Abstract
Malignant rhabdoid tumor (MRT) is a rare and highly aggressive neoplasm of young children. MRT is characterized by inactivation of integrase interactor 1 (INI1). Cyclin-dependent kinase 4 (CDK4), which acts downstream of INI1, is required for the proliferation of MRT cells. Here we investigated the effects of PD 0332991 (PD), a potent inhibitor of CDK4, against five human MRT cell lines (MP-MRT-AN, KP-MRT-RY, G401, KP-MRT-NS, KP-MRT-YM). In all of the cell lines except KP-MRT-YM, PD inhibited cell proliferation > 50 %, (IC50 values 0.01 to 0.6 µM) by WST-8 assay, and induced G1-phase cell cycle arrest, as shown by flow cytometry and BrdU incorporation assay. The sensitivity of the MRT cell lines to PD was inversely correlated with p16 expression (r = 0.951). KP-MRT-YM cells overexpress p16 and were resistant to the growth inhibitory effect of PD. Small interfering RNA against p16 significantly increased the sensitivity of KP-MRT-YM cells to PD (p < 0.05). These results suggest that p16 expression in MRT could be used to predict its sensitivity to PD. PD may be an attractive agent for patients with MRT whose tumors express low levels of p16.
Keywords: PD 0332991, CDK4, p16, rhabdoid tumor, INI1
Introduction
Malignant rhabdoid tumor (MRT) is one of the most aggressive malignancies of early childhood. The majority of MRTs are characterized by biallelic inactivation in the integrase interactor 1 (INI1) tumor suppressor gene, located in chromosome 22q11.2 [1]. The overall survival rate of patients with MRT of kidney is only 20 to 25 % [2]. Therefore, an effective treatment for patients with MRT is urgently needed.
INI1 (also known as SNF5 or BAF47) is a core subunit of all human SWI/SNF complexes. INI1 transcriptionally inhibits the expression of cyclin D, which forms a complex with cyclin-dependent kinase 4 (CDK4), functions as a regulator of cyclin D-CDK4 complex, and induces the expression of the p16 (also known as Ink4a/CDKN2A) [3,4]. Increased p16 expression inhibits cell cycle progression to the S-phase by preventing the activation of cyclin D-CDK4 kinase. In MRT cells, the INI1 gene is mutated or deleted, resulting in a loss of INI1 function [1]. This induces the expression of cyclin D and inhibits p16 expression, which accelerates the transition from the G1 phase to the S phase [5]. These changes result in unregulated cell cycle progression in MRT cells. Therefore, cyclin D-CDK4 kinase is an important therapeutic target for MRT.
PD 0332991 (PD) is a small, highly selective inhibitor of CDK4. As a result, PD inhibits the proliferation of cancer cells that express and activate CDK4. PD has been shown to be effective against colon cancer, breast cancer [6–8], rhabdomyosarcoma [9], multiple myeloma [10], mantle cell lymphoma [11], and glioblastoma [12]. However, it is unknown whether PD is effective against MRT cells. In this study, we found that the inhibition of the proliferation of MRT cells by PD was inversely related to p16 expression.
Materials and Methods
Cell Lines and Cell Culture
MRT cell lines, G401, MP-MRT-AN (AN), KP-MRT-RY (RY), KP-MRT-NS (NS), and KP-MRT-YM (YM) cell lines, were cultured in RPMI1640 medium containing 10% fetal bovine serum (FBS) and were subcultured as previously described [13]. The HeLa human uterine cervix carcinoma cell line was used as a positive control of p16 expression.
Reagents
PD was kindly provided by James Christensen (Pfizer, San Diego, CA, USA). A stock solution of the compound was prepared in dimethyl sulfoxide (DMSO, Sigma. St. Louis, MO, USA) and stored at −80 °C. PD was used at final concentrations from 0 to 10 µM, according to previous reports [6,9,11,14].
WST-8 assay
Cells were seeded in normal growth medium into 96-well cell plates. After 24 h, the culture medium was replaced with culture medium containing PD or DMSO. Cells were cultured and treated in triplicate. Cell proliferation was determined 8 days after the treatment by WST-8 assay using a Cell Counting Kit-8 (Dojin East, Tokyo, Japan) as described previously [15].
Cell cycle analysis
After 48 h incubation with PD or DMSO, the cells were harvested. Flow cytometry analysis was analyzed as described previously [16]. For the BrdU incorporation assay, cells were seeded in 96-well plates, incubated for 24 h, and then PD or DMSO was added. After an additional 48 h, BrdU incorporation was measured with a BrdU labeling and detection kit I (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer’s instructions and examined with a microplate reader (Multiscan JX, Dainippon Pharmaceutical). BrdU incorporation was calculated as OD405-OD490, where OD490 was used as a reference.
Immunoblotting
Cell lysates were purified, adjusted to equal protein concentrations, separated by SDS-PAGE, and transferred as previously described [17]. The membrane was immunoblotted using anti-p16 polyclonal antibody (clone16P04; 1:200; Neomarker, Union City, CA, USA), anti-CDK4 monoclonal antibody (#2906; 1:1000; Cell signaling, Beverly, MA, USA), anti-cyclin D polyclonal antibody (sc-753; 1:200; Santa Cruz Biotechnology), anti-Rb monoclonal antibody (#9309; 1:2000; Cell signaling), and anti-β-actin antibody (clone AC-15; 1:2500, Sigma Chemical Co., St. Louis, MO, USA). Antibody binding was detected with an enhanced chemiluminescence detection system (Amersham, St. Louis, MO, USA).
Immunoprecipitation
After 24 h incubation with culture medium containing PD or vehicle alone, the cells were harvested. Lysates were prepared as described above, incubated for 30 min with the appropriate amount of Rb (4H1) monoclonal antibody (#9309; 1:100; Cell Signaling Technology, Beverly, MA, USA) and protein A/G-plus agarose (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4 °C overnight, and washed 5 times with lysis buffer. The membrane was transferred and blocked as described above and incubated with phospho-Rb (ser780) polyclonal antibody (#9307; 1:1000; Cell Signaling Technology,) or Rb (4H1) monoclonal antibody (#9309; 1:2000; Cell Signaling Technology).
Real-time Polymerase Chain Reaction
Cells were harvested at the log-growth phase. Total RNA was extracted, and transcribed into cDNA as described above [17]. For the experiments in which PD was added, total RNA was extracted after an additional 1 or 2 days. cDNAs for p16, proliferating cell nuclear antigen (PCNA), cyclin A, and glucose-6-phosphate dehydrogenase (GAPDH) were amplified with a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with SYBR-GREEN 1 (TAKARA BIO, INC., Otsu, Japan). The primer sets (Supplementary Table 1) were selected to amplify with very nearly the same efficiency. Target mRNA levels were normalized to GAPDH mRNA levels and calculated by the ΔΔcycle threshold (CT) method [18].
Analysis of enzyme activity for CDK4
CDK4 kinase activity was measured with the non-radioisotopic C2P system (Sysmex, Kobe, Japan) as described previously [19]. Protein lysate was applied to a well of a dot-blot device. To measure kinase activity, CDK molecules were immunoprecipitated from 100 µg of lysate total protein with 2 µg anti-CDK4 antibody and 20 µl protein A Sepharose beads (Amersham) for 1 h at 4 °C. The thiophosphate of ATP-γ-S was transferred to the protein substrate during the on-bead kinase reaction under continuous shaking at 37 °C for 30 min. The introduced thiophosphate was labelled further with 5- iodoacetamidofluorescein for 90 min in the dark at room temperature and blotted onto a polyvinylidene fluoride membrane for 1 h at 37 °C. Fluorescent images of the dot-blot device membrane were evaluated with a Molecular Imager FX image analyzer (Bio-Rad, Hercules, CA, USA), and the fluorescence intensity of the dots was quantified with the Quantity One program (Bio-Rad).
RNA interference of p16
p16 was knocked down by RNA interference (RNAi) as described previously [18]. In brief, double stranded RNA against p16 (p16 small interfering RNA [siRNA]) targeting exon 2 of p16 (NCBI GenBank ID: 1029) was purchased from Invitrogen Corp. (Grand Island, New York, USA). Six hours after the transfection, the medium was replaced with fresh medium with PD or vehicle alone. Two days after the medium was replaced, the cells were harvested and the expression of p16 was evaluated by immunoblotting. Four days after the medium was replaced, cell proliferation was determined colorimetrically by WST-8 assay as described above.
Statistical Analysis
Values are expressed as the mean ± SE. Two-sided Student’s t-test was used to statistically evaluate the differences. P < 0.05 was considered to indicate a statistically significant difference. The levels of p16 in each cell line on immunoblotting was analyzed with Image J 1.43 (NIH, Bethesda, MD, USA) and was normalized to the level of β-actin. The relation between the sensitivity of the MRT cell lines to PD and the p16 expression of the cell lines was analyzed by a Pearson’s correlation coefficient test. In the immunoprecipitation experiments, the level of Phospho-Rb was normalized to the level of Rb.
Results
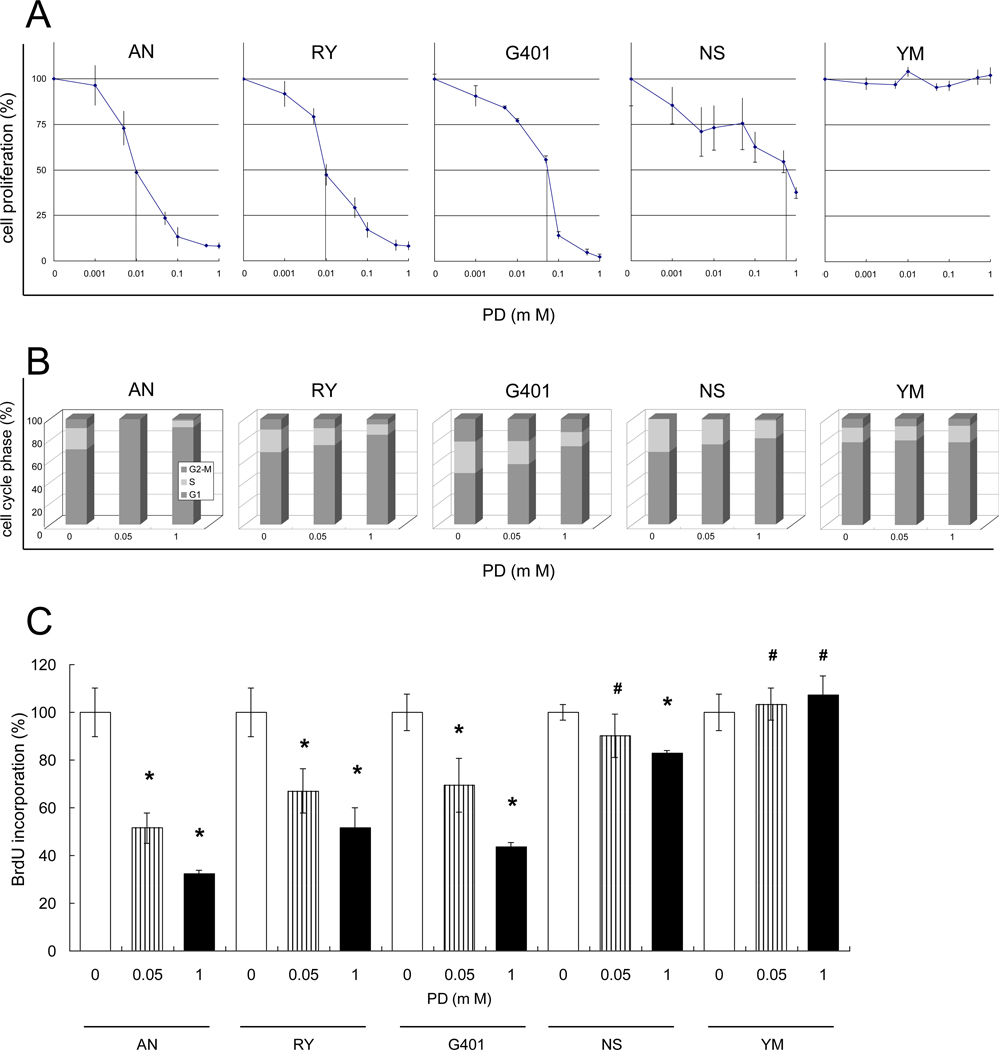
PD inhibited proliferation of AN, RY, G401 and NS MRT cells by suppressing the cell cycle progression at the G1-phase
The AN, RY, G401 and NS cell lines were effectively inhibited by PD in a concentration-dependent manner in a WST-8 assay (Fig. 1A). The 50 % inhibition concentrations (IC50) were 0.01 µM, 0.01 µM, 0.06 µM, and 0.6 µM, respectively. In contrast, the YM cell line was resistant to PD (IC50 >10 µM). The flow cytometry results show that PD at concentrations between 0 to 1.0 µM induced G1 arrest in the AN, RY, G401 and NS cell lines in a concentration-dependent manner, but had no effect on YM cells (Fig. 1B). The BrdU incorporation results were consistent with the WST-8 and flow cytometry results: PD reduced BrdU incorporation (indicating G1 arrest) in the AN, RY, G401 and NS cell lines, but not in the YM cell line (Fig. 1C). PD, even at a concentration of 0.05 µM, significantly reduced BrdU incorporation in the AN, RY, and G401 cell lines (p < 0.05).
Fig. 1.
PD inhibits cell proliferation of most MRT cell lines by suppressing the cell cycle progression at the G1-phase. A, Cells were seeded and allowed to attach for 24 h. PD dissolved in DMSO was added at the indicated concentrations in triplicate cultures. Cell proliferation was determined 8 days after the treatment using a WST-8 assay. Proliferation is expressed as a percent of the proliferation in DMSO, used as a control. Values are the mean of results from three wells. Bars, ±SE. B, Cells were seeded, allowed to attach for 24 h and PD in DMSO (0.05 or 1 µM) or DMSO alone was added. Cell cycle phase was determined after a further 48 h by flow cytometry. C, Cells were seeded, allowed to attach for 24 h, and PD in DMSO (0.05 or 1 µM) or DMSO alone was added to triplicate cultures. The cells were incubated with BrdU solution at 37 °C for 2 h. The proportion of cells in active replication was determined 48 h after adding PD using the BrdU incorporation assay. We obtained similar results in separate experiments and values are the mean of results from three wells. bars, ±SE. *, p < 0.05 relative to the DMSO control. #, p > 0.05 relative to the DMSO control.
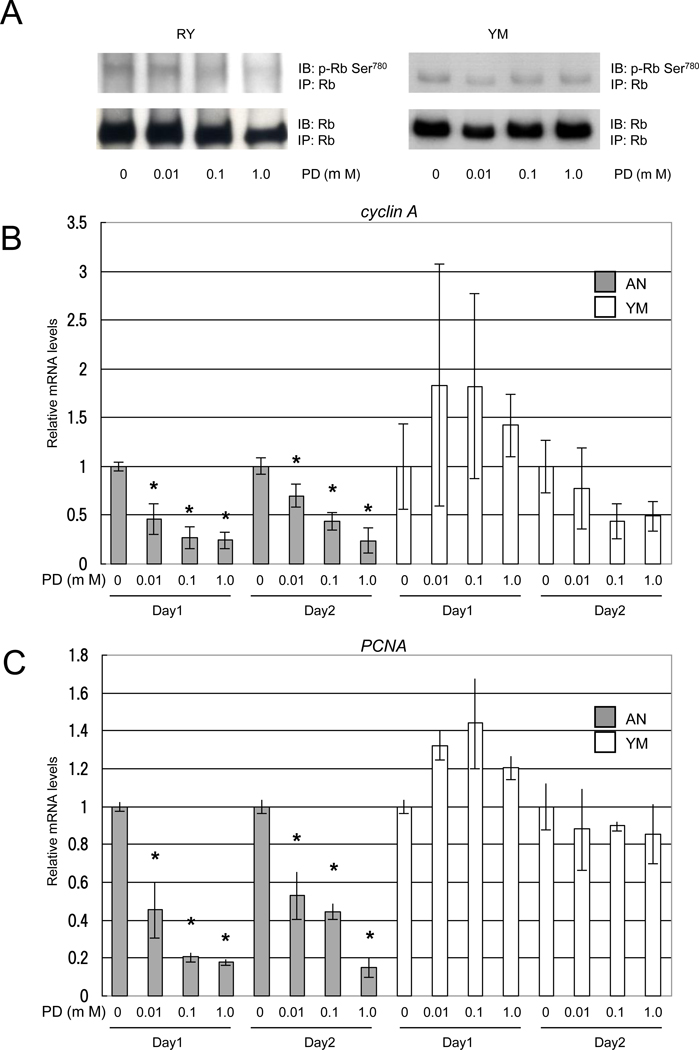
PD dephosphorylates serine 780 residue of Rb and transcriptionally reduces cyclin A and PCNA expressions in PD-sensitive MRT cell lines
PD dephosphorylated serine 780 of Rb (Rbser780) in a concentration-dependent manner in PD-sensitive RY cells but not in PD-resistant YM cells (Fig. 2A). The expressions of cyclin A and PCNA, which are regulated by E2F transcriptional factors, were significantly reduced by PD in a concentration-dependent manner in PD-sensitive AN cells (p < 0.05), but not in PD-resistant YM cells (Fig. 2B and C).
Fig. 2.
PD dephosphorylates Rbser780 and transcriptionally reduces cyclin A and PCNA expressions in a PD-sensitive MRT cell line. A, Cells were harvested 24 h after treatment with PD or vehicle alone, and protein was extracted. The lysates were immunoprecipitated with Rb antibody, separated on a 7.5 % SDS-polyacrylamide gel and probed with Rbser780 or Rb antibody. B and C, Cells were harvested 24 or 48 h after treatment with PD or DMSO, and total RNA was extracted. The mRNA levels were measured for the cyclin A (B) and PCNA (C) genes by real-time PCR and normalized to GAPDH mRNA expression. Similar results were obtained in three separate experiments and values are the mean of results from three wells. The results are shown as relative mRNA levels relative to cells treated with DMSO. bars, ±SE. *, p < 0.05 relative to the DMSO control.
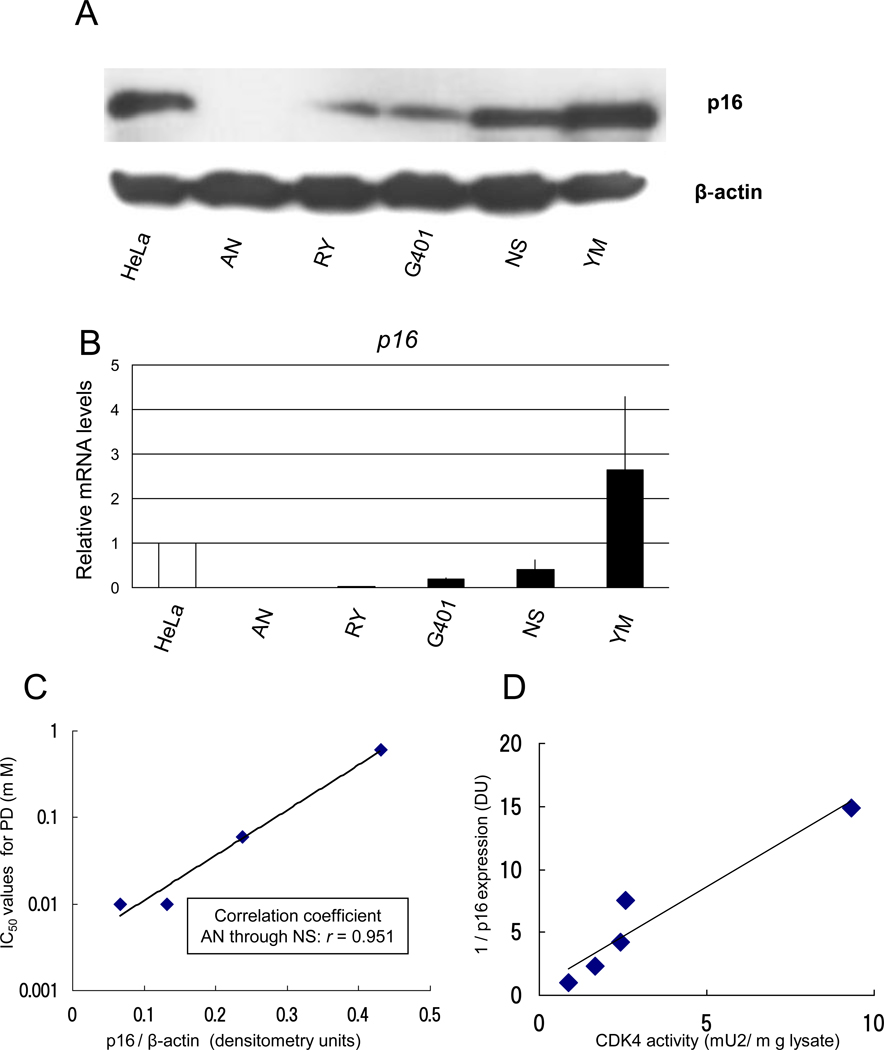
p16 expression of MRT cell lines was inversely correlated with their PD sensitivity and CDK4 activity
Next, we analyzed the relation between IC50 for PD and expression of p16 in the MRT cell lines, because not only PD but also p16 is a strong inhibitor of CDK4. An immunoblot analysis showed that the levels of p16 expression varied in the MRT cell lines (p16/β-actin AN: 0.067, RY: 0.13, G401: 0.24, NS: 0.43, YM: 1.02 densitometry units [DU]; Fig. 3A). Similar results were obtained in three separate experiments. The order of expression of p16 mRNA in the MRT cell lines (AN < RY < G401 < NS < YM; Fig. 3B) was the same as the order of p16 protein expression. The IC50 values were strongly correlated with the levels of p16 expression in the AN, RY, G401 and NS cell lines (correlation coefficient; r = 0.951; Fig. 3C). The YM cell line was excluded from the analysis because its IC50 value could not be estimated. Furthermore, CDK4 activity in the MRT cell lines was inversely correlated with p16 expression (Fig. 3D).
Fig. 3.
p16 expression of the MRT cell lines is inversely correlated with their PD sensitivity and CDK4 activity. A, Cells were harvested at the log-growth phase and the lysates were adjusted to have equal protein concentrations. The lysates were separated on a 10 % SDS-polyacrylamide gel and probed with p16 or β-actin antibody. The HeLa cells were used as a positive control of p16 proteins. We obtained similar results in three separate experiments. B, Cells were seeded in normal growth medium and were harvested at the log-growth phase. Extraction of total RNA, analysis of p16 mRNA, and normalization by GAPDH mRNA were performed as described in Materials and Methods. The HeLa cells were used as a positive control. The results are shown as relative mRNA levels relative to the HeLa cells. Values are the mean of results from three wells. bars, ±SE. Similar results were obtained in three separate experiments. C, The levels of p16 and βactin were calculated by Image J densitometry. The correlation coefficient between p16 /βactin levels on immunoblotting and IC50 values of PD on WST-8 assay was 0.951 in AN through NS. D, CDK4 activity was determined by a modified in vitro kinase activity assay using a C2P system (Sysmex).
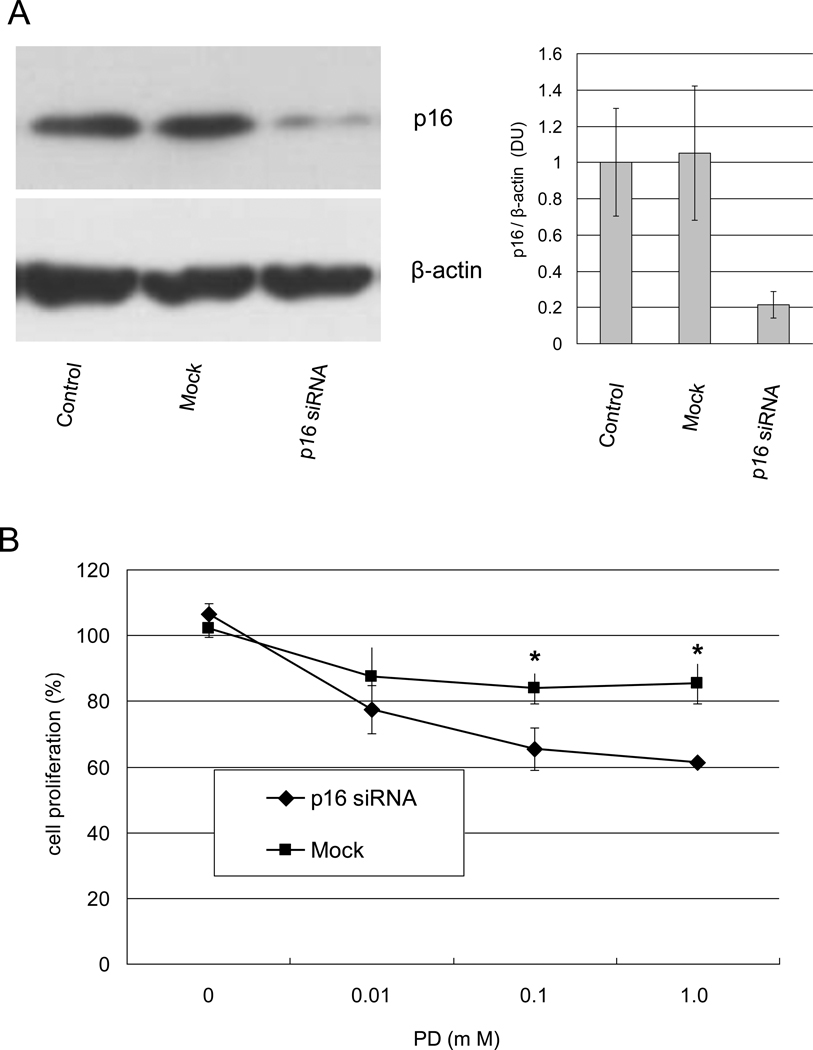
siRNA of p16 increased the sensitivity of MRT cells to PD
We examined whether knockdown of p16 influenced the sensitivity of MRT cells to PD by RNAi. The YM cell line abundantly expressed p16 and was resistant to PD even at a concentration of 10 µM. p16 expression was not reduced in a mock control. In contrast, p16 knockdown reduced p16 protein expression by 79 % in the YM cell line (Fig. 4A). p16 knockdown without PD slightly induced proliferation of the YM cell line although the difference was not significant (Fig. 4B). In addition, YM cells with p16 knockdown were significantly more sensitive to PD than mock transfected cells (p < 0.05), even at concentrations of 0.1 and 1.0 µM.
Fig. 4.
Knockdown of p16 by RNAi increases sensitivity of YM cells to PD. YM cells were seeded in 100 mm cell plates or 96-well plates and allowed to attach for 24 h. The medium was replaced with medium containing Lipofectamine 2000 with or without p16 RNAi. After an additional 6 h, each well was replaced with fresh RPMI1640 medium containing 10% FBS with or without PD in quadruplicate cultures. Immunoblotting analysis was performed after 2 days incubation (A). WST-8 was performed after 4 days incubation Proliferation is expressed as a percent of the proliferation in Mock without DMSO, used as a control. (B). We obtained similar results in separate experiments. Values are the mean of results from three wells. bars, ±SE. *, p < 0.05 relative to the Mock control.
Discussion
We thought that PD could be a promising agent for treating MRT cells. Previous studies have shown that loss of INI1 function is an indicator of MRT. In MRT cells, mutation or deletion of INI1 reduces or abolishes the expression of p16 and induces the expression of cyclin D [1,5]. Both changes result in activated CDK4. This suggested that inhibiting the kinase activity of CDK4 could stop the proliferation of MRT cells. PD inhibits CDK4 by binding to the ATP site [6], and inhibits proliferation of tumor cells that do not harbor mutations of CDK4, cyclin D, and Rb. Mutation of these genes has not previously been reported in MRT cells. Because sequencing of the five MRT cell lines revealed no mutations in the Rb gene (data not shown), we expected that PD was a promising agent for treating MRT cells.
As we expected, PD inhibited the proliferation of most MRT cell lines by inhibiting G1-S cell cycle progression (Fig. 1). PD did not increase the sub-G1 population, indicating that it did not induce apoptosis, even in the PD-sensitive MRT cell lines (data not shown). In addition, PD led to the dephosphorylation of Rbser780, which is a specific target of CDK4, and reduced the expression of cyclin A and PCNA regulated by the E2F transcription factors in the PD-sensitive MRT cell lines, but not in PD-resistant MRT cell line (YM) (Fig. 2). These results are consistent with previous reports on the effect of PD against other cancer cells [6–12]. PD is a highly selective inhibitor of CDK4 (IC50 = 0.01 to 0.6µM), although at more than 10 µM it inhibits a wide variety of tyrosine and serine, threonine kinases [6]. Our results suggest that PD, even at low concentrations that specifically inhibit CDK4 activity, is effective against most MRT cells in vitro.
Why does p16 expression influence the effect of PD against MRT cells? One possibility is that because the proliferation of the MRT cells that express high levels of p16 may be slow, for example the proliferation of YM cells, the growth of the MRT cells was not reduced further by PD. Low expression of p16 induces high CDK4 activity (because p16 is a specific CDK4 inhibitor), which promotes progression from the G1 phase to the S phase. Conversely, high expression of p16 means low CDK4 activity and slow progression to the S phase. CDK4 activity in the MRT cell lines was inversely correlated with p16 expression (Fig. 3D). In fact, the YM cell line, which strongly expresses high levels of p16, had the longest doubling time among our five MRT cell lines (data not shown). We therefore suggest that p16 expression level and CDK4 activity are important for the growth of MRT cells. Another possibility is that, in MRT cells that express high levels of p16, the p16-CDK4-Rb pathway may not be necessary for cell proliferation. Earlier study has shown that p16 expression reduces the BrdU incorporation of PD-treated breast cancer cells and Rb-deficient breast cancer cells are resistant to the growth inhibitory effect of PD [7]. However, in the five MRT cell lines, the sensitivity to PD was independent to Rb expression level (Supplementary Fig. 1). There appears to be intrinsic cell-specific differences in the response to Rb deficiency. Thus, we hypothesize that the proliferation of MRT cells depends on the level of p16 but not Rb and PD inhibits cell proliferation only in MRT cells that express low levels of p16.
Most MRT cells express cyclin D and do not express p16 because of the loss of INI1 function [4,5,20]. However, a small number of MRT cell lines express p16 [21]. Why do some MRT cell lines express p16 even though the INI1 gene is either deleted or mutated in all the MRT cells in this study [17,22]? We can think of three reasons why only the YM cell line overexpressed p16. First, there may be some factor, such as Polycomb group which directly influences p16 expression, overcoming p16 reduction by INI1. One possible change may be a loss of BMI-1, which is a polycomb complex protein that represses transcription of p16 [23,24]. Second, there may be genetic or epigenetic mutations of CDK4, cyclin D and/or Rb, although we did not find any mutation and it is inconsistent with our finding that p16 siRNA increased the sensitivity to PD. Third, there may be another pathway overcoming the CDK4/6-cyclin D-Rb-E2F pathway when PD inhibits activation of CDK4/6, in the YM cells.
PD is currently being used in phase II trials to treat a number of cancers, including multiple myeloma, advanced breast cancer, advanced liposarcoma, recurrent Rb-positive glioblastoma, advanced non-small cell lung cancer, relapsed Mantle cell Lymphoma, and refractory solid tumors.
We expect that evaluation of p16 expression in a MRT will reveal whether PD will be a useful therapeutic agent for children with MRT, although the effects of PD against MRT in vivo must be determined before a clinical trial can be conducted.
In conclusion, we suggest that p16 expression of MRT cells can be used to predict their sensitivity to PD and that if p16 expression of MRT is low, PD may be an effective drug against MRT.
Highlights.
PD 0332991 (PD) could suppress 4 of 5 malignant rhabdoid tumor (MRT) cell lines.
The sensitivity of the MRT cell lines to PD was inversely correlated with p16 expression (r = 0.951).
p16 expression in MRT could be used to predict its sensitivity to PD.
PD may be an attractive agent for patients with MRT whose tumors express low levels of p16.
Supplementary Material
Primers used in real-time PCR. Primers were designed to amplify each mRNA specifically.
Immunoblotting analysis of INI1, p16, CDK4, cyclin D, Rb, and β-actin expressions in the five MRT cell lines. The HeLa cells were used as a positive control of INI1 and p16 proteins. Total cell proteins were adjusted to equal concentrations and were separated on a 7.5 % or 12.5% SDS-polyacrylamide gel and probed with each antibody.
Acknowledgements
We thank Ms. Ryoko Murata for secretarial assistance and Mr. Satoshi Nakayama, Mr. Hideki Ishihara (Sysmex Corp.), and Dr. Toshiyuki Sakai (Department of Molecular-Targeting Cancer Prevention, Kyoto Prefectural University of Medicine) for analyzing CDK4 activity. This study was supported by Grant-in-Aid (17–13) in Cancer Research, from the Ministry of Health, Labour and Welfare of Japan, a Grant-in-Aid from Children's Cancer Association of Japan in 2008, and USPHS grants CA77776 and CA23099 and CA21675 (Cancer Center Support Grant) from the NCI.
Abbreviations
- MRT
Malignant rhabdoid tumor
- INI1
integrase interactor 1
- CDK4
Cyclin-dependent kinase 4
- PD
PD 0332991
- siRNA
small interfering RNA
- PCNA
proliferating cell nuclear antigen
- GAPDH
glucose-6-phosphate dehydrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Versteege I, Sévenet N, Lange J, et al. Truncating mutations of hSNF5/INI1in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson GE, Breslow NE, Dome J, et al. Rhabdoid tumor of the kidney in the National Wilms’ Tumor Study: age at diagnosis as a prognostic factor. J. Clin. Oncol. 2005;23:7641–7645. doi: 10.1200/JCO.2004.00.8110. [DOI] [PubMed] [Google Scholar]
- 3.Zhang ZK, Davies KP, Allen J, et al. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol. Cell. Biol. 2002;22:5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oruetxebarria I, Venturini F, Kekarainen T, et al. p16 is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J. Biol. Chem. 2004;279:3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- 5.Imbalzano AN, Jones SN. Snf5 tumor suppressor couples chromatin remodeling, checkpoint control, and chromosomal stability. Cancer Cell. 2005;7:294–295. doi: 10.1016/j.ccr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Fry DW, Harvey PJ, Keller PR, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 7.Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29:4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 8.Finn RS, Dering J, Conklin D, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saab R, Bills JL, Miceli AP, et al. Pharmacologic inhibition of cyclin-dependent kinase 4/6 activity arrests proliferation in myoblasts and rhabdomyosarcoma-derived cells. Mol. Cancer Ther. 2006;5:1299–1308. doi: 10.1158/1535-7163.MCT-05-0383. [DOI] [PubMed] [Google Scholar]
- 10.Baughn LB, Di Liberto M, Wu K, et al. A novel orally active small molecule potently induces G1 arrest in primary myeloma cells and prevents tumor growth by specific inhibition of cyclin-dependent kinase 4/6. Cancer Res. 2006;66:7661–7667. doi: 10.1158/0008-5472.CAN-06-1098. [DOI] [PubMed] [Google Scholar]
- 11.Marzec M, Kasprzycka M, Lai R, et al. Mantle cell lymphoma cells express predominantly cyclin D1a isoform and are highly sensitive to selective inhibition of CDK4 kinase activity. Blood. 2006;108:1744–1750. doi: 10.1182/blood-2006-04-016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michaud K, Solomon DA, Oermann E, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70:3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugimoto T, Hosoi H, Horii Y, et al. Malignant rhabdoid-tumor cell line showing neural and smooth-muscle-cell phenotypes. Int. J. Cancer. 1999;82:678–686. doi: 10.1002/(sici)1097-0215(19990827)82:5<678::aid-ijc10>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Wang J, Blaser BW, et al. Pharmacologic inhibition of CDK4/6: mechanistic evidence for selective activity or acquired resistance in acute myeloid leukemia. Blood. 2007;110:2075–2083. doi: 10.1182/blood-2007-02-071266. [DOI] [PubMed] [Google Scholar]
- 15.Tamura S, Hosoi H, Kuwahara Y, et al. Induction of apoptosis by an inhibitor of EGFR in neuroblastoma cells. Biochem. Biophys. Res. Commun. 2007;358:226–232. doi: 10.1016/j.bbrc.2007.04.124. [DOI] [PubMed] [Google Scholar]
- 16.Tsuchiya K, Hosoi H, Misawa-Furihata A, Houghton PJ, Sugimoto T. Insulin-like growth factor-I has different effects on myogenin induction and cell cycle progression in human alveolar and embryonal rhabdomyosarcoma cells. Int. J. Oncol. 2007;31:41–47. [PubMed] [Google Scholar]
- 17.Kuwahara Y, Hosoi H, Osone S, et al. Antitumor activity of gefitinib in malignant rhabdoid tumor cells in vitro and in vivo. Clin. Cancer Res. 2004;10:5940–5948. doi: 10.1158/1078-0432.CCR-04-0192. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi K, Tsuchiya K, Otabe O, et al. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem. Biophys. Res. Commun. 2008;365:568–574. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 19.van Nes JG, Smit VT, Putter H, et al. Validation study of the prognostic value of cyclin-dependent kinase (CDK)-based risk in Caucasian breast cancer patients. Br. J. Cancer. 2009;100:494–500. doi: 10.1038/sj.bjc.6604870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsikitis M, Zhang Z, Edelman W, Zagzag D, Kalpana GV. Genetic ablation of Cyclin D1 abrogates genesis of rhabdoid tumors resulting from Ini1 loss. Proc. Natl. Acad. Sci. U S A. 2005;102:12129–12134. doi: 10.1073/pnas.0505300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuwahara Y, Charboneau A, Knudsen ES, Weissman BE. Reexpression of hSNF5 in malignant rhabdoid tumor cell lines causes cell cycle arrest through a p21(CIP1/WAF1)-dependent mechanism. Cancer Res. 2010;70:1854–1865. doi: 10.1158/0008-5472.CAN-09-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katsumi Y, Kuwahara Y, Tamura S, et al. Trastuzumab activates allogeneic or autologous antibody-dependent cellular cytotoxicity against malignant rhabdoid tumor cells and interleukin-2 augments the cytotoxicity. Clin. Cancer Res. 2008;14:1192–1199. doi: 10.1158/1078-0432.CCR-07-1661. [DOI] [PubMed] [Google Scholar]
- 23.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol. Cell. Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in real-time PCR. Primers were designed to amplify each mRNA specifically.
Immunoblotting analysis of INI1, p16, CDK4, cyclin D, Rb, and β-actin expressions in the five MRT cell lines. The HeLa cells were used as a positive control of INI1 and p16 proteins. Total cell proteins were adjusted to equal concentrations and were separated on a 7.5 % or 12.5% SDS-polyacrylamide gel and probed with each antibody.