR analysis.
Reverse transcription-PCRs (RT-PCRs) were carried out on 25 ng of total RNA, with the OneStep RT-PCR kit (Qiagen), according to the supplier's instructions, by using oligonucleotides 6 and 7. As an independent control, the 16S rRNA-specific primers 8 and 9 were used. Prior to RT-PCR, all RNA samples were treated with RNase-free DNase I (Roche). Control experiments, run in the absence of reverse transcriptase reactions, yielded no product.
Mapping of the 5′ end of the transcript.
The 5′/3′ RACE kit (Roche) was used according to the supplier's instructions. Five hundred nanograms of total RNA, previously treated with RNase-free DNase I (Roche), was used to obtain the cDNA by extending primer 10. After the 3′ tailing reaction with a dATP string, the cDNA was amplified by PCR using reverse primer 11 and the forward primer (Oligo dT-Anchor primer) supplied with the kit. The 5′ end of the transcript was then determined by sequencing the PCR product, as described above.
Plasmid constructions.
Orf1 (194 amino acids [aa]) and Orf2 (245 aa) were inactivated, on plasmid pIL2650 (Fig. 1), by generating a frameshift introducing an early stop codon (TGA). Plasmid pIL2650 was digested with BsaI (144 bp downstream of the ATG start codon of orf1), treated with T4 DNA polymerase, and ligated with a BclI linker (CTGATCAG; New England Biolabs). The ligation mixture, digested with BsaI, was transformed into IL1403. Sequence analysis of six transformants revealed two plasmids with one copy of the linker and the expected frameshift, reducing Orf1 to its first 50 aa. One of these was designated pIL2651 (Fig. 1). Plasmid pIL2650 was digested with BsaHI (264 bp downstream of the ATG start codon of orf2), treated with T4 DNA polymerase, and self-ligated. The ligation mixture, digested with BsaHI, was transformed into IL1403. Sequence analysis of the transformants revealed one plasmid with the expected frameshift, reducing Orf2 to its first 105 aa. This plasmid was designated pIL2655 (Fig. 1). orf2, together with its upstream ribosome binding site (RBS) sequence, was cloned on the erythromycin resistance plasmid vector pIL253 (38), under the control of a plasmid promoter. The DNA fragment carrying orf2 was obtained by PCR amplification using plasmid pIL2650 as a template and oligonucleotides 29 and 30 (Table 1). The PCR product was purified on a Qiagen column and cloned at the EcoRI-XbaI sites on plasmid vector pIL253. The resulting construct was designated pIL2617 (Fig. 1). orf1, together with its upstream RBS, promoter, and putative regulatory sequences, was cloned on a chloramphenicol resistance derivative of pIL253, designated pIL871. This plasmid was constructed by deleting a StyI fragment of pIL253, carrying the ermAM gene, and replacing it with a 1.8-kb XhoII fragment of plasmid pGKV259 (44), carrying the cat-86 gene. The DNA fragment carrying orf1 was obtained by PCR amplification using plasmid pIL2650 as a template and oligonucleotides 29 and 30 (Table 1). The PCR product was then cloned at the EcoRI-BamHI sites on plasmid vector pIL871, under the control of a plasmid promoter. The resulting construct was designated pIL2699. Plasmids pIL253 and pIL871, which share the same replication protein, can be maintained in mixture by double selection with erythromycin and chloramphenicol. Under these conditions, both plasmids have the same copy number. This was also observed for plasmids pIL2651 and pIL2699 (data not shown).
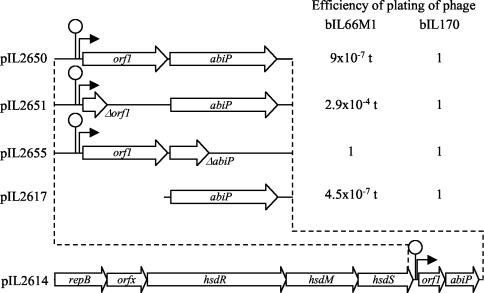
FIG. 1.
Structure and functional characterization of the AbiP determinant. Efficiency of plating is calculated as PFU per milliliter formed on strain IL1403 containing the indicated plasmid divided by PFU per milliliter formed on the control strain, IL1403. t, hardly visible turbid plaques. All segments are cloned on the high-copy-number plasmid pIL253, in the orientation of the replication gene. Results are means from three independent experiments. Lollipop, transcription terminator; bent arrow, promoter.
Nucleotide sequence accession numbers.
The DNA sequence of abiP and that of part of the genome of phage bIL66M1 have been deposited in GenBank (accession no. U90222 and AY249139, respectively).
RESULTS
Cloning and identification of the AbiP determinant.
L. lactis subsp. cremoris strain IL420 is a highly phage resistant strain from our collection. In order to identify the phage resistance mechanism(s) present in this strain, total DNA was partially digested with Sau3AI endonuclease, and fragments of approximately 10 kb were ligated with BamHI-cleaved pIL253 vector DNA (38). The ligation mixture was used to transform L. lactis subsp. lactis IL1403. Emr transformants, resistant to phage bIL66M1 (4), were selected as described in Materials and Methods and then screened for plasmid content. The recombinant plasmid carrying the smallest DNA insert (9.4 kb) was designated pIL352.
Phage bIL66M1, plated on IL1403, formed clear plaques with a mean diameter of 2 mm after overnight incubation. In contrast, when plated on IL1403(pIL352), it formed hardly visible turbid plaques with a strongly reduced efficiency of plating (approximately 10−7). [Efficiency of plating was calculated as PFU per milliliter formed on IL1403(pIL352) divided by PFU per milliliter formed on IL1403.] Phage adsorption was not affected by the presence of pIL352. After 15 min of contact at 30°C, the same efficiency of adsorption (97%) was observed with IL1403 and with IL1403(pIL352). However, one-step growth curves obtained with IL1403 and with IL1403(pIL352) revealed different phage multiplication cycle parameters. Only 0.2% of the adsorbed phages were able to release a progeny on IL1403(pIL352). The burst size was 150 in IL1403 and 35 in IL1403(pIL352). Phages isolated from plaques formed on IL1403(pIL352) were able to grow on IL1403 but not on IL1403(pIL352), indicating that host-controlled modification was not involved. Thus, the phage resistance system encoded by pIL352 is phenotypically similar to previously described abortive phage infection systems and was therefore designated AbiP. The effect of this resistance system was tested on seven additional phages representing the three prevalent lactococcal phage groups (three phages, like bIL66M1, belonging to the 936 group [bIL41, bIL170, and sk1], two belonging to the C6A group [bIL67 and c2], and two belonging to the P335 group [bIL32 and bIL198]). Phages bIL41 and sk1 were sensitive, with efficiencies of plating of 10−7 and 10−8, respectively. The other five phages were resistant to AbiP, suggesting that AbiP is effective only on phages of the 936 group.
Further deletion experiments using various restriction endonucleases demonstrated that the phage resistance determinant was present on a 1.7-kb fragment. Expression of the Abi phenotype was independent of the orientation of the cloned fragment, indicating that an Abi gene(s) and promoter were both present on this fragment. Plasmid pIL253 carrying this fragment was designated pIL2650 (Fig. 1). The cloned fragment was shown to hybridize with a 14-kb plasmid present in strain IL420 (data not shown). This plasmid, designated pIL2614, transferred by electroporation into strain IL1403, was shown to code for both abortive phage infection and a type I restriction-modification (R-M) system (37).
Sequence analysis of the AbiP determinant.
Sequence analysis of the 1,706-bp cloned fragment (GenBank accession no. U90222, from position 8360) revealed two open reading frames (ORFs), designated orf1 and orf2, preceded by the 3′ end of the hsdS gene of a type I R-M system (37) and a transcription terminator but no promoter consensus sequence. Orf1 (194 aa) shares no significant homology with proteins in the databases and presents five putative transmembrane helices (positions 7 to 29, 55 to 77, 82 to 102, 112 to 134, and 155 to 177) (27, 40). Orf2 (245 aa) shares 22% identity with the lactococcal abortive phage infection protein AbiC (19). Alignment of the two proteins reveals three regions of homology separated by an insertion (on AbiC; 344 aa) or by deletion (on Orf2) of two stretches of 34 and 46 amino acids. The two proteins have similar predicted secondary structures, with one (Orf2, positions 15 to 37) or two (AbiC, positions 23 to 45 and 60 to 82) N-terminal transmembrane helices (27, 40) and a large C-terminal loop. However, they differ sharply in isoelectric points (5.7 for Orf2 and 10.1 for AbiC).
Functional analysis of the AbiP determinant.
To determine the role played by orf1 and orf2 in the AbiP phenotype, each ORF was inactivated on plasmid pIL2650 by introduction of a frameshift mutation (see Materials and Methods), and the effect on phage multiplication was studied. Results are presented in Fig. 1. Growth of phage bIL66M1 was 6 orders of magnitude lower on IL1403(pIL2650) than on IL1403. Moreover, plaques formed on IL1403(pIL2650) were turbid and hardly visible. After inactivation of orf1 (pIL2651), phage bIL66M1 still formed hardly visible turbid plaques, but its growth was reduced by only 4 orders of magnitude. In contrast, inactivation of orf2 (pIL2655) fully restored phage growth, suggesting that only orf2 participates in the AbiP phenotype. This result was confirmed by subcloning orf2 on plasmid pIL253 under the control of a plasmid promoter (pIL2617), which conferred the Abi phenotype (Fig. 1). orf2 was thus designated abiP.
The fact that the inactivation of orf1 decreases the efficiency of AbiP might reflect a regulatory function of orf1. To investigate this possibility, the effect of orf1, brought in trans, on the efficiency of the Abi phenotype encoded by pIL2651 was studied. orf1, together with its promoter and the upstream region (which is putatively involved in its regulation), was cloned into pIL871 to yield pIL2699 (see Materials and Methods). Introduction of pIL2699 by transformation into cells carrying pIL2651 did not produce any difference in their AbiP efficiency.
Transcription of abiP.
Northern blot experiments were performed on strains IL1403(pIL2614) and IL1403(pIL2650) by using oligonucleotides 3, 4, and 5 successively as probes. These primers are complementary to sequences located upstream of the putative terminator structure (oligonucleotide 3), in orf1 (oligonucleotide 4), or in abiP (oligonucleotide 5). The experiments revealed a single transcript, corresponding to the sequence 5′ of the transcription terminator, which suggests that the transcription terminator upstream of orf1 and abiP is functional. Use of a PCR fragment covering orf1 and abiP as a probe did not allow detection of any orf1 or abiP transcript. Similar results were obtained with RNAs extracted following phage bIL66M1 infection.
Therefore, expression of abiP was studied by RT-PCR, which is more sensitive than Northern blotting. A low constitutive level of the abiP transcript was detected in both IL1403(pIL2614) and IL1403(pIL2650). This level increased slightly 5 min after infection of strain IL1403(pIL2614) with phage bIL66M1 (Fig. 2). Control experiments performed on 16S rRNA indicated that there were no significant differences in the amounts of RNA samples. Hence, expression of abiP, carried on plasmid pIL2614, is slightly enhanced following phage infection. No such increase was observed when abiP was carried on pIL2650.
FIG. 2.
Expression of abiP from plasmid pIL2614 is enhanced following phage bIL66M1 infection. RT-PCRs using oligonucleotides complementary to abiP (left) and 16S rRNA (right) were conducted, as described in Materials and Methods, on RNAs extracted at 0 and 5 min.
To identify the transcription start site of the abiP transcript, 5′ rapid amplification of cDNA ends (RACE) was performed using oligonucleotide 10, which is complementary to abiP. Amplification products, corresponding to time zero and 5 min after infection of IL1403(pIL2614) or IL1403(pIL2650), were sequenced. Both products revealed the same transcription start site, at nucleotide 8627, upstream of orf1. Thus, the two genes are transcribed as an operon, and transcription is initiated at a promoter with an extended −10 consensus but lacking a −35 consensus sequence (TGATATAAT; positions 8612 to 8620) (26, 28). The amount of the transcript, too small to be detected by Northern blotting, increased slightly in the strain containing the wild-type plasmid pIL2614 following phage infection.
Effect of AbiP on phage DNA replication.
Replication of phage bIL66M1 DNA in the AbiP− strain IL1403 and the AbiP+ strain IL1403(pIL2617) was compared (Fig. 3). Total-cell DNA was extracted at various time intervals following infection of each strain with phage bIL66M1. DNA samples were digested with HincII and quantified with a PhosphorImager after Southern hybridization with total-phage DNA as a probe. A dramatic effect of AbiP on phage DNA replication was observed (Fig. 3A and B). Approximately 10 min after infection, DNA synthesis ceased in AbiP+ cells but strongly increased in AbiP− cells. To assess the effect of the AbiP determinant on phage DNA maturation, the same membranes were hybridized with oligonucleotides 13 and 14, complementary to sequences located downstream and upstream, respectively, of the putative cos sequence (4). In AbiP− cells (Fig. 3C), maturation of phage DNA was revealed by the appearance of two fragments (2.1 and 0.8 kb) originating from the cleavage of the HincII fragment (2.9 kb), which contains the cos sequence. These bands were not observed in AbiP+ cells, suggesting that maturation of phage DNA does not occur.
FIG. 3.
Replication of phage bIL66M1 DNA in AbiP− IL1403 and AbiP+ IL1403(pIL2617) cells. (A) Total-cell DNA was extracted at various time intervals after infection, cleaved with HincII, and analyzed by Southern hybridization. (B) Comparison of kinetics of phage bIL66M1 DNA amounts (expressed in arbitrary units) in AbiP− IL1403 and AbiP+ IL1403(pIL2617) cells. (C) Maturation of phage bIL66M1 DNA in AbiP− IL1403 and AbiP+ IL1403(pIL2617) cells. Only one 2.9-kb fragment containing the cos site is produced in AbiP+ cells, whereas two additional subfragments of 2.1 and 0.8 kb are produced in AbiP− cells.
Effect of AbiP on phage DNA transcription.
Previous transcription studies done on phages of the 936 group (10, 34) have revealed the existence of three temporal classes of transcripts, designated early, middle, and late, respectively. In this work, transcription of each class of transcripts of phage bIL66M1 was compared in the presence and absence of the AbiP determinant.
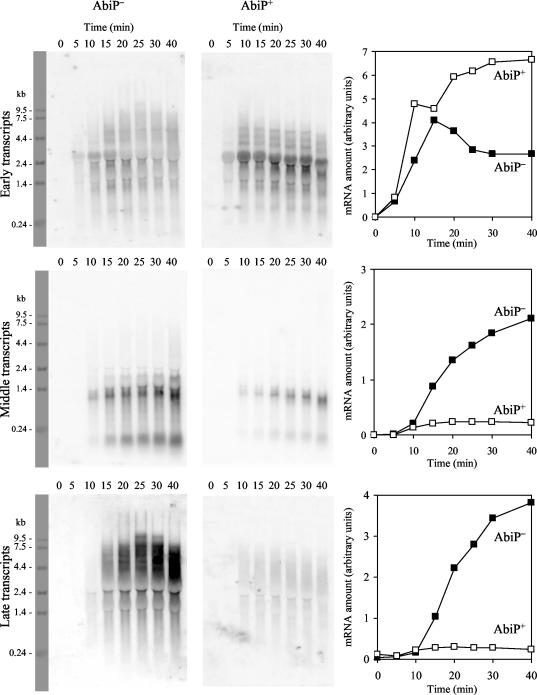
Total-cell RNA was extracted at various time intervals after infection and analyzed by Northern hybridization using a probe complementary to the early, middle, or late transcript. These probes were made of a mixture of DNA fragments obtained by long-range PCR using the closely related phage bIL170, whose DNA sequence is known (15), as a template, with oligonucleotides 15 to 28 (Table 1). In AbiP− cells, amounts of early transcripts increased until 15 min after infection and declined thereafter (Fig. 4). This switch-off is frequent among phages and has already been observed for sk1 (10) and bIL170 (34). It suggests the existence of a temporal control of the amount of early transcripts. Middle and late transcripts were detected approximately 10 min after infection, and their amounts increased regularly through the phage multiplication cycle. Transcription in AbiP+ cells was similar to that in AbiP− cells during the first 10 min following infection (Fig. 4). Later on, transcription was significantly different in both types of cells. The amount of early transcripts increased regularly in AbiP+ cells, whereas it decreased markedly in AbiP− cells. By contrast, amounts of middle and late transcripts remained constant in AbiP+ cells but increased regularly in AbiP− cells. Quantification of the transcripts by use of a PhosphorImager revealed that at 30 to 40 min after infection, AbiP+ cells contained 2.5-fold more early transcripts, 8-fold less middle transcripts, and 12-fold less late transcripts than AbiP− cells. The smaller amounts of middle and late transcripts in AbiP+ cells might be a consequence of the smaller amount of template DNA in these cells (Fig. 3). By contrast, this smaller amount of template DNA did not prevent the constant increase in the amount of early transcripts in AbiP+ cells. This suggests an effect of AbiP on the temporal control of the amount of early phage transcripts.
FIG. 4.
Comparison of kinetics of phage bIL66M1 early, middle, and late transcripts following infection of AbiP− IL1403 or AbiP+ IL1403(pIL2617) cells. Total-cell RNA was extracted at the times indicated above the gels and was hybridized with a mixture of overlapping PCR fragments spanning the early, middle, and late regions. RNA amounts (expressed in arbitrary units) are shown in the graphs on the right.
To better characterize the effect of AbiP on early phage transcripts, the sequence of the first part of the bIL66M1 early region (6,058 bp) was determined and its transcription was studied by Northern blot analysis using oligonucleotide probes. Comparison of the sequence of this region of the bIL66M1 genome revealed that it was homologous to those of sk1 (11) and bIL170 (15) over 53 and 70% of its length, respectively. This is in agreement with the genome conservation already observed in phages of the 936 group, which are homologous over 81 to 94% of their length, with most of the genome diversity observed in the early region (13). Comparison of the bIL66M1 sequence with the transcription map of phage sk1 (10) enabled the identification of promoter PE1 in bIL66M1. By visual inspection, we also identified the putative promoters PE2 and PE3 and the canonical transcription terminator T (Fig. 5A). All the early promoters PE1 to PE3 have the consensus sequence of bacterial vegetative promoters. Transcription study of this region, after infection of AbiP− cells, revealed a complex pattern of transcripts that were temporally controlled, reaching a maximum at 15 min after infection and decreasing later on. In AbiP+ cells, the amount remained constant from 15 to 40 min after infection. An example of the effect of AbiP on the temporal evolution of early transcripts is shown in Fig. 5B, with the 240-nucleotide-transcript initiated at PE1 and terminated at T. These results confirm the existence of a temporal control of the amount of early transcripts, which is abolished in AbiP+ cells.
FIG. 5.
(A) Schematic structure of the early-expressed region of phage bIL66M1. (B) Comparison of amounts of the early transcript E1 in AbiP− IL1403 and AbiP+ IL1403(pIL2617) cells. Total-cell RNA extracted at the time intervals indicated above the gels was hybridized with oligonucleotide 12, specific for orfE1.
DISCUSSION
An abortive phage infection mechanism, designated AbiP, has been identified in L. lactis subsp. cremoris strain IL420. Among the many Abi determinants described in lactococci, AbiP presents the unique feature of affecting both phage DNA replication and transcription.
In strain IL420, the AbiP phenotype is encoded by the native plasmid pIL2614, which also codes for a type I R-M system (37). So far, all lactococcal Abi's whose genetic determinants have been studied are plasmid encoded, except AbiN, which most probably originates from a prophage (36). This can be due to the fact that, as observed for AbiA (17) and AbiP (data not shown), the efficiency of Abi systems is affected by the gene dosage.
The AbiP determinant has been cloned on a 1.7-kb DNA fragment containing two complete ORFs. By mutational analysis, we demonstrated that one of these ORFs, designated abiP, is sufficient to confer the AbiP phenotype. The role of the second ORF (orf1), whose inactivation reduced the efficiency of AbiP, but which was unable to restore full AbiP efficiency when supplied in trans, remains unclear.
Transcriptional analysis of orf1 and abiP revealed that they are part of a poorly transcribed operon and that the amount of transcript increased slightly following phage infection. This increase may be due either to activation of transcription or to stabilization of the transcripts. The first hypothesis is favored by the observation that the operon is transcribed from a promoter lacking a consensus −35 region. This phenomenon was observed only when abiP was present on the wild-type plasmid pIL2614, indicating that other plasmid-encoded genes may be involved. Activation of Abi mechanisms in response to phage infection has been demonstrated in the three Escherichia coli systems that have been studied in detail (32, 39). In lactococci, a phage protein was also shown to drastically increase the efficiency of AbiD1 (6). It will be interesting to determine whether other lactococcal Abi's possess this feature, which may represent a general characteristic of these systems.
On the basis of homology with known proteins, no putative function can be proposed for the orf1- and abiP-encoded proteins. AbiP shares homology only with AbiC, another lactococcal Abi (19). However, the two proteins have different modes of action on phage development: AbiC does not affect phage DNA replication, whereas AbiP blocks DNA production 10 min after infection. Interestingly, AbiD, AbiD1, and AbiF, three other homologous lactococcal Abi's, may also have a different mode of action (20). This suggests that lactococci have evolved paralogous genes to enlarge the already great variety of available phage resistance mechanisms.
AbiP was shown to affect both phage DNA replication and transcription at 10 to 15 min after infection. In AbiP− cells, phage DNA synthesis strongly increased between 15 and 40 min after infection, and maturated forms appeared after approximately 20 min. In AbiP+ cells, phage DNA synthesis stopped 10 to 15 min after infection, and no maturated forms of DNA were detected.
The absence of DNA maturation could result from the absence of either multimeric DNA forms needed for packaging or middle or late phage products needed to resolve DNA structures.
As often observed for phages, and as already described for the highly similar phages sk1 and bIL170 (10, 34), three temporal classes of transcripts were observed for phage bIL66M1. In AbiP− cells, early transcripts, detected approximately 5 min after infection, reached a maximum at 15 min and decreased later on, suggesting the existence of a temporal control of these transcripts. Middle and late transcripts, detected approximately 10 min after infection, increased up to the end of the cycle. In AbiP+ cells, by contrast, amounts of early transcripts increased up to 40 min, whereas the amounts of middle and late transcripts remained much lower. These observations are most simply explained by the hypothesis that the reduced amounts of middle and late transcripts are a consequence of the reduced amount of the DNA template. By contrast, despite the strongly reduced amount of the DNA template in AbiP+ cells, the amount of early transcripts is clearly greater than that in AbiP− cells. To explain this unexpected observation, we propose that AbiP hinders the temporal control of the amount of early transcripts, which normally takes place during phage development.
Since mechanisms of transcription control and DNA replication in lactococcal phages are poorly understood, it is difficult to propose a mode of action for AbiP. However, the observation that AbiP disturbs both phage DNA replication and temporal control of early phage transcription raises the question of how a single gene can affect two biological functions. AbiP could be a multifunctional protein that is able to act independently on phage DNA replication and temporal control of early transcription. AbiP could also interact only with phage DNA replication, which, in turn, would affect transcription—or the converse could be true. There are at present no arguments for or against one of these hypotheses. However, it is noteworthy that in the E. coli phage λ, which shares a general genome structure with lactococcal phages and which is the paradigm for small circular double-stranded DNA phage, DNA replication and transcription are functionally linked (3, 41).
Acknowledgments
S. Domingues was supported by the Fundação Para a Ciência e Tecnologia (Lisbon, Portugal).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anba, J., E. Bidnenko, A. J. Hillier, D. Ehrlich, and M.-C. Chopin. 1995. Characterization of the lactococcal abiD1 gene coding for phage abortive infection. J. Bacteriol. 177:3818-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranska, S., M. Gabig, A. Wegrzyn, G. Konopa, A. Herman-Antosiewicz, P. Hernandez, J. B. Schvartzman, D. R. Helinski, and G. Wegrzyn. 2001. Regulation of the switch from early to late bacteriophage λ DNA replication. Microbiology 147:535-547. [DOI] [PubMed] [Google Scholar]
- 4.Bidnenko, E., S. D. Ehrlich, and M.-C. Chopin. 1995. Phage operon involved in sensitivity to the Lactococcus lactis abortive infection mechanism AbiD1. J. Bacteriol. 177:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bidnenko, E., S. D. Ehrlich, and M. C. Chopin. 1998. Lactococcus lactis phage operon coding for an endonuclease homologous to RuvC. Mol. Microbiol. 28:823-834. [DOI] [PubMed] [Google Scholar]
- 6.Bidnenko, E., M. C. Chopin, S. D. Ehrlich, and J. Anba. 2002. Lactococcus lactis AbiD1 abortive infection efficiency is drastically increased by a phage protein. FEMS Microbiol. Lett. 214:283-287. [DOI] [PubMed] [Google Scholar]
- 7.Bouchard, J. D., E. Dion, F. Bissonnette, and S. Moineau. 2002. Characterization of the two-component abortive phage infection mechanism AbiT from Lactococcus lactis. J. Bacteriol. 184:6325-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouchard, J. D., and S. Moineau. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65-75. [DOI] [PubMed] [Google Scholar]
- 9.Boucher, I., E. Emond, E. Dion, D. Montpetit, and S. Moineau. 2000. Microbiological and molecular impacts of AbiK on the lytic cycle of Lactococcus lactis phages of the 936 and P335 species. Microbiology 146:445-453. [DOI] [PubMed] [Google Scholar]
- 10.Chandry, P. S., B. E. Davidson, and A. J. Hillier. 1994. Temporal transcription map of the Lactococcus lactis bacteriophage sk1. Microbiology 140:2251-2261. [DOI] [PubMed] [Google Scholar]
- 11.Chandry, P. S., S. C. Moore, J. D. Boyce, B. E. Davidson, and A. J. Hillier. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26:49-64. [DOI] [PubMed] [Google Scholar]
- 12.Chopin, A., M.-C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 13.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M. C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cluzel, P. J., A. Chopin, S. D. Ehrlich, and M. C. Chopin. 1991. Phage abortive infection mechanism from Lactococcus lactis subsp. lactis, expression of which is mediated by an Iso-ISS1 element. Appl. Environ. Microbiol. 57:3547-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crutz-Le Coq, A. M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 16.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programmes for the Vax. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinsmore, P. K., and T. R. Klaenhammer. 1994. Phenotypic consequences of altering the copy number of abiA, a gene responsible for aborting bacteriophage infections in Lactococcus lactis. Appl. Environ. Microbiol. 60:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinsmore, P. K., and T. R. Klaenhammer. 1997. Molecular characterization of a genomic region in a Lactococcus bacteriophage that is involved in its sensitivity to the phage defense mechanism AbiA. J. Bacteriol. 179:2949-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durmaz, E., D. L. Higgins, and T. R. Klaenhammer. 1992. Molecular characterization of a second abortive phage resistance gene present in Lactococcus lactis subsp. lactis ME2. J. Bacteriol. 174:7463-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forde, A., and G. F. Fitzgerald. 1999. Bacteriophage defence systems in lactic acid bacteria. Antonie Leeuwenhoek 76:89-113. [PubMed] [Google Scholar]
- 21.Garvey, P., G. F. Fitzgerald, and C. Hill. 1995. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from lactococcal plasmid pNP40. Appl. Environ. Microbiol. 61:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasson, M. 1983. Plasmid complements of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glatron, M. F., and G. Rapoport. 1972. Biosynthesis of the parasporal reclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie 54:1291-1301. [DOI] [PubMed] [Google Scholar]
- 24.Hill, C., I. J. Massey, and T. R. Klaenhammer. 1991. Rapid method to characterize lactococcal bacteriophage genomes. Appl. Environ. Microbiol. 57:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keity, S., and M. Rosenberg. 1987. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J. Biol. Chem. 262:6389-6395. [PubMed] [Google Scholar]
- 27.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, A., R. A. Malloch, N. Fujita, D. A. Smillie, A. Ishihama, and R. S. Haynard. 1993. The −35 recognition region of Escherichia coli σ70 is inessential for initiation of transcription at an “extended −10” promoter. J. Mol. Biol. 232:406-418. [DOI] [PubMed] [Google Scholar]
- 29.Labrie, S., and M. Moineau. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Moineau, S., S. Pandian, and T. R. Klaenhammer. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. Microbiol. 60:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molineux, I. J. 1991. Host-parasite interactions: recent developments in the genetics of abortive phage infections. New Biol. 3:230-236. [PubMed] [Google Scholar]
- 33.O'Connor, L., M. Tangney, and G. F. Fitzgerald. 1999. Expression, regulation, and mode of action of the AbiG abortive infection system of Lactococcus lactis subsp. cremoris UC653. Appl. Environ. Microbiol. 65:330-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parreira, R., S. D. Ehrlich, and M. C. Chopin. 1996. Dramatic decay of phage transcripts in lactococcal cells carrying the abortive infection determinant AbiB. Mol. Microbiol. 19:221-230. [DOI] [PubMed] [Google Scholar]
- 35.Pillidge, C. J., and A. W. Jarvis. 1988. DNA restriction maps and classification of the lactococcal bacteriophages c2 and sk1. N. Z. Dairy Sci. Technol. 23:411-416. [Google Scholar]
- 36.Prévots, F., S. Tolou, B. Delpech, M. Kaghad, and M. Daloyau. 1998. Nucleotide sequence and analysis of the new chromosomal abortive infection gene abiN of Lactococcus lactis subsp. cremoris S114. FEMS Microbiol. Lett. 159:331-336. [DOI] [PubMed] [Google Scholar]
- 37.Schouler, C., F. Clier, A. L. Lerayer, S. D. Ehrlich, and M. C. Chopin. 1998. A type IC restriction-modification system in Lactococcus lactis. J. Bacteriol. 180:407-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 39.Snyder, L. 1995. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol. Microbiol. 15:415-420. [DOI] [PubMed] [Google Scholar]
- 40.Sonnhammer, E. L. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (ed.), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 41.Taylor, K., and G. Wegrzyn. 1995. Replication of coliphage lambda DNA. FEMS Microbiol. Rev. 17:109-119. [DOI] [PubMed] [Google Scholar]
- 42.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Twomey, D. P., P. J. De Urraza, L. L. McKay, and D. J. O'Sullivan. 2000. Characterization of AbiR, a novel multicomponent abortive infection mechanism encoded by plasmid pKR223 of Lactococcus lactis subsp. lactis KR2. Appl. Environ. Microbiol. 66:2647-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Vossen, J. M., D. van der Lelie, and G. Venema. 1987. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 53:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson, M., J. Doskow, and S. Lindsey. 1991. RNA blots: staining procedures and optimization of conditions. Nucleic Acids Res. 19:679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams, J. G., and P. J. Mason. 1985. Hybridization in the analysis of RNA, p139-160. In B. D. Hames and S. J. Higgins (ed.), Nucleic acid hybridization—a practical approach, IRL Press, Oxford, United Kingdom.