Abstract
Neural tissue is arisen from presumptive ectoderm via inhibition of bone morphogenetic protein (BMP) signaling during Xenopus early development. Previous studies demonstrate that ectopic expression of dominant negative BMP4 receptor (DNBR) produces neural tissue in animal cap explants (AC) and also increases the expression level of various genes involved in neurogenesis. To investigate detail mechanism of neurogenesis in transcriptional level, we analyzed RNAs increased by DNBR using total RNA sequencing analysis and identified several candidate genes. Among them, xCITED2 (Xenopus CBP/p300-interacting transcription activator) was induced 4.6 fold by DNBR and preferentially expressed in neural tissues at tadpole stage. Ectopic expression of xCITED2 induced anterior neural genes without mesoderm induction and reduced BMP downstream genes, an eye specific marker and posterior neural marker. Taken together, these results suggest that xCITED2 may have a role in the differentiation of anterior neural tissue during Xenopus early development.
Keywords: xCITED2, DNBR, neurogenesis, Xenopus
INTRODUCTION
Neural induction is a process of neural cell formation in ectoderm during embryogenesis (Kessler and Melton, 1994; Gould and Grainger, 1997; Hemmati-Brivanlou and Melton, 1997; Sasai and De Robertis, 1997; Munoz-Sanjuan and Brivanlou, 2002; De Robertis and Kuroda, 2004). In Xenopus embryos, Spemann organizer is placed in dorsal mesoderm and generates BMP antagonizers such as chordin, noggin and follistatin (Smith and Slack, 1983; Hemmati-Brivanlou and Thomsen, 1995; Harland and Gerhart, 1997; Zoltewicz and Gerhart, 1997; Faure et al., 2000). These molecules induce neural cells via blocking of bone morphogenetic protein (BMP) signaling (Hemmati-Brivanlou and Thomsen, 1995; Sasal et al., 1995; Wilson and Hemmati-Brivanlou, 1995; Munoz-Sanjuan et al., 2002). Previous studies show that activin treatment or microinjection of dominant negative BMP receptor (DNBR) induces neural cells in animal cap explants (AC) (Suzuki et al., 1994; Hawley et al., 1995; Xu et al., 1995). Moreover, various genes have been demonstrated to be involved in neural development (Karsten et al., 2008). Representatively, Zic3 (one of zic finger proteins, contributes initiating of neurogenesis in early stage), NCAM (neural cell adhesion molecule, pan-neural marker), NeuroD, Otx2 (Orthodenticlehomeobox 2, anterior neural marker), HoxB9 (Homeobox protein Hox-B9, posterior neural marker) and RX1 (eye specific marker) have been used for neural markers (Jacobson and Rutishauser, 1986; Sunshine et al., 1987; Lee et al., 1995; Mizuseki et al., 1998; Nakata et al., 1998; Manzanares et al., 2002; Lunardi and Vignali, 2006; Zaghloul and Moody, 2007).
The CITED protein (CBP/p300-interacting transcription activator) family has 4 subtypes, CITED1 called as MSG1, CITED2 (MGR1), CITED3 and CITED4 (MRG2) (Andrews et al., 2000). All family proteins have CR2 domain which is a highly conserved transcription activating domain (Shioda et al., 1997). Since CITED protein does not have DNA-binding motif, it has been studied as a transcriptional co-activator of CBP (Yahata et al., 2000). CITED1 increases transcriptional activity through interacting with CBP and SMAD4 but CITED2 does not have SMAD4 binding motif. CITED2 enhances transcription with other proteins such as Lhx2 which contains LIM domain (Glenn and Maurer, 1999). Previous studies addressed that CITED family proteins play a role in heart, liver development and anterior-posterior patterning (Goodman and Smolik, 2000). Although CITED2 protein has been studied minutely in mammalian cell, the role of CITED2 is not fully understood during Xenopus early development (Fujii et al., 1998; Schlange et al., 2000).
In this study, we found that Xenopus homologue of CITED2 (xCITED2) was induced by DNBR and preferentially expressed in neural tissues. Over-expression of xCITED2 increased neural genes such as Zic3, NeuroD, neurogenin-1, NCAM and Otx2 in AC, but decreased BMP downstream genes and a posterior neural marker, HoxB9. Taken together, the results suggest that xCITED2 functions in anterior neural induction during Xenopus early development.
MATERIALS AND METHODS
Embryo injection and explant culture
Xenopus laevis embryos were obtained by artificial fertilization (Sive et al., 2010). Developmental stages were designated according to Nieuwkoop and Faber (Nieuwkoop, 1969). Embryos at the one cell stage or two-cell stage were injected in the animal pole with mRNA as descried in the figure legends. Animal caps were dissected from the injected embryos at stage 8~9 and cultured to various stages in 67% Leibovitzs L-15 medium (GIBCO/BRL) with BSA (1 mg/ml), 7 mM Tris-HCl (pH 7.5) and gentamicin (50 µg/ml).
Cloning of xCITED2
The xCITED2 ORF sequence is appeared in NCBI GenBank under the accession number NM_001094820. Open reading frame (ORF) was amplified by PCR using cDNA library of stage 12 Xenopus embryos (Primer forward: 5'-GCGAATTCAATGGCAGACCACATGATGGC-3' reverse: 5'CGTCTAGAACACACCTAACAGCTTACTCTG-3'). The full length of xCITED2 ORF was cloned into EcoRI/XbaI-digested pCS2 vector. For epitope tagging, xCITED2 ORF were cloned into pCS2-HA vector (pCS2-HA-xCITED2).
In vitro transcription
All synthetic mRNAs used for microinjection were produced by in vitro transcription. The xCITED2 cDNA was inserted in the pCS2 vector. The cDNA were linearized and used for in vitro synthesis of capped mRNA using in vitro transcription kit (Ambion) in accordance with the manufacturer's instructions. The synthetic RNA was quantified by ethidium bromide staining in comparison with a standard RNA.
RNA isolation and reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from whole embryo or cultured animal explants with TRIzol reagent (Life Technologies, Inc.) following the manufacturer's instruction. RT-PCR was performed with a Superscript pre-amplification system (Invitrogen). PCR was performed as follows: first, a denaturation step of 94℃ for 5 minutes; second, 94℃ for 1 minute; third, each annealing temperature, for 1 minute ; fourth, 72℃ for 1 minute; fifth, repeat second, third and fourth steps 19-30 cycles of amplification was performed as described at the Xenopus Molecular Marker Resource (XMMR; University of Texas). Primer set for xCITED2 was following forward: CTCATCATCATCAGCACACC, reverse: CGATCACCAAGGACATAAGG. ODC was used as control to normalize the amount of cDNA used.
Western blotting
Embryos were injected at the one cell stage with RNA constructs as described, and frozen at stage 11. They were then homogenized in lysis buffer (50 mM Tris [pH 7.4]), 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 0.1% SDS, 50 mM NaF and 1 mM Na3VO4) containing of 1 mM PSMF, 15 mM glycerophosphate, 1 X proteinase inhibitor cocktail (Calbiochem). Cell lysates were cleared by centrifugation. Proper amount of lysate was boiled in sample buffer, and resolved by electrophoresis in 10% SDS-polyacrylamide gels. HA-tagged xCITED2 proteins were visualized after western blotting using rabbit polyclonal anti HA (Santa Cruz, sc-Y11) using ECL Western blotting detection reagents (Amersham Pharmacia Biotech).
RESULTS
xCITED2 is identified using total RNA sequencing analysis in DNBR-treated animal cap explants
We screened for the expressed RNA in DNBR-injected AC using total RNA sequencing (Jones and Woodland, 1987; Karsten et al., 2008). We analyzed about 22,000 contigs of the expressed sequences (bigger than 300 nucleotides) and found several uncharacterized genes induced by DNBR during early Xenopus development. We interested in the expressed sequence [accession number NM_001094820] which encodes a protein that shared identity of 69% with hCITED2; its homologues were also identified in mouse and chick. In total RNA sequencing data, the expression level of xCITED2 was increased 4.6-fold by DNBR at stage 11, when compared with the data obtained from the untreated control AC.
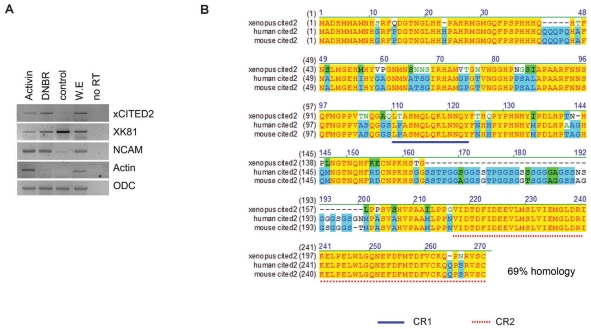
In order to confirm the induction of xCITED2 in DNBR-treated sample, RT-PCR analysis was performed with AC obtained from embryos in various conditions. As shown Fig. 1A, the expression level of xCITED2 was increased in the samples treated with either DNBR or Activin at stage 24, but not in the untreated control AC. The control AC differentiated to epidermis (as shown by enhanced expression of XK81 in Fig. 1A). As we have known, either activin- or DNBR-treated samples differentiated to neural tissues (as shown by NCAM expression in Fig. 1A). We cloned and sequenced the full-length cDNA of xCITED2 into pCS2 vector using PCR amplification. xCITED2 encodes 225 amino acids and contains highly conserved CR-1 and CR-2 domain as indicated in Fig. 1B. CR-2 is a Glu/Asp-rich carboxy-terminal domain consisted of 55 amino acids and it has been known to acts as a transcription activating domain.
Fig. 1.
Blocking of BMP4 signaling induced the expression of xCITED2 in animal cap explants of Xenopus embryos. (A) Animal caps were dissected from stage 8~9 embryos that had been injected at the one cell stage or two-cell stage with 2 ng of DNBR RNAs or animal cap explants dissected from un-injected embryos were treated with activin (50 ng/ml). Animal caps were harvested at stage 24 and RT-PCR analysis was performed. Pan-neural marker: NCAM, mesoderm marker: Actin, epidermis marker: XK81. ODC serves as mRNA loading control. (B) Alignment of xenopus xCITED2 with human and mouse homologues of CITED2. Yellow indicated that Amino-acids which are conserved between xenopus CITED2 (gene bank number: NP_001088289.1) and other members (human: NP_006070.2, mouse: NP_034958.2). All family members contain transcription activating domain (CR-2 domain) in C-terminal region. This diagram was generated by the VectorNTI 8.0.
xCITED2 is preferentially expressed in neural tissues
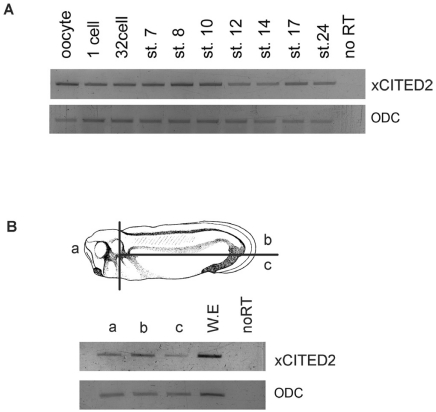
To investigate temporal expression of xCITED2 during Xenopus early development, we performed RT-PCR analysis with embryos of several developmental stages (Fig. 2A). Transcripts of xCITED2 were present from oocyte until tadpole stage. We then determined spatial expression of xCITED2 at stage 28. Each part was dissected as shown in Fig. 2B and immediately performed RT-PCR analysis (Fig. 2B). xCITED2 was highly expressed in head and dorsal regions.
Fig. 2.
Temporal and spatial expression pattern of xCITED2 in Xenopus development. (A)Temporal Expression pattern of xCITED2 was analyzed using RT-PCR at various stages as indicated. ODC serves as loading control. xCITED2 was expressed from Oocyte and maintained until tail-bud stages. (B) Spatial expression pattern of xCITE2 was also analyzed by RT-PCR with dissected parts as indicated from stage 28 embryos.
Together, DNBR induced expression of xCITED2 in AC and its spatial expression pattern in whole embryos suggest that xCITED2 has a role in neural development of Xenopus embryos.
Ectopic expression of xCITED2 induces neural genes in animal cap explants
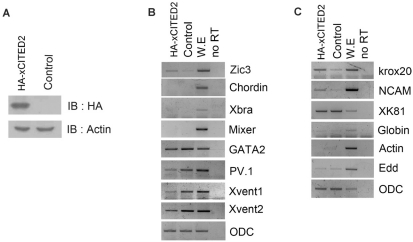
To perform gain of functional studies, we constructed HA-tagged xCITED2 because specific antibody for xCITED2 was commercially not available. Expression level of HA-xCITED2 was checked by western blotting using anti-HA probe (Fig. 3A). Over-expression of HA-xCITED2 induced a neural specific gene, Zic3 without organizer (chordin), mesoderm (Xbra) and endoderm (Mixer) gene induction in AC, but expression levels of BMP downstream genes (PV.1, Xvent1 and Xvent2) were reduced at stage 12. Consistently, neural specific genes (NCAM and Krox20) were expressed without either dorsal (Actin) or ventral (Globin) mesoderm, and endoderm (Edd) formation by xCITED2 in AC at stage 24. The results suggest that xCITED2 play a role in neurogenesis without inducing dorsal mesoderm.
Fig. 3.
xCITED2 induced neurogenesis in Xenopus Animal cap. (A) Expression level of HA-xCITED2 was confirmed by western-blot analysis. 1 ng of HA-xCITED2 RNAs were injected at 1 cell stage embryo and harvested at stage 12. Immuno-bloting was performed using anti-HA probe. (B) 1 ng of xCITED2 RNAs were injected at 1 cell stage and dissected animal cap at stage 8 and incubated in animal cap media until stage 12 or stage 24 (C). RT-PCR was performed to analyze various genes expression. ODC was used as loading control, ventral marker: Xvent1/2, GATA2, PV.1, endoderm marker: mixer and Edd, mesoderm marker: Xbra, Actin, organizer marker: chordin and neural marker: Zic3, NCAM, Krox20.
xCITED2 induces anterior neural tissue
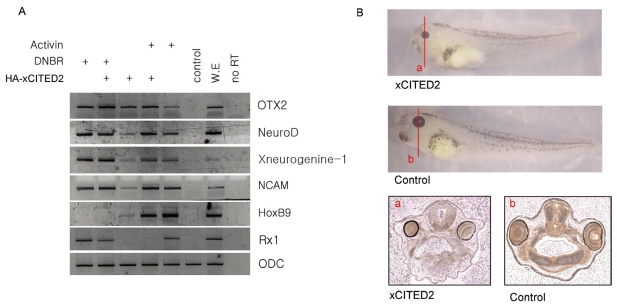
We found that xCITED2 was induced by DNBR and over-expression of xCITED2 induced neural genes in AC. Because DNBR-induced neural tissue is anterior, we examined whether xCITED2-induced neural tissue was also anterior. As shown in Fig. 4, xCITED2 induced an anterior specific neural marker; OTX2. OTX2 was similarly expressed in either DNBR or activin-treated AC. Overexpression xCITED2 further increased OTX2 transcripts in either DNBR or activin-treated AC. At the same conditions, xCITED2 reduced a posterior specific neural marker; HoxB9 which was induced by activin treatment in AC. In addition, other neural markers NCAM, NeuroD and Xneurogenin-1 were also induced, but an eye specific marker; Rx1 was dramatically reduced by co-injection of xCITED2.
Fig. 4.
xCITED2 induced anterior neural gene. (A) 1 ng of xCITED2 RNAs were injected alone or co-injected with 1 ng of DNBR RNAs at 1 cell stage and dissected animal cap at stage 8 and incubated in animal cap media containing activin (50 ng/ml) or none as indicated until stage 24. RT-PCR was performed to analyze various genes expression. Pan-Neural marker: NCAM, NeuroD and Xneurogenine-1, anterior neural marker: OTX2, posterior neural marker: HoxB9, eye specific marker: Rx1. ODC was used as loading control. (B) The lines represent the angle of sectioning for a and b. Transverse section through xCITED2 injected embryo or control embryo.
The results suggest that xCITED2 induces anterior neural tissue in AC of Xenopus embryos.
DISCUSSION
Previous studies have demonstrated that inhibition of BMP4 signaling leads to induce neurogenesis (Hawley et al., 1995). To understand the mechanism of neurogenesis, we investigated DNBR-induced genes using total RNA sequencing analysis. We analyzed total 21,891 expressed sequences and found 506 transcripts were up-regulated more than 3-fold. Up-regulated transcripts included a number of well-known genes involved in neurogenesis such as Zic, OTX and Sox family genes. In addition, we found several uncharacterized genes in neurogenesis processes (data not shown).
In this works, we examined the xCITED2 which was one of uncharacterized genes. xCITED2 has been well known to interact with CBP/p300 and act as a co-transcriptional activator. Although physiological function of xCITED2 has been fully understood in mammalian cell, the role of xCITED2 in early development has been elusive. During chick development, CITED2 is expressed in an anterior region of primitive streak, presomitic and lateral plate mesoderm, in the head-fold (future forebrain) and head mesoderm (Schlange et al., 2000). This spatial expression pattern suggests that CITED2 may functions in neurogenesis. In this work, we showed that xCITED2 was expressed in head and dorsal region and overexpression of xCITED2 resulted in anterior neural induction in AC. In addition, we constructed xCITED2-deltaCR2 construct which does not contain the CR-2 domain. Over-expression of xCITED2-deltaCR2 did not induce any neural genes (data not shown). This suggests that xCITED2 functions to induce anterior neural genes as a transcriptional co-activator.
Interestingly, CBP/p300 which is major binding partner of xCITED2 is expressed in anterior neural region in dorsal side during Xenopus early development (Fujii et al., 1998). Although Yoichi Kato et al. demonstrated that functional inhibition of CBP/p300 induces neutralization, co-expression of xCITED2 and CBP/p300 together has not been examined and may function in neurogenesis because physiological function of CBP/p300 depends on its binding partner (Kato et al., 1999). Further studies involving relationship between xCITED2 and CBP/p300 will help to understand the neurogenesis of vertebrate embryos.
ACKNOWLEDGEMENTS
We thank Dr. Makoto Asashima (Dept. of Life Science, University of Tokyo) for activin protein. This research was supported by Basic Science Research Program (KRF-2005-C00115, KRF-2005-015-C00390 and KRF-2009-0077052) and by Priority Research Centers Program (2010-0029642) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology.
References
- 1.Andrews JE, O'Neill MJ, Binder M, Shioda T, Sinclair AH. Isolation and expression of a novel member of the CITED family. Mech Dev. 2000;95:305–308. doi: 10.1016/s0925-4773(00)00362-2. [DOI] [PubMed] [Google Scholar]
- 2.De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faure S, Lee MA, Keller T, ten Dijke P, Whitman M. Endogenous patterns of TGFbeta superfamily signaling during early Xenopus development. Development. 2000;127:2917–2931. doi: 10.1242/dev.127.13.2917. [DOI] [PubMed] [Google Scholar]
- 4.Fujii G, Tsuchiya R, Itoh Y, Tashiro K, Hirohashi S. Molecular cloning and expression of Xenopus p300/CBP. Biochim Biophys Acta. 1998;1443:41–54. doi: 10.1016/s0167-4781(98)00179-1. [DOI] [PubMed] [Google Scholar]
- 5.Glenn DJ, Maurer RA. MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone alpha-subunit gene expression. J Biol Chem. 1999;274:36159–36167. doi: 10.1074/jbc.274.51.36159. [DOI] [PubMed] [Google Scholar]
- 6.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 7.Gould SE, Grainger RM. Neural induction and antero-posterior patterning in the amphibian embryo: past, present and future. Cell Mol Life Sci. 1997;53:319–338. doi: 10.1007/PL00000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harland R, Gerhart J. Formation and function of Spemann's organizer. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- 9.Hawley SH, Wünnenberg-Stapleton K, Hashimoto C, Laurent MN, Watabe T, Blumberg BW, Cho KW. Disruption of BMP signals in embryonic Xenopus ectoderm leads to direct neural induction. Genes Dev. 1995;9:2923–2935. doi: 10.1101/gad.9.23.2923. [DOI] [PubMed] [Google Scholar]
- 10.Hemmati-Brivanlou A, Melton D. Vertebrate neural induction. Annu Rev Neurosci. 1997;20:43–60. doi: 10.1146/annurev.neuro.20.1.43. [DOI] [PubMed] [Google Scholar]
- 11.Hemmati-Brivanlou A, Thomsen GH. Ventral mesodermal patterning in Xenopus embryos: expression patterns and activities of BMP-2 and BMP-4. Dev Genet. 1995;17:78–89. doi: 10.1002/dvg.1020170109. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson M, Rutishauser U. Induction of neural cell adhesion molecule (NCAM) in Xenopus embryos. Dev Biol. 1986;116:524–531. doi: 10.1016/0012-1606(86)90153-3. [DOI] [PubMed] [Google Scholar]
- 13.Jones EA, Woodland HR. The development of animal cap cells in Xenopus: a measure of the start of animal cap competence to form mesoderm. Development. 1987;101:557–563. [Google Scholar]
- 14.Karsten SL, Kudo LC, Geschwind DH. Gene expression analysis of neural cells and tissues using DNA microarrays. Curr Protoc Neurosci. 2008;Chapter 4:Unit 4.28. doi: 10.1002/0471142301.ns0428s45. [DOI] [PubMed] [Google Scholar]
- 15.Kato Y, Shi Y, He X. Neuralization of the Xenopus embryo by inhibition of p300/CREB-binding protein function. J Neurosci. 1999;19:9364–9373. doi: 10.1523/JNEUROSCI.19-21-09364.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessler DS, Melton DA. Vertebrate embryonic induction: mesodermal and neural patterning. Science. 1994;266:596–604. doi: 10.1126/science.7939714. [DOI] [PubMed] [Google Scholar]
- 17.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 18.Lunardi A, Vignali R. Xenopus Xotx2 and Drosophila otd share similar activities in anterior patterning of the frog embryo. Dev Genes Evol. 2006;216:511–521. doi: 10.1007/s00427-006-0064-9. [DOI] [PubMed] [Google Scholar]
- 19.Manzanares M, Nardelli J, Gilardi-Hebenstreit P, Marshall H, Giudicelli F, Martínez-Pastor MT, Krumlauf R, Charnay P. Krox20 and kreisler co-operate in the transcriptional control of segmental expression of Hoxb3 in the developing hindbrain. EMBO J. 2002;21:365–376. doi: 10.1093/emboj/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- 21.Muñoz-Sanjuán I, Bell E, Altmann CR, Vonica A, Brivanlou AH. Gene profiling during neural induction in Xenopus laevis: regulation of BMP signaling by post-transcriptional mechanisms and TAB3, a novel TAK1-binding protein. Development. 2002;129:5529–5540. doi: 10.1242/dev.00097. [DOI] [PubMed] [Google Scholar]
- 22.Muñoz-Sanjuán I, Brivanlou AH. Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci. 2002;3:271–280. doi: 10.1038/nrn786. [DOI] [PubMed] [Google Scholar]
- 23.Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic family and its role in neural and neural crest development. Mech Dev. 1998;75:43–51. doi: 10.1016/s0925-4773(98)00073-2. [DOI] [PubMed] [Google Scholar]
- 24.Nieuwkoop PD. The formation of the mesoderm in urodelean amphibians. I. Induction by the endoderm. Wilhelm Roux Arch Entwickl Mech Org. 1969;162:341–373. doi: 10.1007/BF00578701. [DOI] [PubMed] [Google Scholar]
- 25.Sasai Y, De Robertis EM. Ectodermal patterning in vertebrate embryos. Dev Biol. 1997;182:5–20. doi: 10.1006/dbio.1996.8445. [DOI] [PubMed] [Google Scholar]
- 26.Sasal Y, Lu B, Steinbelsser H, De Robertis EM. Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus. Nature. 1995;378:419. doi: 10.1038/378419d0. [DOI] [PubMed] [Google Scholar]
- 27.Schlange T, Andrée B, Arnold H, Brand T. Expression analysis of the chicken homologue of CITED2 during early stages of embryonic development. Mech Dev. 2000;98:157–160. doi: 10.1016/s0925-4773(00)00454-8. [DOI] [PubMed] [Google Scholar]
- 28.Shioda T, Fenner MH, Isselbacher KJ. MSG1 and its related protein MRG1 share a transcription activating domain. Gene. 1997;204:235–241. doi: 10.1016/s0378-1119(97)00551-9. [DOI] [PubMed] [Google Scholar]
- 29.Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2010. [Google Scholar]
- 30.Smith JC, Slack JM. Dorsalization and neural induction: properties of the organizer in Xenopus laevis. J Embryol Exp Morphol. 1983;78:299–317. [PubMed] [Google Scholar]
- 31.Sunshine J, Balak K, Rutishauser U, Jacobson M. Changes in neural cell adhesion molecule (NCAM) structure during vertebrate neural development. Proc Natl Acad Sci U S A. 1987;84:5986–5990. doi: 10.1073/pnas.84.16.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki A, Thies RS, Yamaji N, Song JJ, Wozney JM, Murakami K, Ueno N. A truncated bone morphogenetic protein receptor affects dorsal-ventral patterning in the early Xenopus embryo. Proc Natl Acad Sci U S A. 1994;91:10255–10259. doi: 10.1073/pnas.91.22.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson PA, Hemmati-Brivanlou A. Induction of epidermis and inhibition of neural fate by Bmp-4. Nature. 1995;376:331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]
- 34.Xu RH, Kim J, Taira M, Zhan S, Sredni D, Kung HF. A dominant negative bone morphogenetic protein 4 receptor causes neuralization in Xenopus ectoderm. Biochem Biophys Res Commun. 1995;212:212–219. doi: 10.1006/bbrc.1995.1958. [DOI] [PubMed] [Google Scholar]
- 35.Yahata T, de Caestecker MP, Lechleider RJ, Andriole S, Roberts AB, Isselbacher KJ, Shioda T. The MSG1 non-DNA-binding transactivator binds to the p300/CBP coactivators, enhancing their functional link to the Smad transcription factors. J Biol Chem. 2000;275:8825–8834. doi: 10.1074/jbc.275.12.8825. [DOI] [PubMed] [Google Scholar]
- 36.Zaghloul NA, Moody SA. Alterations of rx1 and pax6 expression levels at neural plate stages differentially affect the production of retinal cell types and maintenance of retinal stem cell qualities. Dev Biol. 2007;306:222–240. doi: 10.1016/j.ydbio.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 37.Zoltewicz JS, Gerhart JC. The Spemann organizer of Xenopus is patterned along its anteroposterior axis at the earliest gastrula stage. Dev Biol. 1997;192:482–491. doi: 10.1006/dbio.1997.8774. [DOI] [PubMed] [Google Scholar]