Abstract
Neurite outgrowth and its maintenance are essential aspects of neuronal cells for their connectivity and communication with other neurons. Recent studies showed that over-expression of either leucine-rich repeat kinase 2 (LRRK2), whose mutations are associated with familial Parkinson's disease (PD), or Rab5b, an early endosomal marker protein, induces reduction in neurite length. Based on our recent findings that LRRK2 co-localizes and interacts with Rab5, we tested the hypothesis that LRRK2 and Rab5 may functionally interplay while controlling neurite outgrowth. Firstly, we confirmed previous reports that over-expression of either the LRRK2 PD-specific G2019S mutant or the Rab5 constitutively active Q79L mutant, but not of dominant negative N133I mutant, significantly reduces neurite outgrowth. Secondly, when over-expression of either LRRK2 wild type (WT) or G2019S was accompanied with over-expression of one of the Rab5 variants (WT, Q79L and N133I), or with down-regulation of Rab5, the reduction extent of its neurite length was similar to that of cells over-expressing LRRK2 alone, regardless of Rab5's status. Finally, we observed similar patterns of neurite length regulation in embryonic rat hippocampal neuron cultures. Taken together, our results suggest that LRRK2 and Rab5 functionally coordinate their regulation of neurite outgrowth and that LRRK2 is a more critical factor than Rab5.
Keywords: neurite outgrowth, LRRK2, Parkinson's disease, Rab5, PC12 cells
INTRODUCTION
Leucine-rich repeat kinase 2 (LRRK2) is a large protein of 285 kDa that is composed of functional GTPase and kinase domains as well as several protein-protein interaction domains such as WD40 and leucine-rich repeat (LRR) (Paisan-Ruiz et al., 2004; Zimprich et al., 2004; Gloeckner et al., 2006; West et al., 2007). Missense mutations of LRRK2 have been reported to cause Parkinson's disease (PD), the second most common neurodegenerative disease, in an autosomal dominant manner (Paisan-Ruiz et al., 2004; Zimprich et al., 2004). Among these mutations, G2019S is the most prevalent mutation that exhibits kinase activity stronger than that of the wild type (West et al., 2005; Jaleel et al., 2007; Luzon-Toro et al., 2007; West et al., 2007; Gandhi et al., 2009) and R1441C may disrupt LRRK2's homodimerization (Deng et al., 2008; Gandhi et al., 2008; Klein et al., 2009). Since G2019S has been found in both familial and sporadic cases of PD (Gilks et al., 2005; Lesage et al., 2007), elucidation of the normal and pathogenic functions of LRRK2 will be critical for our understanding of the etiology of PD.
LRRK2 has been proposed to cause increase of protein aggregation and neuronal toxicity (Smith et al., 2005; Greggio et al., 2006; Smith et al., 2006; West et al., 2007; Heo et al., 2010), shortening of neurite length in primary neuron cultures (MacLeod et al., 2006; Plowey et al., 2008) and regulation of endocytosis rate (Shin et al., 2008). In addition, LRRK2 has been reported to interact with multiple cellular proteins including Rab5b (Shin et al., 2008), parkin (Smith et al., 2005), α/β-tubulin heterodimers (Gandhi et al., 2008), FADD [Fas-associated protein with death domain, (Ho et al., 2009)], heat shock protein 90 [Hsp90, (Wang et al., 2008)], the dishevelled family members [DVL1-3, (Sancho et al., 2009)] and others (Dachsel et al., 2007). Among these, Rab5b is an isoform of Rab5, which is a member of the Rab family, a small GTPase family. Rab5 is present in early endosomes and regulates endocytosis by controlling endosome fusion and motility (Gorvel et al., 1991; Stenmark et al., 1994). In addition, Rab5 is known to be localized in presynaptic vesicles (de Hoop et al., 1994; Shin et al., 2008) and affects endocytosis rates (Wucherpfennig et al., 2003; Shin et al., 2008). Rab5 has been reported to negatively regulate neurite outgrowth in rat pheochromocytoma PC12 cells treated with nerve growth factor [NGF, (Liu et al., 2007)]. Over-expression of Rab5 wild type (WT) or of the constitutively active GTPase mutant, Q79L, inhibited neurite outgrowth whereas over-expression of the dominant negative GTPase mutant, N133I, enhanced neurite development (Liu et al., 2007). Recently, Rab5 has been reported to regulate caspase-8-mediated cell motility (Torres et al., 2010) and to spatially control branching within dendritic arbors (Satoh et al., 2008).
Based on our previously finding that LRRK2 interacts with Rab5b and that this interaction regulates endocytosis of synaptic vesicles (Shin et al., 2008), we investigated here whether this interaction also affected each partner's ability to regulate neurite outgrowth using PC12 cells treated with NGF as well as primary hippocampal neuronal cultures. Our results suggest that LRRK2 is a more critical factor than Rab5 in regulating neurite outgrowth although both proteins functionally coordinate regulation of neurite outgrowth.
MATERIALS AND METHODS
Plasmids and other reagents
cDNAs of wild type and mutants of LRRK2 and Rab5b were cloned into modified pcDNA3.1 plasmids (Invitrogen, Carlsbad, CA) with Myc and Flag tags, respectively (Shin et al., 2008). The shLRRK2 plasmids containing LRRK2 specific sequences (5' CTGATCCAGTTAAAGAATATGGTTGTGCC 3') were purchased ORIGENE (Rockville, Maryland). The sequences of siRNAs for Rab5b are 5' AGGCAUAUGCAGAUGACAA 3' (Rab5 siRNA-1) and 5' GCACGAAAGCUAAGACAUA 3' (Rab5 siRNA-2). Since both Rab5 siRNAs showed similar neurite lengths, we used only siRab5-1 (data not shown). Construction of plasmids containing each gene for LRRK2 G2019S, R1441C and Rab5 Q79L, N133I were previously reported (Shin et al., 2008).
Cell culture, transient transfection and western analysis
PC12 or rat primary hippocampal neuron cells were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, 5% horse serum and 1% antibiotics or Neurobasal medium (Invitrogen, cat no. 21103-049), respectively. To induce neurite outgrowth, PC12 cells were seeded at a density of 40% on the cover glass and treated with nerve growth factor (NGF, Invitrogen,) at 50 ng/ml for 7 days with replenishment of NGF every two days. For primary neuronal culture, rat E17 primary dissociated hippocampal neurons were prepared and cultured as previously described (Shin et al., 2008). The indicated plasmids and/or siRNAs were transiently transfected by Lipofectamine 2000 (Invitrogen) at the third day of NGF treatment for PC12 cells or at the fifth day after seeding of primary neuronal cells. The amounts of plasmids for transfections were as follows: 4µg for transfection of LRRK2 or shLRRK2 plasmids, 2µg for transfection of Rab5 plasmids, 4 and 1µg for cotransfection of LRRK2 or shLRRK2 and Rab5 plasmids, respectively, or 100 nM for all siRNAs. If necessary, the amounts of plasmid DNAs were kept constant at 5µg by addition of vector plasmids. Cells were fixed for microscopic analysis after incubation for 7 more days.
To test protein expression level, PC12 cells were transiently co-transfected with the indicated plasmids and pMACS4.1 expressing CD4Δ (Miltenyi Biotec, Bergisch Gladbach, Germany) and harvested after 2~3 days of incubation. Co-transfected cells were sorted by MACS (Magnetic-activated cell sorting, Miltenyi Biotec, Bergisch Gladbach, Germany). MACS utilizes CD4 antibodies coupled to micromagnetic beads which specifically detect CD4Δ expressed on the surface of transfected cells. A magnetic field was applied to sort cells bound to the CD4 antibodies with magnetic beads. The protein expression level was determined by immunoblotting of each cell lysates.
Immunofluorescence staining
To identify transfected cells with LRRK2 and/or Rab5, cells were stained with antibodies against the corresponding proteins or their specific fusion tags. To detect over-expression of LRRK2 or Rab5, antimyc (Sigma, cat no. M5546) or anti-flag (Sigma, cat no. F3165) antibodies were used, respectively. To detect cells transfected with shLRRK2 plasmids or Rab5 siRNAs, anti-LRRK2 (Norvus, cat no. 300-268) or anti-Rab5 (Santa Cruz, cat no. sc-598) antibodies were used, respectively, and cells expressing the corresponding proteins at low level in comparison to neighboring cells were selected for neurite analysis (Fig. 1). In addition, to identify cells transfected with siRNAs, shDNA or vector plasmids alone, GFP plasmids were co-transfected and GFP antibodies (a gift from Dr. Eunjoon Kim of KAIST, Korea) were used for their detection. Alexa 488 goat anti-mouse IgG (Molecular Probe, cat no. A11001) or Alexa 555 goat anti-rabbit IgG (Molecular probe, cat no. A21422) were used as secondary antibodies to visualize positive cells.
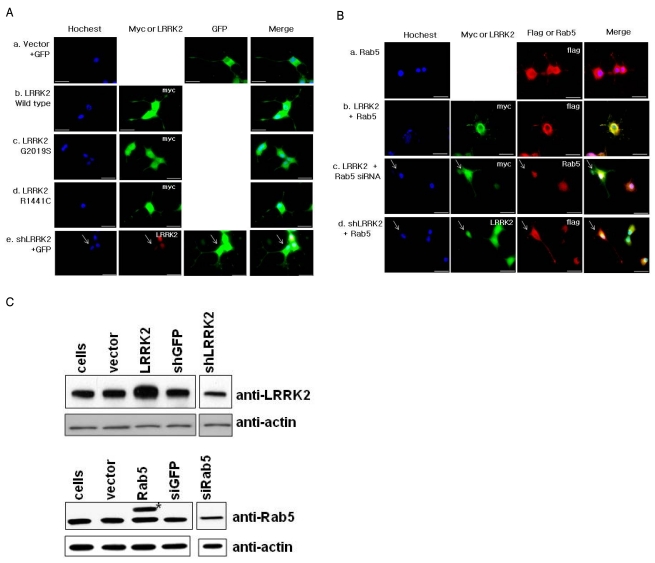
Fig. 1.
(A) Representative fluorescence microscopic images of PC12 cells transfected with vector (a), myc tagged-LRRK2 wild type (b), -G2019S (c), -R1441C (d) and shLRRK2 (e) plasmids. (B) Representative fluorescence microscopic images of PC12 cells transfected with flag tagged Rab5 (a), myc tagged LRRK2 WT with flag tagged Rab5 plasmids (b), myc tagged LRRK2 WT with Rab5 siRNAs (c) and shLRRK2 with flag tagged Rab5 plasmids (d). PC12 cells were seeded on coverglass and treated with NGF (50 ng/ml) for 10 days. Cells were transfected with the indicated plasmid on the third day and fixed on the 10th day for microscopic analysis. Total amounts of DNA used were kept constant by addition of vector plasmids for all samples. For each case, nuclear staining was shown by Hoechst staining and proteins of interest were stained by the antibody against the tag (over-expression) or protein itself (down-expression). To identify cells down-expressing LRRK2 or Rab5, cells weakly stained by specific antibodies relative to adjacent cells were selected and used for analysis of neurite length (indicated by arrows). GFP plasmids were used to visualize neurites, if needed. The scale bar is 20µm. (C) Expression levels of LRRK2 and Rab5 in PC12 cells. PC12 cells were transfected by myc-LRRK2, flag-Rab5, shLRRK2 plasmids or Rab5 siRNA-1s (siRab5) and harvested for 3 or 2 days for LRRK2 or Rab5 analysis, respectively. The weak expression of LRRK2 and low transfection efficiency of the PC 12 cell line made it impossible to directly detect LRRK2 expression in western analysis. Therefore, the MACS (Miltenyi Biotec, Bergisch Gladbach, Germany) method was applied to sort positively transfected cells. PC12 cells transiently co-transfected with the indicated plasmids and pMACS4.1 expressing CD4Δ (Miltenyi Biotec) and sorted by application of magnetic field which detected cells bound to CD4 antibodies coupled to micromagnetic beads. Expression levels of both proteins are compared to the cellular actin level using anti-LRRK2, -Rab5 or -actin antibodies. *Indicates exogenously expressed flag tagged Rab5.
Assay for neurite outgrowth
To analyze neurite outgrowth patterns, transfected and immunostained cells for all samples were observed under a fluorescence microscope (Olympus model BX61) set up under the same condition. Five fields on the coverglass were randomly selected and their images taken. Then, every cell in the images satisfying the experimental conditions such as over- or down-expression of specific proteins was selected for further neurite analysis. The number of cells analyzed was in the range of 400~1,800. The selected cells' total and the longest neurite lengths were analyzed using a computer-assisted image analysis program [Meta-Morph software Program, Molecular Devices, Downingtown, PA, USA (Klimaschewski et al., 2002)]. Values for each cell's total and the longest neurite lengths were divided by its cell body diameter to compensate differences of their soma sizes since cells with larger cell bodies generally contain longer neurites. From these values, the average and the standard error of the mean (SEM) for each experimental condition were calculated and shown as relative values to the vector control. Since the data pattern of the total and the longest neurite lengths were very similar to each other, only the pattern of the total neurite length is shown.
RESULTS
To address whether LRRK2 and Rab5 coordinately regulate neurite outgrowth, we cultured NGF-treated PC12 cells with a combination consisting of over- or down-expression of LRRK2 and/or Rab5. The resulting cells were analyzed by immunofluorescence using anti-LRRK2 and/or anti-Rab5 antibodies. When necessary, anti-GFP or other specific antibody was used to identify transfected cells. A representative immunostaining pattern for each condition is shown in Fig. 1 and quantitative analyses of neurite lengths of transfected cells are shown in Fig. 2. We assessed neurite outgrowth according to two factors, total lengths of neurites and the longest neurite length of each counted cell using MetaMorph, a computer-assisted program. Analyses using these two factors revealed that both neurite length patterns were similar to each other. Therefore, we showed only total neurite length pattern (Fig. 2).
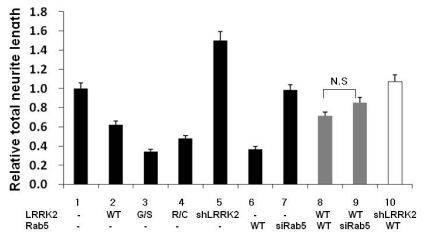
Fig. 2.
Analysis of the neurite lengths of PC12 cells transfected with the indicated plasmids and/or siRNAs. Cells were transfected and treated as explained in Fig. 1. Cells in five randomly selected fields were analyzed. Whether cells were transfected with the indicated plasmids and/or siRNAs was confirmed by the expression pattern of the proteins after staining with specific antibodies. Cells clearly showing over- or down-expression of corresponding proteins were selected. Every selected cell was subjected to neurite length analysis that was carried out using an automated image analysis program (MetaMorph Program, Molecular Devices, Downingtown, PA, USA) with the same parameter set for all samples. The resulting total neurite length of each cell is shown as average with standard error of mean (SEM). The Y axis is arbitrally set as 1 for the neurite length of cells transfected with vector plasmids. G/S and R/C indicate LRRK2 G2019S and R1441C mutants, respectively. N.S. means no significant difference between the indicated groups by ANOVA test (p>0.05). Lanes 2, 3, 4, 5, 6, 8 and 9, but not lanes 7 and 10, showed statistically significant differences compared to the control, lane 1 (p<0.05).
We observed that PC12 cells over-expressing the LRRK2 pathogenic mutant, G2019S, had dramatically reduced neurite length while PC12 cells over-expressing the wild type LRRK2 modestly reduced neurite length (Fig. 1A & 2 lanes 1~3). When over-expressed, another pathogenic mutant R1441C exhibited neurite length between those of WT and G2019S (Fig. 1A & 2 lanes 1~4). As previously reported, down-regulation of LRRK2 expression by specific shLRRK2 plasmids enhanced neurite outgrowth (Fig. 1A and 2 lanes 1 & 5). Our results corroborate previous studies performed with rat primary cultures (MacLeod et al., 2006) and human neuroblastoma SH-SY5Y cells (Plowey et al., 2008). In addition, our results showed that over-expression of Rab5 strongly inhibits neurite outgrowth, in agreement with a previous study [Fig. 1B & 2, (Liu et al., 2007)]. Our side-by-side comparison showed that the inhibitory effect by Rab5 was much greater than that by LRRK2 wild type (WT) and was similar to that by G2019S (Fig. 1 & 2). In addition, Rab5 siRNA showed little difference from the vector control in neurite length (Fig. 2 lanes 1 & 7).
Notably, when both of LRRK2 and Rab5 were over-expressed, they exhibited neither additional nor synergistic effect on neurite length. Instead, the extent of reduction in neurite lengths was similar to that by LRRK2 over-expression alone (Fig. 2 lanes 2 vs 8). This finding suggests that these two proteins functionally interact and co-regulate neurite outgrowth, which is further supported by co-localization of these over-expressed proteins in both cell body and neurites [Fig. 1Bb, (Shin et al., 2008)]. Next we tested the effect of over-expressing one protein while knocking down expression of the other. Toward this, we used Rab5 siRNA-1 (siRab5) and shLRRK2 plasmid, a plasmid containing a short-hairpin RNA (shRNA) sequence against LRRK2 (ORIGENE), to down-regulate Rab5 and LRRK2, respectively. As shown in Fig. 1C, these methods efficiently knock-down the expression of the corresponding proteins in PC12 cells. As control, we confirmed that either GFP siRNA or shGFP plasmid did not detectably affect neurite outgrowth by immunoflulorescence staining and computer analysis (Fig. 3 and data not shown). Cells transfected with either mixture of myc-LRRK2 plasmids and Rab5 siRNAs or mixture of flag-Rab5 and shLRRK2 plasmids, were identified by staining with antibodies against myc and Rab5, or LRRK2 and flag, respectively (Fig. 1Bc & 1Bd). Cells exhibiting over-expression of one protein and simultaneously reduced expression of the other protein were selected and analyzed for neurite lengths. Cells over-expressing LRRK2 with down-expression of Rab5 showed no significant differences from cells over-expressing both LRRK2 and Rab5 in terms of neurite length (Fig. 1B and Fig. 2 lanes 8 vs 9). In contrast, cells over-expressing Rab5 with down-expression of LRRK2 showed neurite length shorter than cells down-expressing LRRK2 alone and much longer than the cells over-expressing Rab5 alone (Fig. 1 and Fig. 2 lanes 5, 6 & 10). Taken together, our results indicate that LRRK2 expression level more critically determines neurite length than the Rab5 expression level.
Fig. 3.
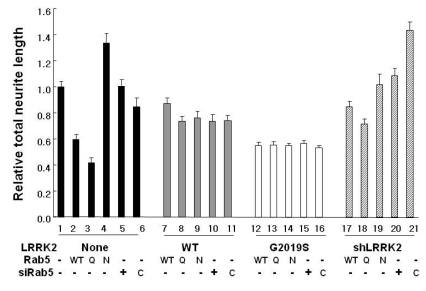
Neurite analysis of PC12 cells after over-expression of Rab5 wild type (WT), Q79L (Q) or N133I (N) with over- or down-expression of LRRK2. Total neurite length of each condition is shown as an average with SEM. All procedures were carried out as described in Fig. 2. C indicates GFP siRNAs used as a negative control.
Rab5 is a member of a small GTPase family recycling between active GTP- and inactive GDPbound forms. The active Rab5 negatively regulates neurite outgrowth (Liu et al., 2007). To investigate how the active or inactive status of Rab5 affects neurite outgrowth regulated by LRRK2, we utilized the Rab5b's constitutively active Q79L and the dominant negative N133I proteins and performed an extensive analysis (Fig. 3). In agreement with the previous study, in NGF-treated PC12 cell, overexpression of Q79L and N133I proteins showed reduction and extension of the neurite outgrowth, respectively [Fig. 3, lanes 3 & 4 (Liu et al., 2007)], although the neurite length differences among samples in this particular set were smaller than the one observed in Fig. 2 (Compare lanes 1, 6 & 8 in Fig. 2 to lanes 1, 2 & 7 in Fig. 3). It is interesting that Rab5 siRNAs showed neurite length similar to that of the vector control or the siGFP control whereas cells expressing Rab5 N133I considerably extended their neurite lengths (Fig. 3, lanes 1, 4, 5 & 6). This may indicate that the active status, but not the concentration, of Rab5 is critical for regulation of neurite outgrowth.
When either LRRK2 WT or G2019S was co-expressed with one of Rab5 WT, constitutively active and dominant negative forms, their neurite lengths were similar to those of cells expressing either LRRK2 WT or G2019S protein alone, respectively, regardless of which form of Rab5 was co-expressed (Fig. 3, lane 2~4 vs lanes 7~9 & 12~14). This was also the case when siRab5 was used instead of over-expression of Rab5 (Fig. 3, lane 5, 6 vs lanes 10, 11 & 15, 16). This is striking in that LRRK2 can disable Rab5's active signal to regulate neurite outgrowth. In contrast, cells down-regulating LRRK2 while over-expressing Rab5 WT or Q79L exhibited neurite length of each sample in the middle between the one observed under downregulation of LRRK2 alone and the one seen with over-expressing the corresponding Rab5 alone (Fig. 3, lanes 2, 3 vs lanes 17, 18). Down-regulation of LRRK2 with Rab5 N133I over-expression resulted in shorter neurite length than down-regulation of LRRK2 alone and over-expression of Rab5 N133I alone, both of which showed considerable extension of neurite (Fig. 3, lanes 4, 20 & 21 and Fig. 2, lane 5). No additional or synergistic effect by combination of down-regulation of LRRK2 with Rab5 N133I over-expression was observed, suggesting again co-regulation of neurite outgrowth by LRRK2 and Rab5. It is not clear why cells down-expressing LRRK2 with Rab5 N133I over-expression showed no difference from vector control instead of extension in neurite length. The result in Fig. 3 strongly suggests that both cellular concentration and kinase activity of LRRK2 are important for negative neurite outgrowth co-regulated by Rab5.
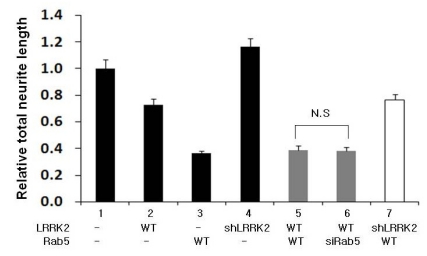
Furthermore, we carried out a similar set of functional analyses using hippocampal E17 primary neuronal cells over-expressing LRRK2 with over- or down-expression of Rab5 and found similar patterns of neurite outgrowth regulation as those in PC12 cells (Fig. 2 & 4). Specifically, neither overnor down-expression of Rab5 showed any difference in neurite length as long as LRRK2 was co-expressed (Fig. 4 lanes 5 & 6). However, the over-expression of LRRK2 with over- or down-expression of Rab5 in primary neuronal cultures resulted in relatively shorter neurites than over-expression of LRRK2 alone (Fig. 4 lanes 2, 5 & 6) whereas the same sets in PC12 cells resulted in neurites similar to or slightly longer than that when LRRK2 alone was over-expressed (Fig. 2 & 3). It may be due to differences of cell types used or of protocols to induce neurite outgrowth. However, the results here strongly suggested that, as long as LRRK2 was over-expressed, the identity or cellular concentration of Rab5 was not critical to regulate neurite outgrowth.
Fig. 4.
Analysis of neurite length of rat hippocampal neuronal cells transfected with the indicated plasmids and/or siRNAs. Rat E17 primary dissociated hippocampal neuronal cells were cultured and the indicated plasmids and/or siRNAs were transiently transfected on the fourth day after seeding. Cells were fixed for microscopic analysis after incubation for 7 more days and analyzed for total neurite length as explained in Fig. 2. ns means no significant difference between the indicated groups by ANOVA test (p>0.05). Lanes 2, 3, 5, 6 and 7, but not lanes 4, showed statistically significant differences compared to the control, lane 1 (p<0.05).
Taken together, our results suggest that LRRK2 and Rab5 co-regulate neurite outgrowth and LRRK2 is a more critical factor than Rab5 to determine the neurite length.
DISCUSSION
In this study, we confirmed previous findings that LRRK2, especially LRRK2 G2019S, and Rab5 negatively regulate neurite outgrowth (MacLeod et al., 2006; Liu et al., 2007; Plowey et al., 2008). Our data also showed that LRRK2 WT decrease neurite length although the effect is less prominent than that of G2019S. LRRK2 WT's effect on neurite length was ill defined in previous reports (MacLeod et al., 2006; Plowey et al., 2008). In addition, there is also a report that LRRK2 WT transgenic mice exhibited no difference in neurite length from that of non-transgenic control mice (Parisiadou et al., 2009). It might be due to difference of cell lines used in each study.
In this report, we provide several lines of evidence suggesting that Rab5 and LRRK2 co-regulate neurite outgrowth and that LRRK2 is a more critical factor than Rab5. Firstly, over-expression of both proteins clearly showed no additional or synergistic effect (Fig. 2~4). While Rab5 either WT or Q79L over-expression shortened neurites significantly greater than LRRK2 WT, co-expression of Rab5 with LRRK2 WT proteins reduced neurite lengths similar to those observed with LRRK2 alone (Fig. 2 & 3). In fact, co-expression of LRRK2 WT or G2019S with one of the Rab5 (WT, Q79L and N133I) exhibited a pattern of neurite length similar to the one observed with LRRK2 WT or G2019S expression alone regardless of Rab5's identity (Fig. 3). This was more striking when compared with co-expression of LRRK2 WT or G2019S with Rab5 N133I to Rab5 N133I over-expression alone. The cells co-expressing LRRK2 WT or G2019S with Rab5 N133I reduced neurite length to the level of the corresponding LRRK2 over-expression alone whereas the cells over-expressing Rab5 N133I alone considerably extended neurite lengths (Fig. 3). Secondly, down-expression of Rab5 using Rab5 siRNA did not affect neurite lengths (Fig. 2 & 3). Thirdly, down-regulation of LRRK2 expression with over-expression of Rab5 overrided the over-expression effect of Rab5 WT or of its constitutively active Q79L, resulting in longer neurites compared to over-expression of corresponding Rab5 alone, but shorter than down-regulation of LRRK2 alone (Fig. 2 & 3). This suggested that when cellular LRRK2 concentration was low, Rab5 could partially function as a regulator of neurite outgrowth. Taken together, our results suggest that regulation of neurite outgrowth via LRRK2 and Rab5 is not effected independently, but possibly through a shared mechanism.
Recently, we have reported that LRRK2 overexpression decreases the endocytosis rate of synaptic vesicles (Shin et al., 2008). Since LRRK2 also inhibits endocytosis of the NGF receptor, TrkA, which is the first step of neurite outgrowth following NGF treatment, it corroborates the fact that LRRK2 over-expression negatively regulates neurite outgrowth. In addition, Rab5 inactivation appears to be essential for endosomes containing TrkA to become signaling endosomes whose signals result in neurite outgrowth (Liu et al., 2007). Since LRRK2 overexpression with combination of different form of Rab5s resulted in neurite length similar to that of LRRK2 expression alone regardless of either cellular concentration or active status of Rab5 (Fig. 2~4), LRRK2 might function in other steps following endocytosis of TrkA-containing vesicles and inhibit the negative effect of Rab5. This notion is further supported by the previous reports showing that inhibition of neurite outgrowth, but not of endocytosis, is dependent on the active level of LRRK2 kinase (MacLeod et al., 2006; Shin et al., 2008). In other words, over-expression of the LRRK2 G2019S mutant, expressing kinase activity stronger than that of wild type induced shorter neurites, but no apparent difference in endocytosis rate compared to wild type (MacLeod et al., 2006; Shin et al., 2008). It may be interesting to investigate whether LRRK2 kinase and/or GTPase activity regulates Rab5 activity via signal transduction.
A small GTPase protein is active as a signal transmitter only when it binds to GTP. The binding of the GTPase protein to GTP or GDP is facilitated by GEF (guanine nucleotide-exchange factors) or GAP (GTPase activating protein), respectively. Recently, homodimerization of the GTPase protein has been reported as another mechanism to activate GTPase protein without GEF (Gasper et al., 2009). In fact, both LRRK2 and Rab5 contain an active GTPase domain and homodimerizations of both Rab5 and LRRK2 were reported (Daitoku et al., 2001; Deng et al., 2008; Greggio et al., 2008; Klein et al., 2009). Since LRRK2 and Rab5 interact with each other (Shin et al., 2008) it is possible that their interaction interrupts signal transmission by the GTPase domain of Rab5 but still maintains LRRK2's kinase activity that regulates neurite outgrowth. Another possibility is that LRRK2 kinase phosphorylates Rab5, resulting in Rab5's conformational change and activity. We are actively pursuing this possibility.
ACKNOWLEDGEMENTS
This work was supported by the financial supports from IRGB (to W. Seol) funded by Inje University in Korea, Mid-career Researcher Program (2009-0083924) and Leading Foreign Research Institute Recruitment Program (2009-00505) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and technology (MEST, to W. Seol). We would like to thank Dr. Eunjoon Kim (KAIST, Korea) for anti-GFP antibody and members of our laboratory for their help and discussion.
References
- 1.Dachsel JC, Taylor JP, Mok SS, Ross OA, Hinkle KM, Bailey RM, Hines JH, Szutu J, Madden B, Petrucelli L, Farrer MJ. Identification of potential protein interactors of Lrrk2. Parkinsonism Relat Disord. 2007;13:382–385. doi: 10.1016/j.parkreldis.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daitoku H, Isida J, Fujiwara K, Nakajima T, Fukamizu A. Dimerization of small GTPase Rab5. Int J Mol Med. 2001;8:397–404. doi: 10.3892/ijmm.8.4.397. [DOI] [PubMed] [Google Scholar]
- 3.de Hoop MJ, Huber LA, Stenmark H, Williamson E, Zerial M, Parton RG, Dotti CG. The involvement of the small GTP-binding protein Rab5a in neuronal endocytosis. Neuron. 1994;13:11–22. doi: 10.1016/0896-6273(94)90456-1. [DOI] [PubMed] [Google Scholar]
- 4.Deng J, Lewis PA, Greggio E, Sluch E, Beilina A, Cookson MR. Structure of the ROC domain from the Parkinson's disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc Natl Acad Sci U S A. 2008;105:1499–1504. doi: 10.1073/pnas.0709098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi PN, Chen SG, Wilson-Delfosse AL. Leucine-rich repeat kinase 2 (LRRK2): a key player in the pathogenesis of Parkinson's disease. J Neurosci Res. 2009;87:1283–1295. doi: 10.1002/jnr.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J Neurosci Res. 2008;86:1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 8.Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, Shaw K, Bhatia KP, Bonifati V, Quinn NP, Lynch J, Healy DG, Holton JL, Revesz T, Wood NW. A common LRRK2 mutation in idiopathic Parkinson's disease. Lancet. 2005;365:415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 9.Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O'Neill E, Meitinger T, Kolch W, Prokisch H, Ueffing M. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet. 2006;15:223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 10.Gorvel JP, Chavrier P, Zerial M, Gruenberg J. rab5 controls early endosome fusion in vitro. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 11.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Greggio E, Zambrano I, Kaganovich A, Beilina A, Taymans JM, Daniels V, Lewis P, Jain S, Ding J, Syed A, Thomas KJ, Baekelandt V, Cookson MR. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J Biol Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heo HY, Park JM, Kim CH, Han BS, Kim KS, Seol W. LRRK2 enhances oxidative stress-induced neurotoxicity via its kinase activity. Exp Cell Res. 2010;316:649–656. doi: 10.1016/j.yexcr.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Ho CC, Rideout HJ, Ribe E, Troy CM, Dauer WT. The Parkinson disease protein leucine-rich repeat kinase 2 transduces death signals via Fas-associated protein with death domain and caspase-8 in a cellular model of neurodegeneration. J Neurosci. 2009;29:1011–1016. doi: 10.1523/JNEUROSCI.5175-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein CL, Rovelli G, Springer W, Schall C, Gasser T, Kahle PJ. Homo- and heterodimerization of ROCO kinases: LRRK2 kinase inhibition by the LRRK2 ROCO fragment. J Neurochem. 2009;111:703–715. doi: 10.1111/j.1471-4159.2009.06358.x. [DOI] [PubMed] [Google Scholar]
- 17.Klimaschewski L, Nindl W, Pimpl M, Waltinger P, Pfaller K. Biolistic transfection and morphological analysis of cultured sympathetic neurons. J Neurosci Methods. 2002;113:63–71. doi: 10.1016/s0165-0270(01)00473-3. [DOI] [PubMed] [Google Scholar]
- 18.Lesage S, Janin S, Lohmann E, Leutenegger AL, Leclere L, Viallet F, Pollak P, Durif F, Thobois S, Layet V, Vidailhet M, Agid Y, Durr A, Brice A, Bonnet AM, Borg M, Broussolle E, Damier P, Destee A, Martinez M, Penet C, Rasco O, Tison F, Tranchan C, Verin M. LRRK2 exon 41 mutations in sporadic Parkinson disease in Europeans. Arch Neurol. 2007;64:425–430. doi: 10.1001/archneur.64.3.425. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Lamb D, Chou MM, Liu YJ, Li G. Nerve growth factor-mediated neurite outgrowth via regulation of Rab5. Mol Biol Cell. 2007;18:1375–1384. doi: 10.1091/mbc.E06-08-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luzon-Toro B, Rubio de la Torre E, Delgado A, Perez-Tur J, Hilfiker S. Mechanistic insight into the dominant mode of the Parkinson's disease-associated G2019S LRRK2 mutation. Hum Mol Genet. 2007;16:2031–2039. doi: 10.1093/hmg/ddm151. [DOI] [PubMed] [Google Scholar]
- 21.MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, Long CX, Lobbestael E, Baekelandt V, Taymans JM, Sun L, Cai H. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancho RM, Law BM, Harvey K. Mutations in the LRRK2 Roc-COR tandem domain link Parkinson's disease to Wnt signalling pathways. Hum Mol Genet. 2009;18:3955–3968. doi: 10.1093/hmg/ddp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satoh D, Sato D, Tsuyama T, Saito M, Ohkura H, Rolls MM, Ishikawa F, Uemura T. Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat Cell Biol. 2008;10:1164–1171. doi: 10.1038/ncb1776. [DOI] [PubMed] [Google Scholar]
- 27.Shin N, Jeong H, Kwon J, Heo HY, Kwon JJ, Yun HJ, Kim CH, Han BS, Tong Y, Shen J, Hatano T, Hattori N, Kim KS, Chang S, Seol W. LRRK2 regulates synaptic vesicle endocytosis. Exp Cell Res. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 29.Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, Dawson VL, Dawson TM, Ross CA. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci U S A. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. Embo J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torres VA, Mielgo A, Barbero S, Hsiao R, Wilkins JA, Stupack DG. Rab5 mediates caspase-8-promoted cell motility and metastasis. Mol Biol Cell. 2010;21:369–376. doi: 10.1091/mbc.E09-09-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Xie C, Greggio E, Parisiadou L, Shim H, Sun L, Chandran J, Lin X, Lai C, Yang WJ, Moore DJ, Dawson TM, Dawson VL, Chiosis G, Cookson MR, Cai H. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J Neurosci. 2008;28:3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 35.Wucherpfennig T, Wilsch-Brauninger M, Gonzalez-Gaitan M. Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release. J Cell Biol. 2003;161:609–624. doi: 10.1083/jcb.200211087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]