Abstract
Serratia liquefaciens MG1 contains an N-acylhomoserine lactone-mediated quorum-sensing system that is known to regulate swarming motility colonization. In this study, we describe for S. liquefaciens MG1 the development of a novel biofilm consisting of cell aggregates and differentiated cell types, such as cell chains and long filamentous cells. Furthermore, quorum sensing is shown to be crucial for normal biofilm development and for elaborate differentiation. A mutant of S. liquefaciens MG1 that was incapable of synthesizing extracellular signal formed a thin and nonmature biofilm lacking cell aggregates and differentiated cell chains. Signal-based complementation of this mutant resulted in a biofilm with the wild-type architecture. Two quorum-sensing-regulated genes (bsmA and bsmB) involved in biofilm development were identified, and we propose that these genes are engaged in fine-tuning the formation of cell aggregates at a specific point in biofilm development.
N-Acylhomoserine lactone (AHL)-mediated quorum sensing (QS) controls diverse phenotypic traits in various gram-negative proteobacteria (for a recent review, see reference 29). However, despite the proposed role of QS systems in bacterial colonization of surfaces, to date only a limited number of organisms are known to require a functional AHL-mediated QS system for formation of biofilms. In Pseudomonas aeruginosa, Burkholderia cepacia, and Aeromonas hydrophila, functional AHL regulatory systems are necessary for formation of microcolony structures (5, 13, 18). In Pseudomonas putida IsoF, AHL-mediated QS affects the architecture of the biofilm, and a QS mutant forms a heterogeneous microcolony-based biofilm rather than the homogeneous biofilm typical of the wild type (26). In these organisms, a specific QS-regulated determinant for biofilm formation has been identified only in P. putida IsoF. An AHL-regulated long-chain fatty acid coenzyme ligase was proposed to alter the fatty acid composition of the cell membrane, changing the surface properties of the cell and ultimately the biofilm structure (26).
Serratia liquefaciens is an opportunistic pathogen which is capable of colonizing a wide variety of surfaces in water, soil, the digestive tracts of rodents, plants, insects, fish, and humans (10). S. liquefaciens secretes a broad range of hydrolytic enzymes, and strain MG1 can differentiate into specialized swarmer cells capable of rapid surface motility (6). Swarming motility in S. liquefaciens MG1 is AHL regulated and is directed through specific control of the swrA gene. This gene encodes a peptide synthetase that catalyzes synthesis of the biosurfactant serrawettin W2, which reduces surface tension and allows swarming motility to occur (17). QS is therefore involved in at least one surface colonization process in S. liquefaciens MG1. SwrI and SwrR, homologues of the I and R proteins characteristic of AHL-mediated systems, have also been reported in this strain (7). SwrI synthesizes the signal molecules N-butanoyl-l-homoserine lactone (C4-HSL) and N-hexanoyl-l-homoserine lactone at a 10:1 ratio (7). The signal molecule (C4-HSL or N-hexanoyl-l-homoserine lactone) is hypothesized to bind the SwrR transcriptional regulator and affect the expression of at least 28 proteins, as shown by two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) (9). This suggests that a number of other phenotypes are also regulated by AHL-mediated QS in S. liquefaciens MG1.
In this study, we demonstrated that the surface-colonizing bacterium S. liquefaciens MG1 forms a unique type of biofilm structure consisting of cell aggregates, differentiated filaments, and cell chains. We showed that normal biofilm formation in S. liquefaciens MG1 requires a functional QS system, and we identified two AHL-regulated genes that are proposed to fine-tune the formation of cell aggregates at a specific point in biofilm development.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth.
The bacterial strains used in this study are listed in Table 1. All strains were routinely grown at 30°C in Luria-Bertani broth or Difco minimal broth (DMB) supplemented with 0.2% (wt/vol) glucose and 0.5% (wt/vol) Casamino Acids. For complementation of strains MG44 and MG3633 to MG3651 in the static biofilm assay and flow chamber experiments, 250 nM C4-HSL was added to the medium. In the complementation flow chamber experiments with strains MG3646(pBsmA), MG3646(pBsmAOp), and MG3651(pBsmB), 250 nM C4-HSL and 30 μg of gentamicin ml−1 were added to the medium.
TABLE 1.
Strains and plasmids
| Strain(s) or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Promega |
| MV1190 (λ-pir) | Δ(lac-proAB) Δ(srl-recA)306::Tn10[F′ traD36 proAB lacIq Δ(lacZ)M15] thi supE; lysogenized with λ-pir phage | 11 |
| HB101 | recA thi pro leu hsdRM+ Smr | 15 |
| Serratia liquefaciens strains | ||
| MG1 | Wild type, Apr Tcr | 8 |
| MG44 | MG1, swrI gene disrupted with a streptomycin cassette, Smr | 7 |
| MG3633 to MG3651 | MG44, disruption in a C4-HSL responsive gene by a mini-Tn5 transposon carrying promoterless luxAB and a kanamycin marker, Kmr | 17 |
| MG3659 | gfpmut3*-T0-T1-tagged MG1, Kmr | This study |
| MG3663 | gfpmut3*-T0-T1-tagged MG44, Kmr | This study |
| Plasmids | ||
| pBluescript II SK | Cloning vector, Apr | Stratagene |
| pBR322 | Cloning vector, Apr Tcr | New England Biolabs |
| pUCGM | Gmr | 24 |
| pJBA28 | Apr Kmr | 2 |
| pBRGM | pBR322 bearing the 855-bp gentamicin cassette from pUCGM | This study |
| pML001 | pBluescript II SK bearing a 1.4-kb bsmA fragment | This study |
| pML002 | pBluescript II SK bearing a 1.2-kb bsmB fragment | This study |
| pML003 | pBluescript II SK bearing a 2.8-kb bsmA-nifS fragment | This study |
| pBsmA | pBRGM bearing a 1.4-kb bsmA fragment | This study |
| pBsmB | pBRGM bearing a 1.2-kb bsmB fragment | This study |
| pBsmAOp | pBRGM bearing a 2.8-kb bsmA-nifS fragment | This study |
| RK600 | ori ColE1 RK2-Mob+ RK2-Tra+ Cmr; helper plasmid in triparental matings | 15 |
Tcr, tetracycline resistant; Apr, ampicillin resistant; Smr, streptomycin resistant; Kmr, kanamycin resistant; Gmr, gentamicin resistant.
DNA manipulation and nucleotide sequencing.
Plasmid DNA was prepared by using the Wizard Plus Minipreps DNA purification system (Promega). Cloning, chemical transformation of Escherichia coli, and electroporation of S. liquefaciens MG1 were performed by using standard procedures (3). Genomic DNA was extracted from cultures of S. liquefaciens strains MG3646 and MG3651 by using the XS buffer protocol (28). Identification of transposon mutated genes was carried out by using a combination of two protocols. The first protocol was an adaptor ligation PCR protocol described by Siebert et al. (25) and modified by Tillett (27), in which the specific mini-Tn5 transposon primers P6 (5′-GCCCGGTCGCATTACACCTT-3′) and P7.2 (5′-CGCTTCATCACTTCGGTCTGAGA-3′) were used. The second protocol involved shotgun ligating BamHI-digested chromosomal DNA into pBluescript II SK. Clones bearing inserts carrying the Tn5 kanamycin resistance marker and chromosomal DNA were selected and sequenced. Both strands of DNA were sequenced by using a primer walking strategy in a thermocycling reaction with BigDye terminators. Sequences were analyzed at the Automated Sequencing Facility at the University of New South Wales. DNA sequences were compared to other sequences in the GenBank database by using the on-line open reading frame (ORF) finder and BLASTP programs at the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/).
Construction of plasmids pBsmA, pBsmAOp, and pBsmB.
Plasmids pBsmA, pBsmAOp, and pBsmB were constructed by amplifying bsmA, the bsmA-nifS operon, and bsmB from S. liquefaciens MG1 with Pwo polymerase (Roche) by using primers F2ML (5′-TTACCCACCAGAAGCTTGAAGG-3′) (HindIII) and R2ML (5′-CCAGGTTGGCTCTAGATTTCCC-3′) (XbaI), primers F2ML and R7ML (5′-ATCGTCATGGTCTAGAAGGTCG-3′) (XbaI), and primers F5ML (5′-CTGGCAACATATCTAGATTACCC-3′) (XbaI) and R4ML (5′-GGTTCGGTGAGAATTCAGGAG-3′) (EcoRI), respectively; these primers were engineered to code for restriction endonuclease sites (underlined), and the enzyme that cleaved each sequence is indicated in parentheses. Plasmid pBluescript II SK and the PCR-amplified fragments (containing either bsmA, bsmB, or the bsmA-nifS operon) were digested with the corresponding restriction endonuclease sites engineered into the primers and purified, and the fragments were ligated into pBluescript II SK to obtain pML001 (bearing bsmA), pML002 (bearing bsmB), and pML003 (bearing the bsmA-nifS operon). Plasmid pBRGm was then constructed by ligating the 855-bp SmaI fragment from pUCGm containing the gentamicin resistance marker into the SspI site of pBR322. The inserts from pML001, pML002, and pML003 were then transferred to plasmid pBRGm to obtain plasmids pBsmA, pBsmB, and pBsmAOp, respectively.
Construction of gfp-tagged S. liquefaciens strains MG3659 and MG3663.
S. liquefaciens strains MG1 and MG44 were tagged with gfpmut3*-T0-T1 by transposition with pJBA28 to create S. liquefaciens strains 3659 and 3663, respectively. The transposition procedure was carried out as described previously (2).
Microtiter plate biofilm screening.
Nineteen previously isolated mutants bearing Tn5::luxAB::npt insertions in QS-regulated genes (17) were screened for biofilm formation in 24-well polystyrene microtiter dishes (Sarstedt Inc., Newton, N.C.) as previously described (21, 22). DMB supplemented with 0.2% glucose and 0.5% Casamino Acids was used, and biofilms were allowed to form for 24 h at 30°C with shaking and then assessed by microscopy for microcolony formation at the base of the wells.
Luminescence assays.
Induction of the mutated genes in MG3646 and MG3651 was determined by measuring luxAB expression after addition of C4-HSL. Samples were taken hourly during growth and measured for luminescence with a Wallac Victor2 1420 multilabel counter.
Flow chamber biofilm experiments.
Biofilms of S. liquefaciens strains were grown by using DMB supplemented with 0.05% (wt/vol) glucose and Casamino Acids. The flow chamber consisted of a glass base (10 by 5 cm) with two glass slides (7.5 by 2.5 cm) glued on top with a UV-curing glue (3042B; Three Bond Co., Ltd., Tokyo, Japan). A 1-mm gap was left between the slides, and a glass coverslip (6 by 2.4 cm) was glued on top of the slides, resulting in an all-glass flow tunnel. Silicone tubing was connected to each side of the flow cell tunnel and sealed with silica gel. All experiments were carried out at room temperature (approximately 21°C), and the flow chamber was sterilized by autoclaving prior to inoculation. The flow chamber was inoculated by injecting the appropriate strain into the flow channel with a small syringe. Flow into the chamber was halted for 1 h, after which sterile medium was pumped through the flow chamber with a peristaltic pump at a flow rate of 0.18 ml/min. Development of biofilms was monitored by light microscopy and fluorescence microscopy. To monitor biofilm development by fluorescence microscopy, gfp-tagged strains MG3659 (wild type) and MG3663 (swrI mutant) were used. Images of S. liquefaciens biofilms that were fixed with 2% glutaraldehyde and stained with 0.01% acridine orange were obtained with a confocal microscope. To ensure that glutaraldehyde had no effect on biofilm structure, we compared biofilms fixed and stained as described above and biofilms formed with the gfp-tagged strains (strains MG3659 and MG3663). Glutaraldehyde fixation had no effect on biofilm structure (data not shown). Thus, fixation and staining were the preferred methods of image capture since biofilms formed by the gfp-tagged strains lost green fluorescent protein fluorescence over time.
Quantification of bacterial aggregates formed in biofilms.
To quantify bacterial aggregation, triplicate 48-h-old biofilms of the S. liquefaciens wild type and the S. liquefaciens swrI mutant were fixed and stained as described above. Confocal images (magnification, ×400) were captured from 10 random points in each flow chamber, and total aggregates were counted.
Biofilm COMSTAT analysis.
For statistical evaluation of biofilm structures, triplicate flow cells were prepared for each strain, and 10 image stacks (2-μm intervals) per flow chamber were obtained for the mature biofilm at 72 h postinoculation. The images were analyzed with the biofilm computer program COMSTAT (12).
Microscopy.
Biofilms from the microtiter plate assay were examined with an Olympus IMT2 inverted scanning confocal laser microscope (Olympus Optical Co. Ltd., Tokyo, Japan) equipped with a 40× phase-contrast lens whose numerical aperture was 0.6 under normal light. Biofilm development was observed with an Olympus CH-2 light microscope and with a Leica DMLB epifluorescence microscope (Leica Microsystems, Wetzlar, Germany), both equipped with a 40× lens whose numerical aperture was 0.65. Scanning confocal laser microscopy images of the flow chambers were obtained with an Olympus GB200 microscope fitted with a piezoelectric z stage. The microscope was equipped with a 40× lens whose numerical aperture was 0.95 and a 60× oil immersion lens whose numerical aperture was 1.4. Image scanning was carried out with the 488-nm laser line of an argon laser. Captured images were further processed for display by using Photoshop software (Adobe, Mountain View, Calif.).
Nucleotide sequence accession numbers.
The nucleotide sequences of the bsmA-nifS operon and bsmB have been deposited in the GenBank database under accession numbers AF537272 and AF537273, respectively.
RESULTS
S. liquefaciens MG1 forms a biofilm with a novel structure in flow chambers.
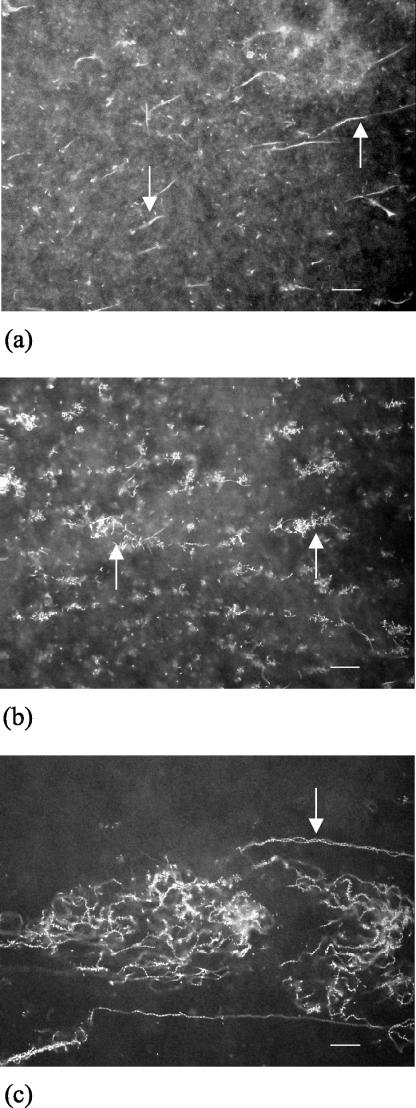
The development of the S. liquefaciens MG1 biofilm is distinct from the development of biofilms formed by model biofilm-forming bacteria, such as P. aeruginosa and E. coli. As a control, we confirmed that P. aeruginosa forms a biofilm consisting of microcolonies in our flow chamber setup, which allowed us to make direct comparisons between the two biofilm structures (data not shown). During development of the S. liquefaciens MG1 biofilm, elaborate cellular differentiation and structural differentiation were clearly observed. Specifically, at 24 h, long filamentous cells were observed (Fig. 1a); at 48 h, aggregation of vegetative bacteria with the filamentous cells was observed (Fig. 1b); and at 72 h, intertwining cell chains where there was biofilm maturation (Fig. 1c) were observed.
FIG.1.
Confocal xy images of S. liquefaciens MG1 biofilm at different times during development. (a) Formation of filamentous cells at 24 h; (b) aggregation with the filamentous cells at 48 h; and (c) mature biofilm of intertwining cell chains at 72 h. The arrows indicate examples of filamentous cells (a), aggregates (b), and intertwining cell chains (c). Bars = 20 μm.
S. liquefaciens swrI mutant forms a biofilm that is morphologically distinct from and thinner than the biofilm formed by the wild type.
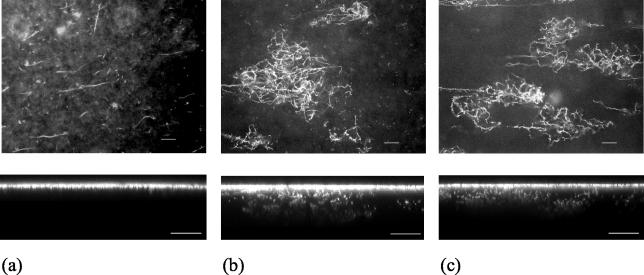
Compared to the wild-type biofilm, striking differences in biofilm architecture were observed in the swrI mutant biofilm. At 24, 48, and 72 h, the swrI mutant biofilm was identical to the 24-h wild-type biofilm consisting of long filamentous cells (Fig. 2a). This suggests that development in the mutant biofilm stalled after 24 h. In contrast, the wild-type biofilm exhibited six- to sevenfold more aggregation at 48 h (number of aggregates per field of view for the wild type, 18.6 ± 4.34 [average ± standard deviation]; number of aggregates per field of view for the swrI mutant, 2.3 ± 1.9) and contained intertwining cell chains at 72 h (Fig. 2b). The contrast between the mature wild-type and swrI mutant biofilms was further supported by COMSTAT analysis (Table 2). The swrI mutant biofilm was on average five times thinner than the wild-type biofilm, and the maximum biofilm thickness of the swrI mutant biofilm was approximately 19 μm, compared to approximately 52 μm for the wild-type biofilm. The total biovolume of the wild-type biofilm was approximately four times greater than that of the swrI mutant biofilm, confirming the greater biofilm-forming ability of the wild type. The wild-type biofilm also had slightly greater substratum coverage than the swrI mutant biofilm. Addition of 250 nM C4-HSL to the flow medium of the swrI mutant biofilm restored the developmental process to that of the wild-type biofilm, as shown in Fig. 2c and Table 2 (COMSTAT analysis). These data were replicated on more than 10 separate occasions, and we therefore concluded that proper biofilm formation by S. liquefaciens MG1 requires a functional QS system.
FIG. 2.
Confocal xy (top panels) and xz (bottom panels) images of 72-h flow chamber biofilms formed by the S. liquefaciens MG44 swrI mutant strain (a), wild-type strain MG1 (b), and swrI mutant strain MG44 in the presence of C4-HSL (c). Bars = 20 μm.
TABLE 2.
COMSTAT analysis of mature biofilms formed by S. liquefaciensa
| Strain | Mean thickness (μm) | Maximum thickness (μm) | Biovolume (μm3/μm2) | Substratum coverage (%) |
|---|---|---|---|---|
| S. liquefaciens MG1 | 34.99 ± 8.10 | 51.60 ± 12.11 | 34.76 ± 7.91 | 99.07 ± 1.41 |
| S. liquefaciens MG44 | 7.21 ± 0.85 | 18.93 ± 1.55 | 8.46 ± 0.82 | 94.42 ± 1.96 |
| S. liquefaciens MG44 with C4-HSL | 28.21 ± 4.76 | 37.27 ± 4.94 | 29.69 ± 4.83 | 99.36 ± 0.78 |
The values are means ± standard deviations.
Identification of S. liquefaciens C4-HSL-regulated biofilm genes.
In an effort to identify the specific C4-HSL-regulated genes required for biofilm formation by S. liquefaciens MG1, a set of 19 C4-HSL-responsive reporter-transposon S. liquefaciens MG44 mutants (17) were screened for reduced biofilm formation in a microtiter plate. Two of the 19 transposon mutants screened, MG3646 and MG3651, were defective in biofilm formation. Included in the 19 transposon mutants was the swrA surfactant mutant. Although this mutant is deficient in swarming colonization (17), it adhered and formed a normal biofilm in the static biofilm assay and was not studied further. The growth rates of both MG3646 and MG3651 were similar to that of the parent strain (data not shown), indicating that reduced biofilm formation was not a result of poor growth.
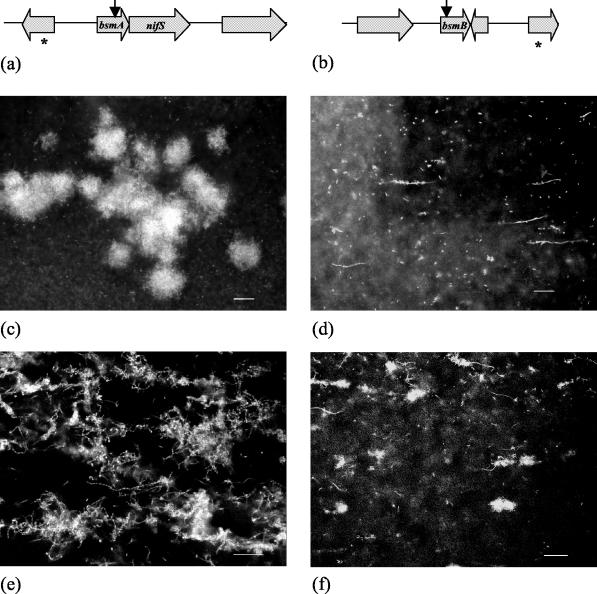
Sequence information from the site of the transposon insertion in MG3646 indicated that the transposon had disrupted a 645-bp ORF, and the deduced sequence of the protein encoded by this ORF exhibited 61% identity and 76% similarity (over 152 amino acid residues) to the sequence of a hypothetical protein (GenBank accession number NP_230828) from Vibrio cholerae. This ORF was designated bsmA (for “biofilm structure mutant A”). Downstream within the same putative operon and overlapping by 4 nucleotides is a gene coding for a protein with 61% identity and 76.5% similarity to a putative NifS homologue (GenBank accession number NP_230829) from V. cholerae (Fig. 3a). Measurement of promoter activity through detection of luminescence after addition of C4-HSL revealed that bsmA is induced 2.8-fold at the transition from the logarithmic phase to the stationary phase. While addition of C4-HSL enhanced transcription of bsmA, some expression still occurred in the absence of C4-HSL, indicating that other regulatory elements also contribute to regulation of this gene.
FIG. 3.
(a and b) Genetic maps of the site of Tn5 insertion and surrounding ORFs in biofilm mutant strains MG3646 (a) and MG3651 (b). The vertical arrow indicates the site of Tn5 insertion, and the asterisk indicates an incomplete ORF. (c to f) Confocal xy images of 72-h flow chamber biofilms formed by S. liquefaciens MG3646 (c), S. liquefaciens MG3651 (d), S. liquefaciens MG3646(pBsmA) (e), and S. liquefaciens(pBsmB) (f). Bars = 20 μm.
Analysis of the sequence data indicated that the sequence of transposon mutant MG3651 was disrupted in a 531-bp ORF designated bsmB (Fig. 3b). BLASTP analysis of the translated protein sequence of bsmB revealed no similarity to sequences in the GenBank database. This suggests that a novel gene involved in biofilm formation was present. Fifty-three base pairs downstream from bsmB is a 318-bp ORF in the opposite orientation, suggesting that bsmB is not in an operon. Promoter activity assays for bsmB in the presence and absence of C4-HSL revealed that this gene is highly dependent on a functional QS system for expression and is maximally induced 9.8-fold at the transition from the logarithmic phase to the stationary phase in the presence of C4-HSL.
Biofilm formation by MG3646 and MG3651.
Biofilms of mutants MG3646 and MG3651 were established in flow chambers to determine how each mutation affected biofilm structure. Biofilms formed by MG3646 and MG3651 in the absence of C4-HSL were identical to the swrI mutant biofilm, with no cell chains and few aggregates (data not shown). However, in the presence of C4-HSL, the MG3646 mutant displayed a biofilm phenotype different from that of the swrI mutant and that of the wild type (Fig. 3c). This indicated that advancement of a biofilm developmental process had occurred through addition of C4-HSL, which was attributed to expression of other AHL-controlled biofilm genes, but was incomplete due to the transposon mutations. In the presence of C4-HSL the bsmA mutant formed a biofilm with large aggregates (Fig. 3c). Closer inspection revealed that aggregation with the filamentous cells was uncontrolled, which resulted in the formation of large aggregates. In contrast, the bsmB mutant biofilm resembled the biofilm of the swrI mutant and displayed little or no aggregation with the filamentous cells (Fig. 3d).
Complementation of MG3646 and MG3651.
Biofilms formed by MG3646(pBsmA) in the presence of 250 nM C4-HSL had the wild-type biofilm structure and contained filamentous cells, cell chains, and aggregates, although there were some differences (Fig. 3e). Large aggregates and gross adhesion to filamentous cells and cell chains by vegetative bacteria were observed. Biofilms formed by MG3646 bearing the bsmA-nifS operon in trans formed biofilms identical to that of MG3646 bearing pBsmA, suggesting that nifS plays no role in biofilm morphology (data not shown). Biofilms formed by MG3651(pBsmB) in the presence of 250 nM C4-HSL exhibited aggregation with the filamentous cells, although formation of intertwining cell chains was not observed (Fig. 3f).
DISCUSSION
Biofilm development has been the focus of intense interest recently, and the ultimate goal is to develop methods for biofilm prevention, control, or eradication. In cases where AHL signaling has been found to be important for biofilm formation, the biofilms consist of mushroom-like microcolonies separated by water channels (5, 13, 18, 26). Until now, there has been no evidence for AHL regulation of cellular differentiation in biofilm development, and only one study has linked an AHL-regulated determinant to biofilm architecture (26). In S. liquefaciens MG1, surface colonization through swarming motility has previously been shown to require a C4-HSL-regulated peptide synthetase that directs the formation of a surfactant (7, 17). Similarly, in this study we demonstrated that colonization through biofilm formation is also dependent on the C4-HSL regulatory system, and we identified specific genes in the QS regulon necessary for biofilm formation.
In this study we found that biofilms of S. liquefaciens MG1 consist of different morphotypes, including differentiated filamentous cells and cell chains. This is in contrast to other previously well-described biofilms of microorganisms such as P. aeruginosa and E. coli, whose biofilms consist of undifferentiated cells packed together into microcolonies (19, 20, 23). Previous observations of filamentous cells or cell chains have been reported; however, the regulation or form of cellular differentiation has not been addressed previously (4, 16). Given the change in S. liquefaciens MG1 biofilm architecture that leads to a mature biofilm over 72 h, it is proposed that biofilm development follows a genetically encoded program. This is clearly demonstrated by the differentiation of specific cells into filamentous cells and cell chains. The filament formation process does not occur when S. liquefaciens MG1 is grown in liquid culture, leading to the conclusion that this process is a surface-based phenomenon (data not shown).
Furthermore, in this study we demonstrated that the production of C4-HSL by S. liquefaciens MG1 is crucial for the formation of a normal biofilm. Statistical analysis by COMSTAT supported the observation that C4-HSL is necessary for formation of a thick biofilm. Importantly, the swrI mutant biofilm exhibited wild-type levels for all COMSTAT parameters tested after addition of the signal. Clearly, C4-HSL is necessary to drive the biofilm through its developmental program. Because COMSTAT was specifically designed for analysis of microcolony-like biofilms, not all of the parameters in the COMSTAT program are applicable for analysis of the S. liquefaciens MG1 biofilm. As a result, to distinguish specific differences between the wild-type and swrI mutant biofilms, we also relied on careful microscopic observation. Microscopic inspection of the swrI mutant biofilm showed that although this biofilm contained filamentous cells at 24 h, very little aggregation of the filamentous cells with undifferentiated rod-shaped cells had occurred at 48 h, and no cell chains were apparent at 72 h. It can be inferred that C4-HSL is required for aggregation with filamentous cells and population differentiation into cell chains. We propose that aggregation is an important developmental step for cell chain differentiation, possibly by providing a framework for cell chain support. To support the significance of the QS system in biofilm formation, attempts were made to mutate swrR. However, all efforts to generate an swrR mutant have failed, suggesting that such a mutation is lethal.
Two transposon mutants bearing insertions in a C4-HSL-regulated gene (the genes which were mutated were bsmA and bsmB) were identified as poor biofilm formers. Biofilms formed by both the bsmA and bsmB mutants in the flow chamber contained no cell chains and had abnormal aggregate formation. The excess aggregation by the bsmA mutant and no aggregation by the bsmB mutant suggest that aggregation is a highly regulated process, with a gene product necessary for activating aggregation (i.e., BsmB) and a gene product necessary to control the size of aggregation (i.e., BsmA). Similarly, fruiting body formation in Myxococcus xanthus requires activators and repressors for proper structure formation (14). The involvement of both C4-HSL-regulated genes in aggregation indicates that QS is involved at a specific stage of biofilm development and is crucial for continued biofilm maturation.
The BsmA protein had the highest level of similarity (as assessed by BLASTP) with a hypothetical protein from V. cholerae. Interestingly, in V. cholerae the bsmA homologue is part of an operon encoding a NifS homologue. The primary activity of NifS-like proteins is the desulfuration of l-cysteine, yielding l-alanine and elemental sulfur (30). The elemental sulfur is then necessary for the building of iron-sulfur (Fe-S) clusters on proteins. It has previously been suggested that protein posttranslational modification in S. liquefaciens MG1 may be C4-HSL regulated (9). On a 2D-PAGE gel, the isoelectric points of four proteins from the swrI mtuant were reported to shift in response to addition of C4-HSL (9). It is possible that the formation of NifS-mediated Fe-S clusters on these four proteins may affect their isoelectric points, resulting in a shift in their positions on a 2D-PAGE gel.
To ensure that the effects on biofilm formation of the transposon insertions of bsmA and bsmB were not polar effects, we complemented the corresponding strains with bsmA and bsmB. Biofilms formed by MG3646(pBsmA) had the wild-type biofilm structure, but there were some differences. While filamentous cells, cell chains, and aggregates were present in the MG3646(pBsmA) biofilm, the aggregates were larger than those produced in the wild-type biofilm and the cell chains and filamentous cells were heavily coated with vegetative bacteria, suggesting that the product of bsmA is probably an adhesin. The excess adhesion and aggregation seen in the MG3646(pBsmA) biofilm were probably a result of overexpression through the multicopy plasmid used for complementation. As mentioned above, the bsmA gene product appears to control bacterial aggregation with the filamentous cells. If bsmA codes for an adhesion, it is currently unclear how the proposed adhesin mediates the control of bacterial aggregation. However, since MG3646 bearing plasmid pBsmAOp formed a biofilm identical to that formed by MG3646(pBsmA), we concluded that the altered biofilm seen as a result of the transposon mutation in bsmA was due to the disruption of bsmA and not to a polar effect on the downstream nifS-like gene.
Later stages in biofilms formed by MG3651(pBsmB) did not have the full wild-type biofilm structure. As stated above, the bsmB gene is likely to encode a positive effector of bacterial aggregation. The MG3651 transposon mutant was unable to aggregate unless bsmB was supplied in trans, which restored aggregation. However, biofilm development was not able to progress to formation of the characteristic intertwining cell chains, presumably because the plasmid was a multicopy plasmid. Because bsmB is highly dependent on C4-HSL for induction and is induced 10-fold, other C4-HSL-regulated gene expression involved in biofilm formation is diluted, and this prevents further biofilm development. Based on the ORFs surrounding bsmB and the ability of bsmB in trans to restore the mutant biofilm partially, it is highly unlikely that a polar effect on a gene downstream of bsmB is responsible for the biofilm deficiency of MG3651. This is particularly evident since a putative gene 53 bp downstream from bsmB is present in the orientation opposite that of bsmB, indicating that bsmB is not in an operon and is expressed monocistronically.
In this study, we found that S. liquefaciens MG1 forms has a novel differentiated biofilm structure and that biofilm development is dependent on C4-HSL. We also identified two AHL-regulated genes important for normal biofilm formation. Current work in our laboratory is aimed at determining whether there are other C4-HSL-controlled genetic determinants for normal biofilm development. Furthermore, studies are being directed at understanding the functions inherent to the highly differentiated biofilm formed by S. liquefaciens MG1. This includes the expression of virulence traits, the role of the AHL-controlled colonization genes in attachment, and the resistance to predation offered by the elongated cells at the biofilm surface. Previous work has demonstrated that S. liquefaciens MG1 cells longer than 15 μm are resistant to grazing by protozoans, suggesting that filamentous cells and cell chains are likely to be a defense mechanism against protozoan predation in the environment (1).
Acknowledgments
This work was supported by grants from the Australian Research Council, the Centre for Marine Biofouling and Bio-Innovation, and the Danish Technical Research Council.
REFERENCES
- 1.Ammendola, A., O. Geisenberger, J. B. Andersen, M. Givskov, K.-H. Schleifer, and L. Eberl. 1998. Serratia liquefaciens swarm cells exhibit enhanced resistance to predation by Tetrahymena sp. FEMS Microbiol. Lett. 164:69-75. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjørn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. A., R. Brent, R. F. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 4.Dalton, H. M., L. K. Poulsen, P. Halasz, M. L. Angles, A. E. Goodman, and K. C. Marshall. 1994. Substratum-induced morphological changes in a marine bacterium and their relevance to biofilm structure. J. Bacteriol. 176:6900-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 6.Eberl, L., G. Christiansen, S. Molin, and M. Givskov. 1996. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J. Bacteriol. 178:554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eberl, L., M. K. Winson, C. Sternberg, G. S. A. B. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20:127-136. [DOI] [PubMed] [Google Scholar]
- 8.Givskov, M., L. Olsen, and S. Molin. 1988. Cloning and expression in Escherichia coli of the gene for extracellular phospholipase from Serratia liquefaciens. J. Bacteriol. 170:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Givskov, M., J. Östling, L. Eberl, P. W. Lindum, A. B. Christensen, G. Christiansen, S. Molin, and S. Kjelleberg. 1998. Two separate regulatory systems participate in control of swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:742-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimont, P. A. D., and F. Grimont. 1978. The genus Serratia. Annu. Rev. Microbiol. 32:221-248. [DOI] [PubMed] [Google Scholar]
- 11.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersbøll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 13.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser, D. 1998. How and why myxobacteria talk to each another. Curr. Opin. Microbiol. 1:663-668. [DOI] [PubMed] [Google Scholar]
- 15.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram negative eubacteria: regulation of the Pm promoter in the TOL plasmid studied with all controlling elements in the monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence, J. R., D. R. Korber, B. D. Hoyle, J. W. Costerton, and D. E. Caldwell. 1991. Optical sectioning of microbial biofilms. J. Bacteriol. 173:6558-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindum, P. W., U. Anthoni, C. Christophersen, L. Eberl, S. Molin, and M. Givskov. 1998. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. R. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18-28. [DOI] [PubMed] [Google Scholar]
- 19.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 21.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 22.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type 1 pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 23.Reisner, A., J. A. J. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 24.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 25.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steidle, A., M. Allesen-Holm, K. Riedel, G. Berg, M. Givskov, S. Molin, and L. Eberl. 2002. Identification and characterization of an N-acylhomoserine lactone-dependent quorum-sensing system in Pseudomonas putida strain IsoF. Appl. Environ. Microbiol. 68:6371-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tillett, D. 2000. Ph.D. thesis. University of New South Wales, Sydney, Australia.
- 28.Tillett, D., and B. A. Neilan. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobaceria. J. Phycol. 36:251-258. [Google Scholar]
- 29.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 30.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NifS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 90:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]