Abstract
Pyruvate carboxylase (PC) catalyzes the ATP-dependent carboxylation of pyruvate to oxaloacetate, an important anaplerotic reaction in mammalian tissues. To effect catalysis, the tethered biotin of PC must gain access to active sites in both the biotin carboxylase domain and the carboxyl transferase domain. Previous studies have demonstrated that a mutation of threonine 882 to alanine in PC from Rhizobium etli renders the carboxyl transferase domain inactive and favors the positioning of biotin in the biotin carboxylase domain. We report the 2.4 Å resolution X-ray crystal structure of the Rhizobium etli PC T882A mutant which reveals the first high-resolution description of the domain interaction between the biotin carboxyl carrier protein domain and the biotin carboxylase domain. The overall quaternary arrangement of Rhizobium etli PC remains highly asymmetrical and is independent of the presence of allosteric activator. While biotin is observed in the biotin carboxylase domain, its access to the active site is precluded by the interaction between Arg353 and Glu248, revealing a mechanism for regulating carboxybiotin access to the BC domain active site. The binding location for the biotin carboxyl carrier protein domain demonstrates that tethered biotin cannot bind in the biotin carboxylase domain active site in the same orientation as free biotin, helping to explain the difference in catalysis observed between tethered biotin and free biotin substrates in biotin carboxylase enzymes. Electron density located in the biotin carboxylase domain active site is assigned to phosphonoacetate, offering a probable location for the putative carboxyphosphate intermediate formed during biotin carboxylation. The insights gained from the T882A Rhizobium etli PC crystal structure provide a new series of catalytic snapshots in PC and offer a revised perspective on catalysis in the biotin-dependent enzyme family.
Pyruvate carboxylase (PC; EC 6.4.1.1) catalyzes the bicarbonate- and MgATP-dependent carboxylation of pyruvate to oxaloacetate, providing a major anaplerotic route in mammalian tissues (reviewed in (1)). Consequently, PC plays a significant role in a variety of cellular anabolic processes including gluconeogenesis (2), neurotransmitter biosynthesis (3, 4), and lipogenesis (5). In addition, PC activity has been correlated with glucose-mediated insulin release in pancreatic β-cells (6, 7, 8), and several studies have linked PC activity to tumor cell proliferation (9, 10, 11). Deficiencies in PC are characterized by developmental delay (12, 13, 14) and aberrant PC activity is associated with obesity and Type II diabetes (15, 16). Given its central role in metabolism and its implication in a variety of diseases and disorders, there is considerable interest in advancing a more complete description of PC structure and function.
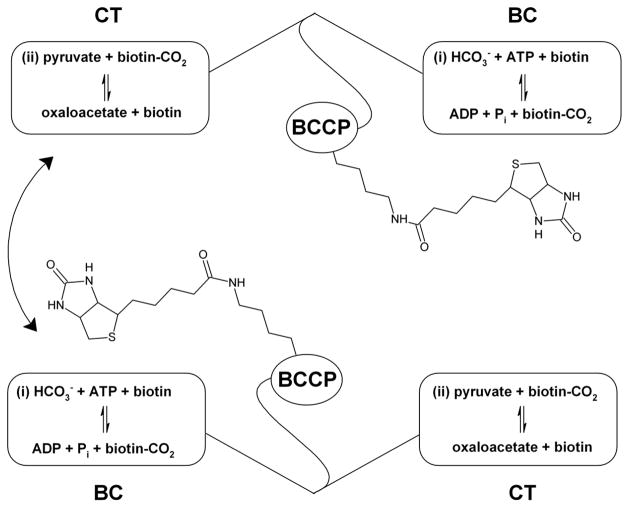
PC is a multi-functional enzyme comprised of four individual domains: a biotin carboxylase (BC) domain, a carboxyl transferase (CT) domain, a biotin carboxyl carrier protein (BCCP) domain and a central allosteric/tetramerization domain. The overall reaction is best described as a nonclassical, two-site Bi Bi Uni Uni ping-pong system where the biotin cofactor, covalently linked by its valerate side chain to a conserved lysine in the BCCP domain, transfers a carboxyl group from bicarbonate in the BC domain to pyruvate in the CT domain. Using bicarbonate as the carboxyl group donor, the tethered biotin is first carboxylated in the BC domain, with the concomitant hydrolysis of MgATP to MgADP and phosphate. Subsequently, the tethered carboxybiotin travels to a neighboring CT domain where the carboxyl group is transferred to pyruvate, producing oxaloacetate (Scheme 1). The reaction is allosterically activated by acetyl-Coenzyme A (acetyl-CoA), primarily by increasing the rate of MgATP hydrolysis in the BC domain (17). Acetyl-CoA is also reported to stabilize the overall enzyme structure by tightening the PC tetramer complex (18, 19, 20).
Scheme 1.
Recently, complete X-ray crystal structures of PC enzymes cloned from Rhizobium etli (RePC) (21) and Staphylococcus aureus (SaPC) (22, 23) have been reported. These structures have led to a more detailed description of PC catalysis, revealing that the tethered biotin travels more than 60 Å across the subunit cleft of the tetramer, transferring a carboxyl group between active sites on opposing polypeptide chains (Scheme 1) (21, 22, 24). This updated model of catalysis, which is strongly supported by both structural and kinetic evidence, explains why only a tetrameric PC will exhibit activities for the full forward and reverse reactions (20, 25). The transfer of carboxybiotin between polypeptide chains implies that changes in the quaternary arrangement of the enzyme can affect catalysis by altering the distance between active sites. The structure of wild-type RePC complexed with the non-hydrolyzable allosteric activator analogue, ethyl-CoA, revealed a highly asymmetrical tetramer, with the active sites on the top face of the tetramer positioned in closer proximity than those on the bottom face (21). In contrast, the X-ray crystal structures of SaPC co-crystallized both with and without acetyl-CoA, exhibited a much more symmetrical tetramer with the active sites nearly equidistant on both faces of the tetramer (22, 23). Since the quaternary arrangement determines the distance that carboxybiotin must travel between active sites in PC, it is of interest to determine the factors responsible for inducing a symmetrical or asymmetrical arrangement in the tetrameric enzyme. The previous RePC and SaPC crystal structures varied in their crystallization conditions, the identity of substrates and activators included for co-crystallization, and in the source organism from which they were cloned. One aim of the current study, therefore, is to clarify which of these factors is responsible for the substantial observed differences in the overall enzyme conformations.
The X-ray crystal structures of SaPC and the C-terminal portion of Homo sapiens PC (HsPC) revealed the binding position for pyruvate and tethered biotin in the active site of the CT domain (22). These structures highlighted a conserved threonine residue in the CT domain active site (Thr882 in RePC, Thr908 in HsPC), positioned between the N-1 of tethered biotin and pyruvate. Kinetic analyses of site-directed mutations led to the proposal that Thr882 facilitates proton shuttling between pyruvate and biotin in the CT domain of PC (22, 26). Surprisingly, while mutating Thr882 to alanine in RePC obliterates PC activity in the CT domain, this mutation had an increased rate over wild-type for the individual partial reactions in the BC domain (26). This observation was explained by proposing that, in the absence of CT domain substrates, the T882A mutation shifts the biotin occupancy equilibrium to favor the placement of biotin in the BC domain. Based on this proposal, we sought to determine the structure of T882A RePC in complex with BC domain substrate analogues in the hopes of capturing the first high-resolution example of a BCCP-BC domain interaction.
Here we describe the asymmetrical structure of T882A RePC co-crystallized with the bona fide activator, acetyl-CoA, at 2.4 Å resolution. Consistent with the hypothesis that the T882A RePC mutant favors placement of the BCCP-biotin in the BC domain, we observe the first high-resolution description of a biotin-dependent enzyme with its BCCP domain directly interacting with the BC domain. This structure reveals that tethered biotin accesses the BC domain from a different position and binds in the active site in a different orientation than free biotin. Despite the interaction between BCCP and BC domains, tethered biotin does not fully access the BC domain active site in the current structure. We propose that the BC domain is structured to regulate access of carboxybiotin to the active site in the reverse reaction. This structure reveals features of biotin carboxylase catalysis that are applicable to the majority of enzymes in the biotin-dependent enzyme family.
MATERIAL AND METHODS
General
IPTG, ampicillin, kanamycin, and chloramphenicol were purchased from Research Products International Corp. Acetyl-CoA was purchased from Crystal Chem, Inc. Restriction enzymes were purchased from New England Biolabs, thyroglobulin was purchased from Calbiochem and all other compounds were purchased from Sigma-Aldrich. Rhizobium etli PC was previously cloned into a modified pET-17b vector for expression in λ(DE3) lysogenized Escherichia coli (21). The T882A mutation was generated by the QuikChange site-directed mutagenesis protocol (Stratagene, La Jolla, CA) as previously described (26).
Site-directed Mutagenesis and Domain Truncation Mutations
The R353A, E248A, E248D, and E248R mutations were generated for the present study using the QuikChange site-directed mutagenesis protocol. For the BC domain truncation, it was determined that Gln465 represents the C-terminal end of the BC domain, based on the X-ray crystal structure of RePC (pdb id = 2QF7). The ΔBC RePC construct, therefore, begins at Gln466. Construction of the ΔBC RePC domain deletion in the pET-17b expression plasmid was as follows: ~2.1 kbp fragment was PCR amplified from the wild-type RePC pET-17b template using the following primers: 5′-CCG CAT GCA TCA GCA GGT CAA GCG C-3′ (forward) and 5′-GCA GCG GCT AGC TCA TCC GCC GTA A-3′ (reverse). Primers were synthesized by Integrated DNA Technologies (Coralville, IA). The PCR amplified fragment was digested with NheI/NsiI and ligated into a PstI/NheI digested modified pET-28a-(His)8-TEV expression vector. This modified pET-28a vector was generated by insertion of two additional N-terminal His residues and a TEV cleavage tag using QuikChange mutagenesis (see Figure S1 for the complete pET-28a-(His)8-TEV multiple cloning region sequence). The ΔBC RePC construct maintains the complete allosteric, CT, and BCCP domains of RePC. The correct gene sequence of all PC constructs was confirmed by complete sequencing of the gene at Functional Biosciences, Inc. (Madison, WI).
RePC T882A protein production and purification
RePC T882A was expressed in E. coli HMS174(DE3) cells from a modified pET-17b vector, co-expressed with the pCY216 vector encoding E. coli biotin protein ligase (BirA) and was produced using a 40 L batch culture in LB medium (containing 200 mg/L ampicillin, 30 mg/L chloramphenicol, and 1 mg/L biotin) as previously described (26). After reaching an Optical Density (600 nm) of 1.0, the culture was induced for 24 h at 16 °C with 1 mM IPTG and 20 mM L-arabinose.
RePC T882A was purified using Ni2+-affinity and dye-affinity chromatography as previously described (21). The harvested cells were disrupted by sonication and centrifuged at 4 °C prior to loading onto a 5 mL Ni2+-charged Hi-Trap chelating HP column (Amersham). The enzyme was eluted from the column using a gradient to 300 mM imidazole, with the predominant PC peak eluting at a concentration of 150 – 250 mM imidazole. Purified protein fractions were subsequently pooled and dialyzed into Buffer “A”, containing 50 mM MES (pH 7.0), 100 mM NaCl, 10 mM MgCl2, 1 mM EGTA and 2 mM DTT prior to loading onto a 10 mL column of Reactive Green-19 dye-affinity resin (Sigma-Aldrich). The column was washed with 50 mL of Buffer A containing 250 mM NaCl prior to being eluted from the column with Buffer A containing a series of ATP and NaCl concentrations (500 mM NaCl + 0 mM ATP; 750 mM NaCl + 0.2 mM ATP; 1 M NaCl + 1 mM ATP and 2 M NaCl + 1 mM ATP). The PC eluted at 2 M NaCl + 1 mM ATP was diluted in Buffer A to 13 mg/mL and dialyzed into storage buffer containing 10 mM HEPES (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 1 mM TCEP, and 1 mM NaN3. The final dialyzed protein was drop frozen in liquid nitrogen for storage at −80 °C. The enzyme concentration was determined spectrophotometrically using the calculated molar extinction coefficient of 118 000 M−1cm−1 at 280 nm (27).
The production and purification protocol for the BC domain truncation (ΔBC RePC) and for all site-directed mutants of RePC are provided in the supplementary materials.
Analysis of RePC Oligomeric State
Size exclusion chromatography was performed on an ÄKTA FPLC system using a Superose 6 10/300 GL size exclusion column from Amersham Pharmacia Biotech (Piscataway, NJ). Between 0.5 – 1 mg ΔBC RePC and wild-type RePC was loaded and eluted at a flow rate of 0.3 mL min−1 in a buffer containing 10 mM Tris (pH 7.4), 150 mM NaCl, 5% (v/v) glycerol, and 2 mM DTT. The eluent also included 250 μM acetyl-CoA when assessing the oligomeric state of ΔBC RePC in the presence of acetyl-CoA. Eluted samples were monitored at 280 nm and the apparent molecular weights were estimated from a calibration curve. The Superose 6 10/300 GL size exclusion column was calibrated using the following set of proteins: carbonic anhydrase (29 kDa), ovalbumin (45 kDa), bovin serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), β-amylase (200 kDa), apoferritin (443 kDa) and thyroglobulin (669 kDa). The size exclusion column was calibrated by fitting the partition coefficient (Kav) against the log molecular weight of each standard protein. Kav is defined as (Ve-Vo)/(Vt-Vo) where Ve is the elution volume, Vo is the void volume and Vt is the geometric column volume.
Enzyme Assays
Pyruvate carboxylation was measured by coupling the full forward reaction to malate dehydrogenase as previously described (26). Protein concentrations used in this assay ranged from 1 – 300 μg in accordance with activity. The reverse reaction of the BC domain was measured by coupling MgADP phosphorylation to glucose-6-phosphate dehydrogenase and hexokinase as previously described, with the exception that the assay was performed in a 1 mL reaction volume (26). Enzyme concentrations ranged from 0.3 – 1 mg for the MgADP phosphorylation reaction. All assays were performed in triplicate and reported errors are the standard deviation resulting from the three trials.
Hybrid Tetramer Enzyme Assays
Purified K1119Q and T882A RePC mutant solutions were mixed together in a 1:1 ratio (mg/mL). The enzyme mixture was subsequently diluted 10-fold (volume:volume) with reaction buffer (50 mM Bis-Tris, 25 mM Tricine, 25 mM glycine, pH 7.5) and allowed to incubate at room temperature for approximately 30–45 min prior to use in the enzymatic assays to ensure the complete disassociation of tetramers. Wild-type RePC enzyme solutions were also diluted 10-fold with reaction buffer and allowed to stand 30–45 min. The kcat and kcat/Km (MgATP) were determined with the diluted wild-type enzyme to account for any loss of enzymatic activity due to dilution inactivation (25). Reactions were initiated with the addition of either the wild-type or the K1119Q:T882A enzyme mix and the initial rates of pyruvate carboxylation were determined at varying concentrations of MgATP (0.03–2.5 mM) using the malate dehydrogenase coupled assay system. All reactions (pH 7.5, 25 °C) contained 15 mM HCO3−, 10 mM pyruvate, 5 mM MgCl2, 0.25 mM acetyl-CoA (unless otherwise indicated), 0.24 mM NADH and malate dehydrogenase (10 U). Data were fit to the Michaelis-Menten equation and errors reported on kcat and kcat/Km (MgATP) were determined from the non-linear least-squares fits.
Protein Crystallization
All crystals of T882A RePC were obtained at room temperature by the hanging-drop vapor diffusion method. For the crystals grown in the presence of acetyl-CoA, the reservoir solution consisted of 14% PEG 4K, 10% MPD (2-methyl-2,4-pentanediol), 450 mM KCl and 100 mM triethanolamine (pH 8.0) and the protein solution consisted of 12 mg/mL RePC purified as described above, 1 mM TCEP, 5 mM ADP, 5 mM phosphonoacetate and 1 mM acetyl-CoA. The protein storage buffer included 10 mM MgCl2. The protein solution and reservoir solution were mixed in a 1:1 ratio to a final volume of 6 μL and the drop was microseeded with pulverized RePC T882A crystals after approximately 24 h. The resulting rod-like RePC T882A + acetyl-CoA crystals (~100 μm × 40 μm × 40 μm) grew within 2–5 days. After 7–10 days, the crystals were transferred to a cryprotectant solution consisting of 16% PEG 4K, 15% MPD, 100 mM triethanolamine (pH 8.0), 500 mM KCl, 1 mM TCEP, 5 mM ADP, 5 mM phosphonoacetate and 1 mM acetyl-CoA and flash cooled in liquid nitrogen. The crystals belonged to the space group C2, with four subunits in the asymmetric unit and the unit cell parameters a = 371 Å, b = 92 Å, c = 261 Å, α = γ = 90°, β = 134.7°.
For the crystals grown in the presence of Na+ L-aspartate, the reservoir solution consisted of 16% PEG 4K, 10% MPD, 200 mM KBr and 100 mM triethanolamine (pH 8.0) and the protein solution consisted of 12 mg/mL RePC purified as described above, 2 mM TCEP, 5 mM ATP-γ-S, 100 mM sodium formate and 100 mM L-aspartate. The protein solution and reservoir solution were mixed in a 1:1 ratio to a final volume of 4 μL. The resulting rod-like RePC T882A + L-aspartate crystals (~80 μm × 30 μm × 20 μm) grew within 2–5 days. After 7–10 days, the crystals were transferred to a cryprotectant solution consisting of 22% PEG 4K, 15% MPD, 100 mM triethanolamine (pH 8.0), 225 mM KBr, 2 mM TCEP, 5 mM ATP-γ-S, 100 mM sodium formate and 100 mM L-aspartate and flash cooled in liquid nitrogen. The crystals belonged to the space group I41, with two subunits in the asymmetric unit and the unit cell parameters a = 264 Å, b = 264 Å, c = 92 Å, α = β = γ = 90°.
Data collection, structure determination and refinement
X-ray diffraction data were collected at the Advanced Photon Source (APS) beamline SBC-19-ID on a ADSC q315 CCD detector, with an X-ray wavelength of 0.979 Å. Diffraction images were processed with HKL2000 (28). The structures were solved by molecular replacement using the individual BC and (CT + allosteric) domains of the wild-type RePC enzyme as the search model with the program Phaser (29). As with the structure of the S. aureus PC tetramer in co-crystallized with acetyl-CoA (23), the structures of T882A RePC could be determined only when the individual domains were used as the search models in the molecular replacement solution. The molecular replacement models were extended by several rounds of manual model building with COOT (30) and refinement with REFMAC using TLS rigid body refinement (31, 32). For each independent molecule in the asymmetric unit, four rigid bodies were defined, corresponding to the BC, CT, allosteric and BCCP structural domains. Water molecules were added to the model in COOT with subsequent manual verification. Data collection and processing statistics are summarized in Table 1.
Table 1.
Data Collection and Refinement Statistics
| PDB ID code |
RePC T882A + acetyl CoA 3TW6 |
RePC T882A + L-Asp 3TW7 |
|---|---|---|
| Space Group | C 2 | I 41 |
| Cell Dimensions | ||
| a, b, c (Å) | 371, 92, 261 | 264, 264, 92 |
| α, β, γ (°) | 90, 134.7, 90 | 90, 90, 90 |
| Resolution range, Å | 50.0–2.4 (2.40–2.46)a | 50.0–3.1 (3.1–3.2)a |
| Redundancy | 3.6 (3.4) | 9.5 (9.1) |
| Completeness (%) | 99.2 (97.0) | 100 (100) |
| Unique Reflections | 241 087 | 82 806 |
| Rmerge (%) | 8.3 (35.1) | 9.7 (31.8) |
| Average I/σ | 19.3 (3.4) | 21.2 (6.0) |
| Rwork | 0.189 (0.216) | 0.250 (0.271) |
| Rfree | 0.235 (0.277) | 0.292 (0.326) |
| No. protein atoms | 31 798 | 14 934 |
| No. water molecules | 793 | – |
| Wilson B-value (Å2) | 38.5 | 43.4 |
|
| ||
| Average B-factors (Å2) | ||
| Protein | 31.0 | 24.2 |
| Ligands | 37.6 | – |
| Solvent | 28.5 | – |
|
| ||
| Ramachandran (%) | ||
| Most favored | 88.0 | 88.9 |
| Additionally allowed | 9.8 | 9.9 |
| Generously allowed | 1.3 | 0.8 |
| Disallowed | 0.9 | 0.3 |
|
| ||
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.019 | 0.013 |
| Bond angles (°) | 2.03 | 1.67 |
Values in parentheses are for the highest resolution bin
Docking of carboxyphosphate analogues
The molecular structures of the carboxyphosphate analogues were built and geometry optimized using AM1 semi-empirical calculations with Spartan (Wavefunction Inc., Irvine, CA). The Lamarckian genetic algorithm of AutoDock 4.2 (33) was used as a search method for the possible binding positions of the analogues in the BC domain active site of chain C of the T882A RePC structure. The ligands and receptor molecule were prepared using AutoDockTools (33). Chain C was constrained as a rigid body receptor, but torsions were allowed for specified bonds in the carboxyphosphate ligands. All hydrogens were added to both ligands and receptor followed by a merging of polar hydrogens. Partial atomic Kollman charges were assigned for the protein receptor, while Gasteiger charges were used for the ligands. Although AutoDock 4.2 defaulted to a charge of 0 for the Mg atom of MgADP, a partial charge was manually input as 1.5+ due to the ligands contained in the coordination sphere of the metal. The grid maps for the docking calculation were created using a grid box centered on the lobe of electron density near the MgADP in chain C. The final dimensions of the box were 5.6 Å × 7.9 Å × 5.6 Å. The initial position of the molecule was randomly assigned, and a total of 50 genetic algorithm runs were performed for each molecule. All other parameters were set to their default values: the maximum number of generations was 2,500,000, the elitism was 1, the probability for a gene to undergo a random change was 0.02, and the probability of crossover was 0.8. The results were analyzed by visualizing all 50 docked conformations for each molecule and comparing the calculated relative binding energies. For each molecule, the docked conformation with the lowest relative binding energy was selected.
RESULTS
Structure of T882A Rhizobium etli PC
T882A RePC with acetyl-CoA and MgADP crystallized in the space group C2, with two dimers in the asymmetric unit. Each of these dimers constitutes one half of an individual tetramer. The final structure model consists of two distinct tetramers generated through rotation about a two-fold axis of crystallographic symmetry. The nucleotide portion of acetyl-CoA is observed on only one face of the tetramer, defined as the top face. The tetramers are comprised of two identical chains on the top face of the tetramer (chains A and C on tetramers 1 and 2, respectively) and two identical chains on the bottom face (chains B and D on tetramers 1 and 2, respectively) (Figure S2). There are few crystal contacts between the neighboring tetramers, with the exception of contacts between a helix-turn-helix motif at the N-terminus of the CT domain (residues 516–538) of chain A and the equivalent residues in chain D. These crystal contacts occur in remote corners of these tetramers and do not influence mobile loops or flexible domains relevant to the active sites. The two tetramers superimpose with an RMSD of 2.9 Å for all atoms (Figure S2) and exhibit only minor differences in the electron density corresponding to the more mobile elements of the structure model. The biotin cofactor, for example, is better ordered in tetramer 1 than it is in tetramer 2. There is an additional lobe of electron density corresponding to phosphonoacetate in the BC active site of chain C (vide infra) that is not observed in any of the other active sites.
While acetyl-CoA is bound exclusively to the top face of the tetramer, the BCCP domains with tethered biotin are disordered on the top face and are observed exclusively on the bottom faces of the tetramers. This ordering of the BCCP domain on only one face of the tetramer is similar to what has been observed in the wild-type RePC structure (21). However, in the wild-type RePC structure, BCCP domains on the top face of the tetramer were ordered while BCCP domains on the bottom face were disordered. To assess whether both faces of the tetramer are capable of catalyzing the overall reaction, a mixed set of hybrid tetramers was generated by diluting together the T882A and K1119Q mutants of RePC. The T882A mutant is catalytically inactive in the CT domain while the K1119Q mutant is unable to be biotinylated at residue 1119 (34). Assuming that the monomers will rehybridize independently of each other and allowing for complete equilibration, a mixed population encompassing all possible hybrid tetramer recombinations from a 1:1 mixture is predicted to have 25% wild-type activity if both faces of the tetramer are active (Figure S3). The kcat for the 1:1 mixture is 22% wild-type activity. This confirms that, despite the asymmetry, both faces of the tetramer are capable of catalyzing pyruvate carboxylation.
As in all previous structures of PC, there is significant conformational flexibility in the BC domain (21, 22, 23, 24). A structural superimposition of all 4 monomers (chains A – D), centered on the CT and allosteric domains, reveals a remarkable 65° rotation of the BC domain between top and bottom chains. This rotation pivots around a hinge centered on residue Asp471 in the BC-allosteric domain boundary and is very similar to the 67° rotation observed in the wild-type RePC structure about this same hinge (21). There are also small conformational differences in the orientation of the BC domain between individual tetramers, with a 6° – 9° rotation between BC domains on the bottom face of the tetramers and BC domains on the top face of the tetramers. The rotation is centered on a hinge at residue Asp471. Comparable rotations have been reported about this same hinge in the SaPC tetramer (22, 23). In RePC, this rotation of the BC domain influences the overall distance between active site pairs on neighboring BC and CT domains. The distance between the active sites, defined as the distance from the β-phosphate of MgADP/MgATP in the BC domain to the divalent metal ion in the CT domain, increases from 63 Å in wild-type RePC structure to 72 Å in the current structure. The distances between active site pairs on the bottom face of the tetramer remain relatively unchanged, at 74 Å in both structures.
Although the overall arrangement of T882A RePC in the presence of acetyl-CoA remains highly asymmetrical between top and bottom faces, the rotations in the BC domains result in a nearly equal spacing of active site pairs between the top and bottom faces of the tetramer. These distances are similar to the distances between active site pairs in the SaPC structure: 76 Å in the presence of acetyl-CoA and 79 Å in its absence (22, 23). The increased spacing of active sites in the current structure relative to the wild-type RePC structure may be the result of several different factors, including the presence of the product MgADP in the BC domain active site, the T882A mutation in the CT domain active site, the crystallization in the presence of the bona fide activator acetyl-CoA, or the ordering of the BCCP domain on the bottom face rather than the top face of the tetramer. Additional structures are needed to determine whether changes in the position and rotation of the BC domain are sensitive to substrates, products and allosteric effectors. Nevertheless, contrary to what was previously proposed (21), the current structure reveals that there is no direct correlation between coenzyme-A binding and the relative positioning of neighboring catalytic domains in RePC.
Asymmetry of the Rhizobium etli PC tetramer is independent of acetyl-CoA
The overall arrangement of the T882A RePC tetramer co-crystallized with acetyl-CoA is highly asymmetrical. A very similar asymmetrical arrangement was observed for the wild-type RePC structure complexed with ethyl-CoA (21). Acetyl-CoA, like ethyl-CoA, is bound exclusively to one face of both T882A RePC tetramers and, like ethyl-CoA, only the nucleotide portion of the molecule is clearly defined in the electron density. The position occupied by the bound nucleotide portions of both ethyl-CoA and acetyl-CoA are indistinguishable. The overall asymmetrical arrangement of RePC is also independent of the binding of substrate (MgATP-γ-S) or product (MgADP) in the active site. It is clear from the current structure that the asymmetrical arrangement originally observed for wild-type RePC was not an artifact of crystallization nor was it the result of an aberrant effect from the non-natural activator, ethyl-CoA.
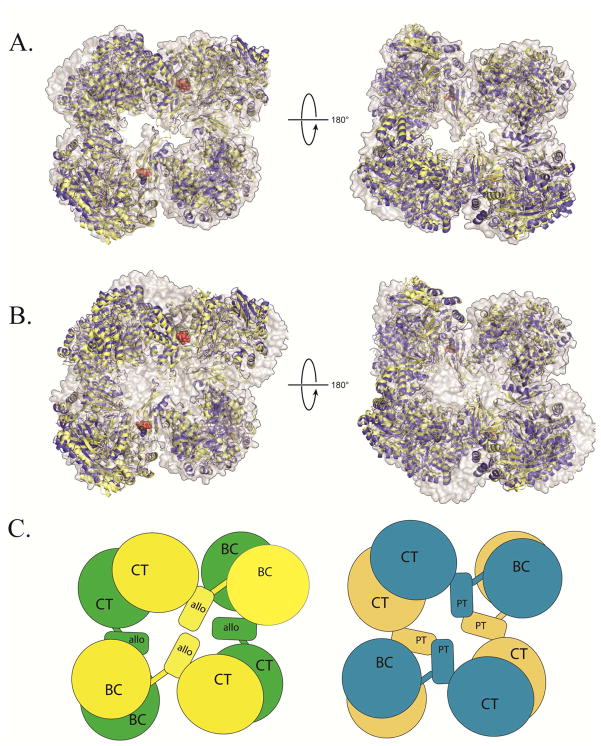
It has been suggested that the allosteric activator is responsible for inducing the asymmetrical conformation of RePC (21). However, recent X-ray crystallographic and cryo-EM structural studies of PC from S. aureus and H. sapiens reveal that both these enzymes are nearly symmetrical, and that this symmetry is independent of acetyl-CoA binding (22, 23, 24). To determine whether asymmetry in the RePC tetramer is a function of coenzyme-A, T882A RePC was co-crystallized with 100 mM L-aspartate in the absence of acetyl-CoA. L-Aspartate is a low affinity allosteric inhibitor of PC from many species, including that from R. etli (data not shown). Even for the best diffracting crystals, the data exhibited high mosaicity and were useable only to 3.1 Å resolution. The refined structure model has relatively high B-factors for residues in the BC domain, preventing an extensive analysis of the structure and precluding a definition of a specific L-aspartate binding site. Nevertheless, the tetrameric arrangement of T882A RePC remains highly asymmetrical in the absence of acetyl-CoA. A superposition of all three RePC structures shows strong similarities, regardless of the presence or absence of allosteric activator (Figure 1A). Conversely, a superposition of T882A RePC and SaPC, both co-crystallized with acetyl-CoA, shows pronounced deviations in their tetramer structures (Figure 1B). Thus, the significant difference in the symmetrical arrangement of PC tetramers cannot be attributed to the allosteric activator. The differences in conformation cannot be attributed to the solution pH, since asymmetrical tetramers have resulted from crystals of RePC grown at pH 5.6 (21) and pH 8.0 (present study), while symmetrical tetramers of SaPC have resulted from crystals grown in the presence and absence of acetyl-CoA at pH 7.5 (22, 23). Therefore, the overall symmetry of the tetramer is proposed to be a species-specific property of the enzyme, with PC from R. etli displaying significant asymmetry while PC from S. aureus and H. sapiens displays much greater internal symmetry (Figure 1C).
Figure 1. Structural superposition of PC X-ray crystal structures from R. etli and S. aureus.
(A) A structural superposition, viewed from the top (left) and bottom (right) face of T882A RePC + acetyl-CoA (yellow ribbons) and T882A RePC + L-aspartate (blue ribbons) with wild-type RePC + ethyl-CoA (2QF7; gray surface representation). For visual clarity, the alignment was centered on residues 450–1050 of chain A for all three structures, corresponding to the structurally rigid allosteric and CT domains. Alignment in this region results in a nearly perfect alignment at one corner of the tetramer and accentuates any remaining deviations in the structures. In the presence and absence of coenzyme-A analogues, all three structures of RePC are closely aligned. (B) A structural superposition, viewed from the top (left) and bottom (right) face of RePC T882A + acetyl-CoA (yellow ribbons) and RePC T882A + L-aspartate (blue ribbons) with wild-type SaPC + acetyl-CoA (3HO8; gray surface representation). The alignment was centered on residues 450–1050 of chain A for all three structures, corresponding to the structurally rigid allosteric and CT domains. The RePC structures do not align well with the SaPC + acetyl-CoA structure, regardless of whether acetyl-CoA is present in the RePC crystallization conditions. (C) A diagrammatic representation of the overall quaternary arrangement of RePC (left; top face = yellow; bottom face = green) and SaPC (right; top face = blue; bottom face = brown) emphasizing the absence of inter-subunit interactions between allosteric domains in RePC and the top-bottom subunit interactions between the central domains (PT = PC tetramerization) in SaPC.
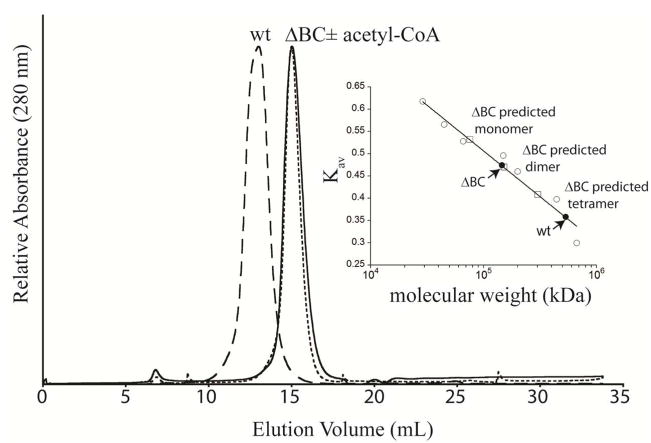
The arrangement of the tetramer in the RePC structures predicts that the tetramer is held together exclusively by BC-BC and CT-CT homodimerization contacts at the four corners of the tetramer (Figure 1C). To confirm that the enzyme in solution adopts a similar conformation to that in the crystal structures, a truncation of the BC domain (ΔBC RePC) was constructed, expressed and purified. The ΔBC RePC construct maintains 50% of the wild-type activity for the oxamate-stimulated decarboxylation of oxaloacetate, demonstrating that the truncated enzyme is properly folded and catalytically competent. Size exclusion chromatography was used to analyze the mobility of ΔBC RePC relative to wild-type. Both in the presence and absence of acetyl-CoA, the ΔBC RePC elutes in a single peak at a molecular weight corresponding to a dimer (Figure 2). Thus, the central allosteric domain of RePC does not contribute to tetramerization.
Figure 2. Size exclusion chromatography of wild-type and BC domain deletion RePC.
Representative chromatograms for wild-type RePC (wt RePC; long dashed line) and the BC domain deletion of RePC (ΔBC). The elution profile of ΔBC RePC is unaffected by the presence (short dashed line) or absence (solid line) of acetyl-CoA. INSET: Standard curve (open circles) and predicted Kav for the ΔBC monomer, dimer and tetramer (open squares). The measured Kav for wild-type and ΔBC is indicated (closed circles), corresponding to a dimer for ΔBC RePC (MW = 150 kDa) and a tetramer for wild-type RePC (MW = 500 kDa).
The BCCP-BC domain interaction
The T882A RePC structure co-crystallized with acetyl-CoA and MgADP captures the interaction between the BCCP domain and the BC domain on the bottom face of tetramers 1 and 2. While both tetramers in the T882A RePC structure reveal the same inter-domain interaction, the electron density describing the BCCP domain is better defined for tetramer 2, and the electron density describing biotin and the B-subdomain lid of BC is better defined for tetramer 1 (Figure S4). In both tetramers, there is considerable flexibility inherent in BCCP and the B-subdomain lid of BC, resulting in high isotropic displacement parameters and incomplete electron density corresponding to these two regions (Figure S4). High B factors and incomplete electron density is commonly observed for both the BCCP domain and the B-subdomain lid of BC in most X-ray crystal structures (21, 22, 23, 35). Although tetramer 1 is illustrated in the accompanying figures, essentially identical domain interactions exist in tetramer 2.
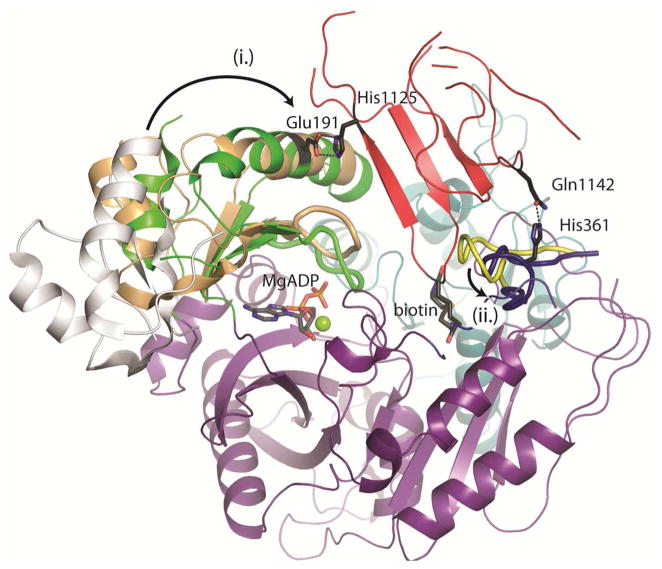
The BCCP domain binds at the interface between two subdomains of BC. The individual subdomains of BC are composed of a central B-subdomain lid dividing distinct N-terminal and C-terminal subdomains (36). The BCCP domain of PC rests against the central α-helix of the B-subdomain lid (Figure 3). This lid closes down on the active site following MgATP binding (35) and is more tightly closed in the current structure than it is in the structure of E. coli BC complexed with MgADP and free biotin (Figure 3). A large surface accessible loop in the C-subdomain, from Thr356 to Gly368, comprises the second half of the BCCP docking site (Figure 3).
Figure 3. BCCP binding in the BC domain of RePC.
A cartoon representation of the BC and BCCP domains on the bottom face (chain B) of T882A RePC. The three subdomains of the BC domain are individually colored: The N-terminal A-subdomain (cyan), the mobile B-subdomain lid (green) and the C-terminal C-subdomain (purple). The BCCP domain (red), biotin and MgADP (CPK colored sticks) are also shown. The BCCP domain docks between the closed B subdomain lid and a large surface loop in the C-subdomain. In (i.), the B-subdomain swings from the open position observed in the overlaid structure of E. coli BC (pdb id = 1BNC; open B-subdomain position colored in gray) to close down over the active site. This closure is tighter in the presence of tethered biotin (B-subdomain colored in green) than it is with free biotin (pdb id = 3G8C; B-subdomain colored in tan). In (ii.), the surface loop (Thr356-Gly368; colored in blue) occupies an alternate position compared with the position it occupies in E. coli BC (pdb id = 1BNC; colored in yellow) in order to accommodate binding of the BCCP domain. The Glu191 – His1125 and His361 – Glu1142 side-chain interactions are illustrated.
The BCCP domain binds at the BC domain in an orientation that inserts the conserved biotinylation motif (MKME; where K represents the biotinylated lysine) within the BC domain. There are very few specific interactions between these domains that contribute to holding the BCCP domain in place. Only two sets of side-chain interactions can be described: Glu191 – His1125 and His361 – Gln1142 (Figure 3). None of these four residues are conserved in PC enzymes. Rather, the bulk of the interdomain interactions are made through non-specific contacts. The calculated buried surface area on BCCP binding in the BC domain is ~1200 Å2, based on the difference in calculated solvent accessible surface area for wild-type RePC chain A and T882A RePC chain B (37). This relatively large buried surface area is a result of both BCCP binding and tight closure of the B-subdomain lid.
Tethered biotin binding in the BC domain
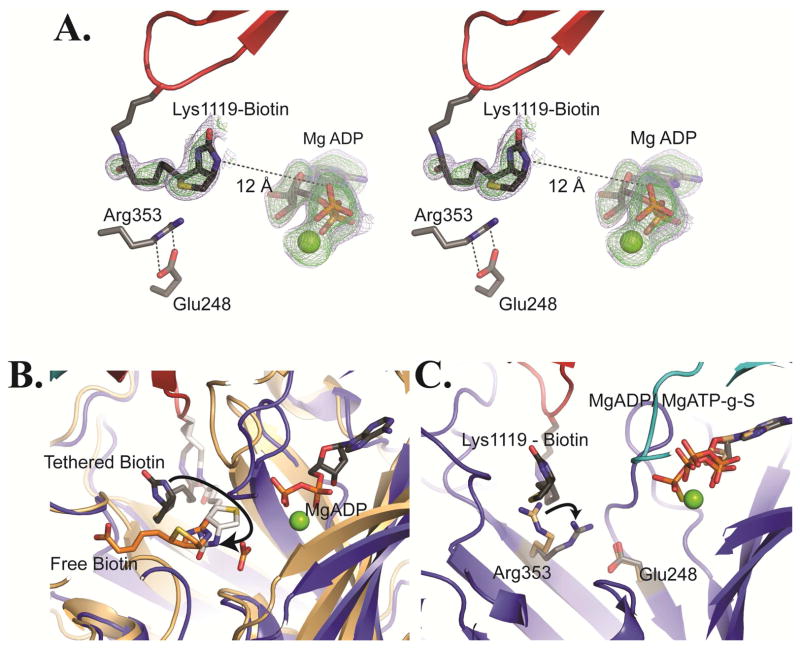
The X-ray crystal structure of T882A RePC reveals electron density for tethered biotin in the BC domains on the bottom faces of each tetramer. The difference density corresponding to biotin is moderate and the modeled atoms have relatively high B-factors, implying that tethered biotin has a high freedom of motion. Similar high B factors have been observed for tethered biotin in the CT domain of SaPC and propionyl-CoA carboxylase (22, 23, 38). An Fo-Fc omit map for BCCP-biotin at a contour level of 2.0σ reveals electron density corresponding to tethered biotin (Figure 4A). Surprisingly, the position occupied by tethered biotin is not within the BC domain active site. Rather, the tethered biotin adopts a conformation with its ureido oxygen pointing back toward the glutamate side-chain of the conserved MKME biotinylation consensus sequence. In this pose, the N-1 position of biotin is located 12 Å from the β-phosphate of MgADP and is not oriented for catalysis. By comparison, in the structure of E. coli BC complexed with free biotin, the N-1 position of biotin is located 7 Å from the β-phosphate of MgADP and is appropriately oriented toward the phosphate oxygens of the nucleotide (39).
Figure 4. Tethered biotin in the BC domain of RePC.
(A) Stereoview of omit Fo – Fc electron density for tethered biotin and MgADP contoured at 2.0σ (blue) and 3.0σ (green) for chain B of T882A RePC. In this position, the N-1 of tethered biotin is positioned 12 Å from the β-phosphate of MgADP. The Arg353 and Glu248 side chains are illustrated as sticks. (B) The actual (gray) and modeled (white) position of tethered biotin in the BC domain active site of the T882A RePC crystal structure (tetramer 1, chain B). The structure is overlaid with that of E. coli BC (pdb id = 3G8C; colored in brown) co-crystallized with free biotin (orange) and bicarbonate. The modeled position demonstrates that tethered biotin can reach the active site and that the N-1 of tethered biotin can occupy a nearly identical position to the N-1 of free biotin in E. coli BC. Free biotin is oriented such that its valerate side-chain points away from the BCCP domain binding site. Consequently, tethered biotin cannot physically adopt the same binding orientation as free biotin. (C) A BC domain overlay of the bottom subunit of wild-typeRePC (chain B; pdb id = 2QF7) with the bottom subunit of tetramer 1 of T882A RePC (chain B). The wild-type RePC structure has ATP-γ-S bound in the active site and does not interact with the BCCP domain. Conversely, the RePC structure of T882A has MgADP bound in the active site and interacts with the BCCP domain. While the position of Glu248 remains unchanged between the two structures, Arg353 swings to interact with Glu248 in the T882A RePC structure. The side chain position of Arg353 in wild-type RePC is colored in brown while its position in the T882A RePC structure is colored in gray.
Despite the non-catalytic position occupied by biotin, the current structure clearly identifies the direction from which tethered biotin accesses the BC domain. This direction is very different from the direction predicted by the orientation of free biotin in the structure of E. coli BC (Figure 4B). Threading from its position in the current structure, tethered biotin can be modeled into the active site by extending the side chain from the defined α-carbon position of Lys1119 using allowable bond lengths and angles. This model demonstrates that tethered biotin is capable of reaching the same final binding region as free biotin in the E. coli BC structure, without requiring changes in the position of the Lys1119 α-carbon on BCCP. However, restrictions on allowable bond lengths and bond angles make it impossible for tethered biotin to position itself in the same orientation as free biotin in the E. coli BC structure. The orientation of tethered biotin in the active site of T882A RePC, therefore, cannot be the same as that of free biotin in the active site of E. coli BC.
Originating from the α-carbon of Lys1119 in the BCCP domain, tethered biotin is capable of accessing the active site. However, in the current structure of T882A RePC, it is precluded from occupying this binding position. There are very few specific contacts between tethered biotin and BC domain side chains. A similar paucity of interactions has been described for tethered biotin in both the exo binding site and the CT active site of PC (22, 40). Only Arg353 is positioned in proximity to tethered biotin in the current structure, with its guanidino group ~4 Å below the thiophene ring of biotin. The side chain of Arg353 is held in this position through a salt bridge with Glu248. Both Arg353 and Glu248 are conserved in the BC domains of all biotin dependent enzymes but are not conserved across the larger ATP grasp superfamily (Figure S5). This pattern of sequence conservation suggests that these residues play a role in the biotin-specific chemistry. The interaction between Arg353 and Glu248 creates a shallow ledge separating biotin from its final binding position in the active site (Figure 4B). Arg353 swings towards Glu248 in the current structure, closing a gate that reduces access of tethered biotin to its binding position in the active site (Figure 4C).
To address whether the observed BCCP-BC interaction is catalytically relevant and to further probe the functions of Arg353 and Glu248 in mediating biotin access to the active site, Arg353 and Glu248 were mutated and the steady state kinetics at saturating substrate concentrations were determined for the mutant-catalyzed reactions in both the forward and reverse directions (Table 2). Both Arg353 and Glu248 are important in catalyzing the full forward reaction, with a substantial loss of pyruvate carboxylation activity observed for all mutations. These same mutations exert much less of an effect on the rate of MgADP phosphorylation with carbamoyl phosphate in the BC domain, with the E248A mutation resulting in a 4-fold rate increase over wild-type. The phosphorylation of MgADP by carbamoyl phosphate has been shown to be stimulated by free biotin and, to an even greater extent, by tethered biotin (41). The increased rate of MgADP phosphorylation in the Glu248 and Arg353 mutations, therefore, is consistent with increased biotin access to the BC domain active site resulting from a disruption of the interaction between Arg353 and Glu248. Detailed kinetics for a R353M mutation are extensively discussed in the accompanying report by Zeczycki et al. (42) and further support the hypothesis that the Arg353-Glu248 pair plays an important role in regulating the accessibility of biotin and carboxybiotin to the BC domain active site.
Table 2.
Kinetic parameters of RePC site-directed enzyme mutants.
| Pyruvate Carboxylation kcat (min−1) |
ADP Phosphorylation with Carbamoyl Phosphate kcat (min−1) |
|
|---|---|---|
| Wild-type | 1830 ± 50 | 3.26 ± 0.05 |
| E248A | 23.8 ± 1.0 | 13.6 ± 0.4 |
| E248D | 50.9 ± 1.8 | 0.33 ± 0.01 |
| E248R | N/D1 | 0.13 ± 0.01 |
| R353A | 1.41 ± 0.13 | 2.23 ± 0.05 |
No activity detected
Phosphonoacetate as a carboxyphosphate mimic in the BC domain active site
The first step of the reaction catalyzed in the BC domain is proposed to be the activation of bicarbonate by MgATP to form a carboxyphosphate intermediate (43) (Figure S6). T882A RePC was co-crystallized with 5 mM ADP and 5 mM phosphonoacetate in an attempt to capture phosphonoacetate in the active site as an isosteric and isoelectronic analogue of carboxyphosphate.
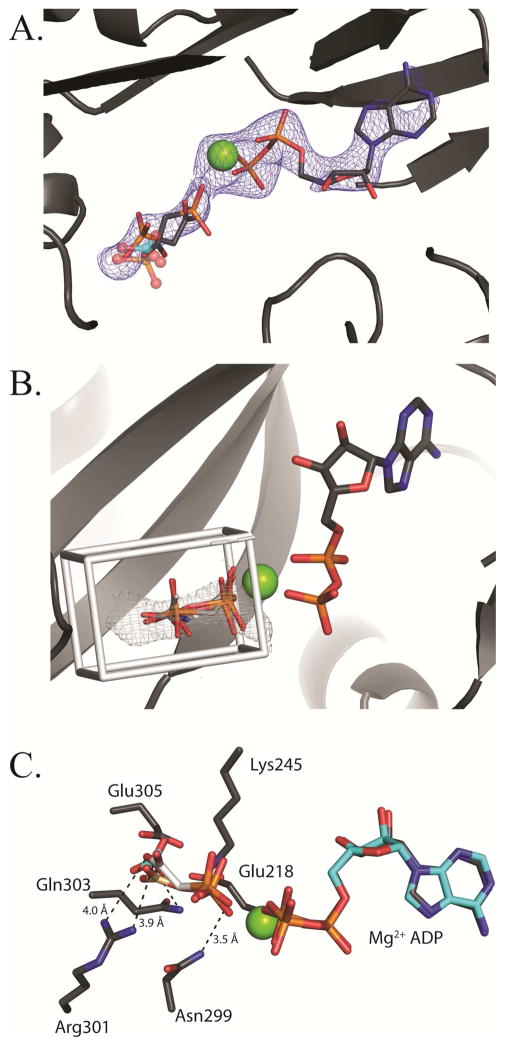
Of the four individual BC domains in the current structure, only one includes a prominent lobe of electron density adjacent to the MgADP nucleotide that can be attributed to bound phosphonoacetate. The density is observed exclusively in the BC domain of chain C, on the top face of tetramer 2. There is no BCCP-biotin bound at this domain. Phosphonoacetate fits into the omit Fo - Fc density contoured at 2.5σ if modeled in two separate conformations: one conformation with the phosphate directed toward MgADP and the opposite conformation with the carboxyl moiety directed toward MgADP (Figure 5A). To demonstrate that the BC domain active site can accommodate the binding of a single phosphonoacetate in multiple conformations, phosphonoacetate and several additional carboxyphosphate analogues were docked into a small docking box surrounding the lobe of electron density (Figure 5B). Docking of phosphonoacetate, carbamoyl phosphate, acetyl-phosphate and carboxyphosphate resulted in a mixed set of conformations, with some molecules docking with the phosphate group directed toward MgADP (acetyl-phosphate and carbamoyl phosphate), away from MgADP (phosphonoacetate) and in a mixture of energetically equivalent conformations (carboxyphosphate) (Table S1). These docking calculations demonstrate that a mixed set of conformations is energetically feasible for the binding of phosphonoacetate in the X-ray crystal structure. The catalytically relevant conformation of carboxyphosphate orients the phosphate moiety toward the β-phosphate of MgADP, following nucleophilic attack on MgATP by bicarbonate. For this orientation, the docked binding position of carboxyphosphate and the modeled binding position of phosphonoacetate in the electron density strongly agree.
Figure 5. Binding of phosphonoacetate in the BC domain active site.
(A) Phosphonoacetate is modeled in two conformations into a lobe of electron density observed in the BC domain active site on the top face (chain C) of tetramer 2. The Fo-Fc omit electron density contoured at 2.5 σ is shown in blue. A structural superposition of E. coli BC with bound bicarbonate (semi-transparent, cyan-colored ball and stick representation; pdb id = 3G8C) and phosphate (semi-transparent, CPK-colored ball and stick representation; pdb id = 1DV1) reveals that both phosphate and bicarbonate share the same binding site. The positions of both the carboxyl and phosphonate moieties of phosphonoacetate align well with the shared binding site for phosphate and bicarbonate. (B) AutoDock 4.2 modeling of carboxyphosphate, phosphonoacetate, acetyl phosphate and carbamoyl phosphate in the active site of BC. The docking grid box was extended to fully encompass the electron density for phosphonoacetate. All calculated docking poses line up closely with the observed Fo-Fc omit electron density. (C) An overlay of docked carboxyphosphate (brown carbon CPK coloring) with one conformation of phosphonoacetate (white carbon CPK coloring). Residues within interacting distance of phosphonoacetate are shown.
DISCUSSION
The primary objective of the current study was to capture a new snapshot of catalysis for PC by crystallizing the T882A mutant of RePC. This approach was motivated by the remarkable observation that a T882A mutation in RePC, which renders the CT domain inactive, results in a substantial increase in the rate of MgATP-hydrolysis and MgADP phosphorylation with carbamoyl phosphate in the BC domain (26). T882A RePC was crystallized in the presence of acetyl-CoA and BC domain substrate analogues, both to confirm the asymmetrical arrangement of the RePC tetramer under a new set of crystallization conditions and also to observe the as yet uncharacterized interaction of the BCCP domain with the BC domain of PC.
Asymmetry of the RePC quaternary structure
Previous X-ray crystal structures of RePC, SaPC and HsPC revealed differences in the overall symmetrical arrangement of the tetrameric enzyme (21, 22, 23). The RePC structure in the presence of ethyl-CoA was highly asymmetrical between the top and bottom faces of the tetramer. The SaPC structures were much more symmetrical between the top and bottom faces, both in the presence and absence of acetyl-CoA. Since carboxybiotin is transferred between active sites on opposing polypeptide chains of PC, the quaternary structure is expected to significantly impact enzyme function. To further investigate what factors may be responsible for altering the quaternary arrangement of PC, RePC was crystallized in the presence and absence of acetyl-CoA. The T882A RePC tetramer remains highly asymmetrical in the presence and absence of acetyl-CoA implying that the overall symmetry of the tetramer is proposed to be a species-specific property of the enzyme.
The asymmetry in RePC prevents any physical interaction between the central allosteric domains on the top and bottom face of the tetramers. Consequently, the RePC tetramer is maintained exclusively through BC-BC and CT-CT homodimerization contacts (Figure 1). A BC domain deletion of RePC forms dimers in the presence and absence of acetyl-CoA, confirming that the central domain of RePC is insufficient to maintain tetramers (Figure 2). Truncation of the BC domain in Bacillus thermodenitrificans PC also results in the formation of active dimers (44). The same truncation in yeast pyruvate carboxylase I isoenzyme is unable to form tetramers and the truncated construct does not co-purify with wild-type tetramers upon co-expression (45). Conversely, there is strong structural evidence to support a role for the central domain in maintaining the tetramers of SaPC and HsPC (22, 23, 24). Similar structural domains have been described in other biotin-dependent enzymes, where they are proposed to play a role in maintaining the quaternary arrangement of the enzyme (38, 46).
While the contribution of the central domain to PC tetramerization appears to vary among species, this domain’s role in binding acetyl-CoA is ubiquitous. Several conserved residues in this domain contribute to the nucleotide binding site for acetyl-CoA and mutation of these residues disrupts allosteric activation (23, 47). Analysis of chimeric enzymes further confirm that allosteric activation is tied to the central domain (48, 49). The central domain of RePC is, therefore, termed the allosteric domain since it houses the acetyl-CoA nucleotide binding site but is not sufficient for tetramerization.
In the T882A RePC structure, the BCCP domain is bound in the BC domain exclusively on the bottom face of the tetramer. In the structure of wild-type RePC, the BCCP domains are ordered exclusively on the top face of the tetramer (21). A recent cryo-EM study of SaPC also revealed a more ordered BCCP on a single face of the tetramer (24). The exclusive ordering of the BCCP domain on only one face of the enzyme is consistent with suggestions that PC and its individual domains are subject to half-of-the-sites reactivity (21, 24, 50, 51, 52). While half-of-the-sites reactivity has been implicated in kinetic studies with E. coli BC, it has not been investigated in detail for PC.
The asymmetry of RePC creates substantial differences in the arrangement of active site pairs between the top and bottom faces of the tetramer. This raises the possibility that the bottom face of the tetramer is locked in a permanently inactive state. Using a mixture of hybrid tetramers, the present study confirms that both the top and bottom faces of the RePC tetramer are active (Table 2 and Figure S3). However, this cannot distinguish between a mechanism with oscillating top/bottom half-of-the-sites reactivity and one with all active site pairs behaving independently and simultaneously. Given the suggestion of half-of-the-sites reactivity for the BC domain of E. coli ACC ((50, 51, 52)) and recent X-ray and cryo electron microscopy structures of PC (21, 24), an oscillating half-of-the-sites mechanism for PC remains a distinct possibility.
Phosphonoacetate as a carboxyphosphate analogue in the BC domain active site
Consistent with half-of-the-sites reactivity, a carboxyphosphate intermediate analogue, phosphonoacetate, is modeled into electron density exclusively on the top face of tetramer 2 (Figure 5). While the putative carboxyphosphate intermediate has never been isolated, several lines of evidence support its role in the biotin carboxylase reaction. The larger family of ATP grasp enzymes, to which BC belongs, all proceed through an acyl-phosphate intermediate (53) and both carbamoylphosphate ((26, 54) and acetyl-phosphate (55) can serve as phosphate donors for the phosphorylation of MgADP to MgATP by PC. In addition, phosphonoacetate is an inhibitor of RePC, sheep liver PC and E. coli BC with respect to both MgATP and HCO3− (56, 57). In the current structure, phosphonoacetate fits into a lobe of electron density immediately adjacent to MgADP in the BC domain (Figure 5). While two Mg2+ ions are expected to bind in the active site, only one Mg2+ is described by the electron density. The absence of a second Mg2+ may contribute to a decreased binding affinity for phosphonoacetate which, in turn, accounts for the relatively weak observed electron density. The B-subdomain lid is closed on the top face of tetramer 2, where electron density corresponding to phosphonoacetate is observed in the active site. Conversely, the lid is disordered on the top face of tetramer 1, where electron density is observed only for MgADP. Since the B-subdomain lid is known to close over the nucleotide substrate (35), lid closure offers further evidence for the binding of phosphonoacetate as a carboxyphosphate analogue in the active site.
Phosphonoacetate appears to bind in two opposing conformations. In one conformation, the phosphonate oxygens are oriented toward MgADP while, in the second, the phosphonate oxygens are oriented in the opposite direction (Figure 5). Multiple binding modes for phosphonoacetate are not surprising given the considerable plasticity associated with this region of the BC domain active site. Both phosphate and bicarbonate ions bind at the same location in X-ray crystal structures of E. coli BC (35, 39), and this binding location aligns with the position of the phosphonate and carboxyl functional groups in the two conformations of phosphonoacetate (Figure 5A). A mixed binding profile for phosphonoacetate is further supported by docking calculations with carboxyphosphate analogues, revealing that both orientations are energetically feasible (Figure 5B and Table S1). The dual conformation is also consistent with the inhibition of RePC by phosphonoacetate, where binding flexibility can explain the relatively low Ki for phosphonoacetate in the MgADP phosphorylation reaction relative to that determined for the pyruvate carboxylation reaction (55). Finally, the flexible conformations for phosphonoacetate in the active site are consistent with recent suggestions that phosphate acts as the general base catalyst to deprotonate biotin (39) (Figure S6). In order for this to occur, the phosphate ion must migrate ~6 Å from the γ-phosphate position of MgATP to occupy its final position near the N-1 of biotin. The active site, therefore, must have sufficient plasticity to facilitate the migration of phosphate.
The BC-BCCP domain interaction in PC
The BC-BCCP domain interaction observed in the T882A RePC structure is the first high resolution description of the interaction between these two domains for any biotin-dependent enzyme. Since all biotin-dependent carboxylase enzymes feature an interaction between the BC and BCCP domains for catalysis, this pose is relevant to a number of other enzymes, including acetyl-CoA carboxylase, methylmalonyl-CoA carboxylase and propionyl-CoA carboxylase. The structure confirms that the BCCP domain of PC binds to the BC domain on its own polypeptide chain prior to translocating to the CT domain on a neighboring polypeptide chain (21, 22, 23, 24). There are very few specific side-chain contacts that contribute to BCCP binding in the BC domain and, where these contacts exist, they are not evolutionarily conserved across PC enzymes. A similar absence of contacts has been described for BCCP domain binding in the CT domain of SaPC and in the CT domain of Roseobacter denitrificans propionyl-CoA carboxylase (22, 38). The lack of significant interdomain contacts facilitates the ready dissociation and translocation of BCCP between the individual catalytic domains of PC.
BCCP binding in the BC domain coincides with the tight closure of the B-subdomain lid. The B-subdomain closes down upon nucleotide binding (35) and subsequently forms a portion of the BCCP docking site. The structure, therefore, predicts that MgATP must bind first in the active site before BCCP can dock and position the tethered biotin in the active site. This is in agreement with the kinetically determined order of substrate binding (58, 59). By virtue of its interaction with BCCP, the B-subdomain lid is closed down more tightly in the presence of BCCP-biotin than it is in the presence of free biotin (Figure 3.i.). Constriction of the B-subdomain lid over the active site facilitates nucleophilic attack on MgATP by bicarbonate and likely contributes to the enhanced catalytic activity observed with BCCP-biotin over free biotin in biotin-dependent enzymes (vide infra). Along with the B-subdomain lid, a surface-accessible loop in the C-subdomain of BC forms the other half of the docking site. This loop represents a 26 amino acid insertion that is unique to the biotin-dependent carboxylases and has been shown to occupy a variety of positions in structures of BC (35). In the current structure, the loop is offset from its position in E. coli BC in order to accommodate the docking of BCCP (Figure 3.ii) (36).
The interaction of BCCP with the BC domain of T882A RePC also offers new insight into the accessibility of tethered biotin to the active site in the BC domain. Recently, Tong and coworkers reported the structure of E. coli BC complexed with free biotin and bicarbonate (39). While this structure is significant, it is unable to explain why free biotin is a relatively poor substrate compared to BCCP-biotin in both E. coli BC and PC (41, 49, 60). The anchor point from which tethered biotin accesses the BC domain active site is clearly defined in the current T882A RePC structure. On this basis, tethered biotin can be rearranged from its defined position in the crystal structure and modeled in an alternate conformation that accesses the active site (Figure 4B). While it can reach the active site, tethered biotin cannot physically achieve the same binding conformation as free biotin. Instead, in order to position the N-1 of tethered biotin in a catalytically relevant position, tethered biotin must bind in a modified orientation relative to free biotin. This altered orientation may account for the substrate preference of tethered biotin over free biotin among biotin-dependent enzymes. Although only Arg353 is positioned to stabilize the enolate of free biotin, several additional residues are in range to provide additional stabilization to the tethered biotin enolate with its alternate binding conformation. For example, the ureido oxygen of tethered biotin is potentially within range of Arg353, Asp399 and Tyr90, all of which may contribute to biotin enolization. In addition, the binding orientation of tethered biotin closes off the active site more completely than free biotin, allowing tethered biotin to better facilitate the nucleophilic attack on MgATP by bicarbonate (Figure 6).
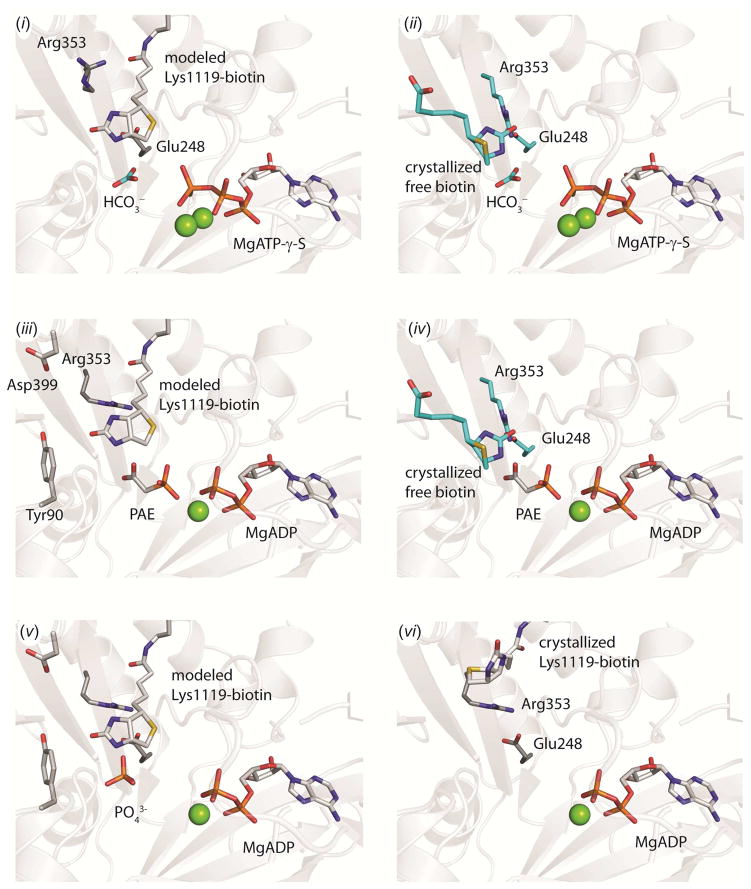
Figure 6. A structural model of catalysis in the BC domain. (i).
Ternary substrate complex with the modeled position of tethered biotin in the active site. The position of HCO3− is illustrated from an overlay with E. coli BC (pdb id = 3G8C, chain A) and MgATP-γ-S is illustrated from an overlay with the BC domain of wild-type RePC (pdb id = 2QF7, chain A). The open side-chain configuration of Arg353 and Glu248 from the structure of wild-type RePC (pdb id = 2QF7, chain A) is also shown. (ii) Ternary substrate complex with free biotin in the active site. The positions of free biotin, HCO3−, Arg353 and Glu248 are from an overlay with E. coli BC (pdb id = 3G8C, chain A) and the position of MgATP-γ-S is from an overlay with the BC domain of wild-type RePC (pdb id = 2QF7, chain A). (iii) Carboxyphosphate-bound intermediate state with modeled tethered biotin and phosphonoacetate (PAE) in the active site. Phosphonoacetate is displayed in a single conformation from an overlay with the BC domain of chain C in the T882A RePC structure. The position of MgADP, Tyr90, Asp399 and Arg353 side-chains are from an overlay with the BC domain of chain B in the T882A RePC structure. (iv) Carboxyphosphate-bound intermediate state with free biotin and phosphonoacetate in the active site. Phosphonoacetate is displayed in a single conformation from an overlay with the BC domain of chain C in the T882A RePC structure. The position of MgADP, free biotin, Arg353 and Glu248 side-chains are from an overlay with the structure of E. coli BC (pdb id = 3G8C, chain A). (v) Phosphate is positioned to serve as the general-base catalyst for the enolization of tethered biotin. The modeled position of tethered biotin in RePC is shown, along with the position of MgADP, Glu248, Asp399, Tyr90 and Arg353 from an overlay with the BC domain of chain B in the T882A RePC structure. (vi) Product-bound state with MgADP in the active site and tethered biotin excluded by the closed conformation of Arg353. The position of all residues and ligands are from the crystal structure of T882A RePC, chain B.
Insight into the mechanism of the BC domain-catalyzed reaction
Together, the X-ray crystal structure of T882A RePC and the accompanying kinetic data with Arg353 and Glu248 mutations suggest that catalysis in PC includes a mechanism to control carboxybiotin access to the BC domain active site. Despite the interaction between BCCP and BC captured in the current structure, tethered biotin is precluded from the active site. Arg353 appears to play a role in limiting carboxybiotin access to the BC domain active site under conditions where the enzyme is not primed for catalysis. RePC strongly favors the carboxylation of pyruvate (full forward reaction) over the decarboxylation of oxaloacetate (full reverse reaction) (26, 55). In a cellular context, pyruvate is regenerated from oxaloacetate by the combined actions of malate dehydrogenase and malic enzyme and there is no metabolic role for the reverse PC-catalyzed reaction. Furthermore, since the active site architecture promotes biotin decarboxylation through stabilization of the biotin enolate, any access of carboxybiotin to the BC domain active site increases the energetically unfavorable decoupling of the BC and CT domain reactions.
Decarboxylation of carboxybiotin occurs at an appreciable rate in both the CT and BC domain active sites of PC, even in the absence of an acceptor substrate. For instance, the active site architecture of the CT domain is sufficient to facilitate rapid decarboxylation of carboxybiotin in the presence of both substrates and substrate analogues (61). When MgADP is bound in the active site and BCCP is bound in the BC domain, the conformation of Arg353 changes to directly interact with Glu248, which may serve to hinder the return and subsequent decarboxylation of carboxybiotin in the BC domain active site. This proposal is supported by the kinetic characterization of Arg353 and Glu248 mutations in the present study and in the accompanying report by Zeczycki et al. (42). It is also consistent with a two-state model proposed for carboxybiotin in PC, based on the biphasic rates observed for the [14C]-carboxyl group transfer from isolated PC-[14C]-carboxybiotin to 2-oxobutyrate (61, 62). According to the model, carboxybiotin in state I is bound in the site of the first partial reaction while, in state II, carboxybiotin is bound outside of the active site but in its near vicinity. These two states remain at equilibrium until 2-oxobutyrate is added, resulting in rapid carboxyl transfer to 2-oxobutyrate in the CT domain. The current structure likely represents state II, with BCCP-biotin precluded from the BC domain active site but poised for translocation to the CT domain upon binding of a CT domain substrate. This model may also apply to other members of the biotin-dependent enzyme family. For example, while E. coli BC catalyzes the hydrolysis of MgATP in the presence of free biotin, it is unable to catalyze this reaction in the presence of BCCP-biotin unless both the CT subunit and the CT substrate, acetyl-CoA, are also present (63). This indicates that a similar predisposition exists in E. coli acetyl-CoA carboxylase to regulate access of tethered biotin to the BC subunit active site and suggests that such a mechanism may have relevance within the broader biotin-dependent enzyme family.
Previous kinetic studies have proposed that the increased rate of MgADP phosphorylation with carbamoyl phosphate in the T882A RePC mutant, compared to the wild-type enzyme, is a direct result of BCCP-biotin favoring placement in the BC domain relative to the CT domain (26). The present study suggests that a salt bridge between Arg353 and Glu248 limits carboxybiotin access to the BC domain active site, thereby promoting efficient coupling between the BC and CT domain half reactions. Notably, MgADP phosphorylation by carbamoyl phosphate proceeds through a phosphoryl transfer mechanism that is known to be independent of biotin and that is contingent on the proper alignment of substrates in the active site. Mutations such as T882A and E248A promote tight closure of the BC domain active site and facilitate phosphoryl transfer by restricting substrate conformational freedom and favoring proper positioning of the substrates for catalysis. The structure of the BCCP-BC domain interaction in T882A RePC reveals that the BC domain active site closes down tightly upon docking the BCCP domain, without tethered biotin directly accessing the active site (Figure 3). Tight closure of the BC domain active site, therefore, requires only that tethered biotin be in the vicinity of the active site and not that it physically enter the active site. The increased rate of MgADP phosphorylation over wild-type for T882A RePC can simply result from the increased occupancy of BCCP in the BC domain and does not require that biotin directly access the active site. In contrast to T882A, mutations in the Arg353 – Glu248 salt bridge are predicted to lead to increased access of tethered biotin to the BC domain active site. The detailed kinetic characterization of mutations in Arg353, discussed in the proceeding report (42), reveals an increased rate for the full reverse reaction compared with the wild-type enzyme, consistent with carboxybiotin gaining greater access to the BC domain active site. When biotin accesses the BC domain active site, it also promotes closure of the B-subdomain lid, as evidenced by the structure of E. coli BC with free biotin (39). Furthermore, the modeled position of tethered biotin in the active site suggests that tethered biotin might further restrict the conformational freedom of MgADP and carbamoyl phosphate (Figure 6). Therefore, mutations such as E248A, which enhance biotin access to the active site, are also expected to promote the phosphoryl transfer reaction.
The T882A RePC crystal structure can be combined with a series of existing structural and kinetic descriptions of biotin carboxylase to assemble a series of structural snapshots that represent catalysis in the BC domain (Figure 6). While the precise position of tethered biotin in the active site is not known, the putative binding position based on the point of origin in BCCP allows for a comparison of the reaction with the free biotin and tethered biotin substrates. In panels (i) and (ii), the ternary substrate complex is poised for nucleophilic attack on MgATP by bicarbonate. In E. coli BC, the rate of MgATP-hydrolysis is 16-fold faster in the presence of tethered-biotin compared to free biotin (41). The alternate binding orientation for tethered biotin closes the active site more completely than free biotin. Tight closure of the B-subdomain lid further seals the active site. Together, this promotes the nucleophilic attack by bicarbonate on MgATP in the full forward reaction and can also account for the increased kcat observed for both MgATP-hydrolysis and MgADP phosphorylation in the presence of tethered biotin.
In panels (iii) and (iv), phosphonoacetate mimics the carboxyphosphate intermediate which is positioned directly between MgADP and biotin. The position of the second Mg2+ is not defined. Both free biotin and tethered biotin are positioned to attack the CO2 electrophile that results from the decomposition of carboxyphosphate, but require a general base catalyst to deprotonate N-1 and generate the biotin enolate intermediate. Recent proposed mechanisms of catalysis invoke the phosphate ion as this general base catalyst (39). For this to occur, the carboxyphosphate intermediate must decompose into CO2 and phosphate. Subsequently, the phosphate must migrate into a position amenable to deprotonation of the biotin N-1 nitrogen. X-ray crystal structures of E. coli BC have revealed that phosphate binds in close proximity to the biotin binding site (35, 39). The two conformations for phosphonoacetate in the T882A RePC structure demonstrate that multiple binding positions are possible for phosphate in the BC domain active site, consistent with a flexible active site pocket that permits phosphate migration between MgADP and biotin. Panel (v) overlays the position of the bound phosphate ion in E. coli BC with the modeled tethered biotin in the T882A RePC structure, revealing that phosphate can occupy a binding position in close proximity to the N-1 of biotin, providing structural support for the proposed role of phosphate as the general base catalyst. Finally, panel (vi) illustrates the current T882A RePC crystal structure, representing the closed state with MgADP bound in the active site and tethered biotin excluded. By impeding the access of carboxybiotin to the BC domain active site in the product bound state, PC ensures a more optimal coupling of MgATP hydrolysis to pyruvate carboxylation. This mechanism is expected to be generally applicable to the larger family of biotin-dependent enzymes.
In summary, structural studies of Rhizobium etli T882A PC demonstrate that the asymmetry in the tetramer is independent of coenzyme-A and that the central allosteric domain does not play a role in maintaining the tetramerization state of the enzyme. The X-ray crystal structure at 2.4 Å resolution provides the first high-resolution description of a biotin-dependent enzyme with the BCCP-biotin bound in the BC domain. It is clear from this structure that tethered biotin must access the active site in an orientation that is different from free biotin, offering a structural explanation for the substrate preference of tethered biotin over free biotin. The structure also suggests a gate-keeping role for Arg353, a flexible residue positioned at the mouth of the active site. Arg353 is proposed to play a role in precluding carboxybiotin access to the active site. The catalytic role of this residue is examined in detail in the accompanying report (42).
Supplementary Material
ABBREVIATIONS
- ATP
adenosine triphosphate
- ATP-γ-S
Adenosine 5′-[γ-thio]triphosphate
- BC
biotin carboxylase
- BCCP
biotin carboxyl carrier protein
- CT
carboxyl transferase
- CoA
coenzyme-A
- DTT
DL-Dithiothreitol
- EGTA
Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HsPC
Homo sapiens pyruvate carboxylase
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- MPD
2-methyl-2,4-pentanediol
- PEG
Poly(ethylene glycol)
- PC
pyruvate carboxylase
- RePC
Rhizobium etli pyruvate carboxylase
- SaPC
Staphylococcus aureus pyruvate carboxylase
- TCEP
Tris(2-carboxyethyl)phosphine
Footnotes
This work was supported by the National Institute of Health grant GM070455 to WWC, JCW, PVA and MSt.M, and NIH award F32DK083898 from the National Institute of Diabetes and Digestive And Kidney Diseases to TNZ. ADL is supported by a GAANN award (Graduate Assistance in Areas of National Need) from the U.S. Department of Education.
Accession codes. The atomic coordinates for RePC T882A co-crystallized with and without acetyl-CoA have been deposited in the Protein Data Bank as entry 3TW6 and 3TW7, respectively.
Supporting information. Supplementary figures S1 – S7 and the detailed methods for protein expression and purification of the BC domain truncation and site-directed mutants of RePC are provided in the supporting information. This material may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Jitrapakdee S, Vidal-Puig A, Wallace JC. Anaplerotic Roles of Pyruvate Carboxylase in Mammalian Tissues. Cell Mol Life Sci. 2006;63:843–854. doi: 10.1007/s00018-005-5410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Large V, Beylot M. Modifications of Citric Acid Cycle Activity and Gluconeogenesis in Streptozotocin-Induced Diabetes and Effects of Metformin. Diabetes. 1999;48:1251–1257. doi: 10.2337/diabetes.48.6.1251. [DOI] [PubMed] [Google Scholar]
- 3.Gamberino WC, Berkich DA, Lynch CJ, Xu B, LaNoue KF. Role of Pyruvate Carboxylase in Facilitation of Synthesis of Glutamate and Glutamine in Cultured Astrocytes. J Neurochem. 1997;69:2312–2325. doi: 10.1046/j.1471-4159.1997.69062312.x. [DOI] [PubMed] [Google Scholar]
- 4.Hazen SA, Waheed A, Sly WS, LaNoue KF, Lynch CJ. Effect of Carbonic Anhydrase Inhibition and Acetoacetate on Anaplerotic Pyruvate Carboxylase Activity in Cultured Rat Astrocytes. Dev Neurosci. 1997;19:162–171. doi: 10.1159/000111202. [DOI] [PubMed] [Google Scholar]
- 5.Freytag SO, Utter MF. Induction of Pyruvate Carboxylase Apoenzyme and Holoenzyme in 3T3-L1 Cells during Differentiation. Proc Natl Acad Sci U S A. 1980;77:1321–1325. doi: 10.1073/pnas.77.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu D, Mulder H, Zhao P, Burgess SC, Jensen MV, Kamzolova S, Newgard CB, Sherry AD. 13C NMR Isotopomer Analysis Reveals a Connection between Pyruvate Cycling and Glucose-Stimulated Insulin Secretion (GSIS) Proc Natl Acad Sci U S A. 2002;99:2708–2713. doi: 10.1073/pnas.052005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen MV, Joseph JW, Ilkayeva O, Burgess S, Lu D, Ronnebaum SM, Odegaard M, Becker TC, Sherry AD, Newgard CB. Compensatory Responses to Pyruvate Carboxylase Suppression in Islet Beta-Cells. Preservation of Glucose-Stimulated Insulin Secretion. J Biol Chem. 2006;281:22342–22351. doi: 10.1074/jbc.M604350200. [DOI] [PubMed] [Google Scholar]
- 8.Hasan NM, Longacre MJ, Stoker SW, Boonsaen T, Jitrapakdee S, Kendrick MA, Wallace JC, MacDonald MJ. Impaired Anaplerosis and Insulin Secretion in Insulinoma Cells Caused by Small Interfering RNA-Mediated Suppression of Pyruvate Carboxylase. J Biol Chem. 2008;283:28048–28059. doi: 10.1074/jbc.M804170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. Altered Regulation of Metabolic Pathways in Human Lung Cancer Discerned by (13)C Stable Isotope-Resolved Metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu KJ, Kleps R, Henderson T, Nyhus L. 13C NMR Study of Hepatic Pyruvate Carboxylase Activity in Tumor Rats. Biochem Biophys Res Commun. 1991;179:366–371. doi: 10.1016/0006-291x(91)91379-q. [DOI] [PubMed] [Google Scholar]
- 11.Forbes NS, Meadows AL, Clark DS, Blanch HW. Estradiol Stimulates the Biosynthetic Pathways of Breast Cancer Cells: Detection by Metabolic Flux Analysis. Metab Eng. 2006;8:639–652. doi: 10.1016/j.ymben.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Marin-Valencia I, Roe CR, Pascual JM. Pyruvate Carboxylase Deficiency: Mechanisms, Mimics and Anaplerosis. Mol Genet Metab. 2010;101:9–17. doi: 10.1016/j.ymgme.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, De Vivo D. Pyruvate Carboxylase Deficiency. In: Pagon RA, Bird TD, Dolan CR, Stephens K, editors. GeneReviews. University of Washington, Seattle; All rights reserved, Seattle (WA): 1993. [Google Scholar]
- 14.Monnot S, Serre V, Chadefaux-Vekemans B, Aupetit J, Romano S, De Lonlay P, Rival JM, Munnich A, Steffann J, Bonnefont JP. Structural Insights on Pathogenic Effects of Novel Mutations Causing Pyruvate Carboxylase Deficiency. Hum Mutat. 2009;30:734–740. doi: 10.1002/humu.20908. [DOI] [PubMed] [Google Scholar]
- 15.Han J, Liu YQ. Reduction of Islet Pyruvate Carboxylase Activity might be Related to the Development of Type 2 Diabetes Mellitus in Agouti-K Mice. J Endocrinol. 2010;204:143–152. doi: 10.1677/JOE-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacDonald MJ, Longacre MJ, Langberg EC, Tibell A, Kendrick MA, Fukao T, Ostenson CG. Decreased Levels of Metabolic Enzymes in Pancreatic Islets of Patients with Type 2 Diabetes. Diabetologia. 2009;52:1087–1091. doi: 10.1007/s00125-009-1319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Branson JP, Nezic M, Wallace JC, Attwood PV. Kinetic Characterization of Yeast Pyruvate Carboxylase Isozyme pyc1. Biochemistry. 2002;41:4459–4466. doi: 10.1021/bi011888m. [DOI] [PubMed] [Google Scholar]
- 18.Osmani SA, Mayer F, Marston FAO, Selmes IP, Scrutton MC. Pyruvate Carboxylase from Aspergillus nidulans Effects of Regulatory Modifiers on the Structure of the Enzyme. Eur J Biochem. 1984;139:509–518. doi: 10.1111/j.1432-1033.1984.tb08035.x. [DOI] [PubMed] [Google Scholar]
- 19.Mayer F, Wallace JC, Keech DB. Further Electron Microscope Studies on Pyruvate Carboxylase. Eur J Biochem. 1980;112:265–272. doi: 10.1111/j.1432-1033.1980.tb07202.x. [DOI] [PubMed] [Google Scholar]
- 20.Attwood PV, Johannssen W, Chapman-Smith A, Wallace JC. The Existence of Multiple Tetrameric Conformers of Chicken Liver Pyruvate Carboxylase and their Roles in Dilution Inactivation. Biochem J. 1993;290(Pt 2):583–590. doi: 10.1042/bj2900583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.St Maurice M, Reinhardt L, Surinya KH, Attwood PV, Wallace JC, Cleland WW, Rayment I. Domain Architecture of Pyruvate Carboxylase, a Biotin-Dependent Multifunctional Enzyme. Science. 2007;317:1076–1079. doi: 10.1126/science.1144504. [DOI] [PubMed] [Google Scholar]
- 22.Xiang S, Tong L. Crystal Structures of Human and Staphylococcus aureus Pyruvate Carboxylase and Molecular Insights into the Carboxyltransfer Reaction. Nat Struct Biol. 2008;15:295–302. doi: 10.1038/nsmb.1393. [DOI] [PubMed] [Google Scholar]
- 23.Yu LP, Xiang S, Lasso G, Gil D, Valle M, Tong L. A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A. Structure. 2009;17:823–832. doi: 10.1016/j.str.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lasso G, Yu LP, Gil D, Xiang S, Tong L, Valle M. Cryo-EM Analysis Reveals New Insights into the Mechanism of Action of Pyruvate Carboxylase. Structure. 2010;18:1300–1310. doi: 10.1016/j.str.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Attwood PV, Geeves MA. Changes in Catalytic Activity and Association State of Pyruvate Carboxylase which are Dependent on Enzyme Concentration. Arch Biochem Biophys. 2002;401:63–72. doi: 10.1016/S0003-9861(02)00039-5. [DOI] [PubMed] [Google Scholar]
- 26.Zeczycki TN, St Maurice M, Jitrapakdee S, Wallace JC, Attwood PV, Cleland WW. Insight into the Carboxyl Transferase Domain Mechanism of Pyruvate Carboxylase from Rhizobium etli. Biochemistry. 2009;48:4305–4313. doi: 10.1021/bi9003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser Crystallographic Software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Winn MD, Isupov MN, Murshudov GN. Use of TLS Parameters to Model Anisotropic Displacements in Macromolecular Refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adina-Zada A, Jitrapakdee S, Surinya KH, McIldowie MJ, Piggott MJ, Cleland WW, Wallace JC, Attwood PV. Insights into the Mechanism and Regulation of Pyruvate Carboxylase by Characterisation of a Biotin-Deficient Mutant of the Bacillus thermodenitrificans Enzyme. Int J Biochem Cell Biol. 2008;40:1743–1752. doi: 10.1016/j.biocel.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Thoden JB, Blanchard CZ, Holden HM, Waldrop GL. Movement of the Biotin Carboxylase B-Domain as a Result of ATP Binding. J Biol Chem. 2000;275:16183–16190. doi: 10.1074/jbc.275.21.16183. [DOI] [PubMed] [Google Scholar]
- 36.Waldrop GL, Rayment I, Holden HM. Three-Dimensional Structure of the Biotin Carboxylase Subunit of Acetyl-CoA Carboxylase. Biochemistry. 1994;33:10249–10256. doi: 10.1021/bi00200a004. [DOI] [PubMed] [Google Scholar]
- 37.Gerstein M. A Resolution-Sensitive Procedure for Comparing Protein Surfaces and its Application to the Comparison of Antigen-Combining Sites. Acta Cryst A. 1992;48:271–276. [Google Scholar]
- 38.Huang CS, Sadre-Bazzaz K, Shen Y, Deng B, Zhou ZH, Tong L. Crystal Structure of the α(6)β(6) Holoenzyme of Propionyl-Coenzyme A Carboxylase. Nature. 2010;466:1001–1005. doi: 10.1038/nature09302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou CY, Yu LP, Tong L. Crystal Structure of Biotin Carboxylase in Complex with Substrates and Implications for its Catalytic Mechanism. J Biol Chem. 2009;284:11690–11697. doi: 10.1074/jbc.M805783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jitrapakdee S, St Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, Mechanism and Regulation of Pyruvate Carboxylase. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanchard CZ, Chapman-Smith A, Wallace JC, Waldrop GL. The Biotin Domain Peptide from the Biotin Carboxyl Carrier Protein of Escherichia coli Acetyl-CoA Carboxylase Causes a Marked Increase in the Catalytic Efficiency of Biotin Carboxylase and Carboxyltransferase Relative to Free Biotin. J Biol Chem. 1999;274:31767–31769. doi: 10.1074/jbc.274.45.31767. [DOI] [PubMed] [Google Scholar]
- 42.Zeczycki TN, Menefee AL, Adina-Zada A, Jitrapakdee S, Wallace JC, Attwood PV, StMaurice M, Cleland WW. Novel Insights into the Biotin Carboxylase Domain Reactions of Pyruvate Carboxylase from Rhizobium etli. Biochemistry.(accompanying manuscript in this series) 2011 doi: 10.1021/bi2012788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knowles JR. The Mechanism of Biotin-Dependent Enzymes. Annu Rev Biochem. 1989;58:195–221. doi: 10.1146/annurev.bi.58.070189.001211. [DOI] [PubMed] [Google Scholar]
- 44.Sueda S, Islam MN, Kondo H. Protein Engineering of Pyruvate Carboxylase: Investigation on the Function of Acetyl-CoA and the Quaternary Structure. Eur J Biochem. 2004;271:1391–1400. doi: 10.1111/j.1432-1033.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- 45.Jitrapakdee S, Surinya KH, Adina-Zada A, Polyak SW, Stojkoski C, Smyth R, Booker GW, Cleland WW, Attwood PV, Wallace JC. Conserved Glu40 and Glu433 of the Biotin Carboxylase Domain of Yeast Pyruvate Carboxylase I Isoenzyme are Essential for the Association of Tetramers. Int J Biochem Cell Biol. 2007;39:2120–2134. doi: 10.1016/j.biocel.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 46.Balsera M, Buey RM, Li XD. Quaternary Structure of the Oxaloacetate Decarboxylase Membrane Complex and Mechanistic Relationships to Pyruvate Carboxylases. J Biol Chem. 2011;286:9457–9467. doi: 10.1074/jbc.M110.197442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adina-Zada A, Hazra R, Sereeruk C, Jitrapakdee S, Zeczycki TN, St Maurice M, Cleland WW, Wallace JC, Attwood PV. Probing the Allosteric Activation of Pyruvate Carboxylase using 2′,3′-O-(2,4,6-Trinitrophenyl) Adenosine 5′-Triphosphate as a Fluorescent Mimic of the Allosteric Activator Acetyl CoA. Arch Biochem Biophys. 2011;509:117–126. doi: 10.1016/j.abb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jitrapakdee S, Adina-Zada A, Besant PG, Surinya KH, Cleland WW, Wallace JC, Attwood PV. Differential Regulation of the Yeast Isozymes of Pyruvate Carboxylase and the Locus of Action of Acetyl CoA. Int J Biochem Cell Biol. 2007;39:1211–1223. doi: 10.1016/j.biocel.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Islam MN, Sueda S, Kondo H. Construction of New Forms of Pyruvate Carboxylase to Assess the Allosteric Regulation by Acetyl-CoA. Protein Eng Des Sel. 2005;18:71–78. doi: 10.1093/protein/gzi011. [DOI] [PubMed] [Google Scholar]
- 50.Mochalkin I, Miller JR, Evdokimov A, Lightle S, Yan C, Stover CK, Waldrop GL. Structural Evidence for Substrate-Induced Synergism and Half-Sites Reactivity in Biotin Carboxylase. Protein Sci. 2008;17:1706–1718. doi: 10.1110/ps.035584.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Queiroz MS, Waldrop GL. Modeling and Numerical Simulation of Biotin Carboxylase Kinetics: Implications for Half-Sites Reactivity. J Theor Biol. 2007;246:167–175. doi: 10.1016/j.jtbi.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 52.Janiyani K, Bordelon T, Waldrop GL, Cronan JE., Jr Function of Escherichia Coli Biotin Carboxylase Requires Catalytic Activity of both Subunits of the Homodimer. J Biol Chem. 2001;276:29864–29870. doi: 10.1074/jbc.M104102200. [DOI] [PubMed] [Google Scholar]
- 53.Galperin MY, Koonin EV. A Diverse Superfamily of Enzymes with ATP-Dependent Carboxylate-amine/thiol Ligase Activity. Protein Sci. 1997;6:2639–2643. doi: 10.1002/pro.5560061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Attwood PV, Graneri BD. Pyruvate Carboxylase Catalysis of Phosphate Transfer between Carbamoyl Phosphate and ADP. Biochem J. 1991;273(Pt 2):443–448. doi: 10.1042/bj2730443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeczycki TN, Menefee AL, Jitrapakdee S, Wallace JC, Attwood PV, StMaurice M, Cleland WW. Activation and Inhibition of Pyruvate Carboxylase from Rhizobium etli. Biochemistry. (accompanying manuscript in this series) 2011 doi: 10.1021/bi201276r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanchard CZ, Amspacher D, Strongin R, Waldrop GL. Inhibition of Biotin Carboxylase by a Reaction Intermediate Analog: Implications for the Kinetic Mechanism. Biochem Biophys Res Commun. 1999;266:466–471. doi: 10.1006/bbrc.1999.1844. [DOI] [PubMed] [Google Scholar]
- 57.Ashman LK, Keech DB. Sheep Kidney Pyruvate Carboxylase. Studies on the Coupling of Adenosine Triphosphate Hydrolysis and CO2 Fixation. J Biol Chem. 1975;250:14–21. [PubMed] [Google Scholar]
- 58.McClure WR, Lardy HA, Cleland WW. Rat Liver Pyruvate Carboxylase. 3 Isotopic Exchange Studies of the First Partial Reaction. J Biol Chem. 1971;246:3584–3590. [PubMed] [Google Scholar]
- 59.Attwood PV, Graneri BD. Bicarbonate-Dependent ATP Cleavage Catalysed by Pyruvate Carboxylase in the Absence of Pyruvate. Biochem J. 1992;287(Pt 3):1011–1017. doi: 10.1042/bj2871011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kondo H, Uno S, Moriuchi F, Sunamoto J, Ogushi S, Tsuru D. Synthesis and Enzymatic Carboxylation of a Biotin-Containing Peptide Representing the Coenzyme Binding Site of E. Coli Acetyl-CoA Carboxylase. Bull Chem Soc Jpn. 1983;56:1176–1180. [Google Scholar]
- 61.Goodall GJ, Baldwin GS, Wallace JC, Keech DB. Factors that Influence the Translocation of the N-Carboxybiotin Moiety between the Two Sub-Sites of Pyruvate Carboxylase. Biochem J. 1981;199:603–609. doi: 10.1042/bj1990603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Attwood PV, Wallace JC. The Carboxybiotin Complex of Chicken Liver Pyruvate Carboxylase. A Kinetic Analysis of the Effects of Acetyl-CoA, Mg2+ Ions and Temperature on its Stability and on its Reaction with 2-Oxobutyrate. Biochem J. 1986;235:359–364. doi: 10.1042/bj2350359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soriano A, Radice AD, Herbitter AH, Langsdorf EF, Stafford JM, Chan S, Wang S, Liu YH, Black TA. Escherichia coli Acetyl-Coenzyme A Carboxylase: Characterization and Development of a High-Throughput Assay. Anal Biochem. 2006;349:268–276. doi: 10.1016/j.ab.2005.10.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.