Abstract
The K1 capsule is an essential virulence determinant of Escherichia coli strains that cause meningitis in neonates. Biosynthesis and transport of the capsule, an α-2,8-linked polymer of sialic acid, are encoded by the 17-kb kps gene cluster. We deleted neuC, a K1 gene implicated in sialic acid synthesis, from the chromosome of EV36, a K-12-K1 hybrid, by allelic exchange. Exogenously added sialic acid restored capsule expression to the deletion strain (ΔneuC), confirming that NeuC is necessary for sialic acid synthesis. The deduced amino acid sequence of NeuC showed similarities to those of UDP-N-acetylglucosamine (GlcNAc) 2-epimerases from both prokaryotes and eukaryotes. The NeuC homologue from serotype III Streptococcus agalactiae complements ΔneuC. We cloned the neuC gene into an intein expression vector to facilitate purification. We demonstrated by paper chromatography that the purified neuC gene product catalyzed the formation of [2-14C]acetamidoglucal and [N-14C]acetylmannosamine (ManNAc) from UDP-[14C]GlcNAc. The formation of reaction intermediate 2-acetamidoglucal with the concomitant release of UDP was confirmed by proton and phosphorus nuclear magnetic resonance spectroscopy. NeuC could not use GlcNAc as a substrate. These data suggest that neuC encodes an epimerase that catalyzes the formation of ManNAc from UDP-GlcNAc via a 2-acetamidoglucal intermediate. The unexpected release of the glucal intermediate and the extremely low rate of ManNAc formation likely were a result of the in vitro assay conditions, in which a key regulatory molecule or protein was absent.
Although most strains of Escherichia coli are harmless commensals, certain isolates can be considered primary pathogens because they possess a variety of virulence determinants that allow the organism to evade host defenses and cause disease. Clinical syndromes vary and include several distinct forms of diarrheal disease, urinary tract infections (32) and, especially in neonates, sepsis and meningitis (28). Despite antimicrobial therapy, meningitis caused by E. coli K1 remains a significant cause of morbidity and mortality in neonates, and neurologic sequelae are common among survivors (34). Most strains of E. coli responsible for these infections synthesize the K1 capsule as an essential virulence factor (24, 26). The K1 capsule is a linear homopolymer of α-2,8-linked N-acetylneuraminic acid (sialic acid; NeuNAc) (24, 28). The capsule provides the organism with an antiphagocytic barrier characterized by the ability of terminal sialic acid residues to inhibit activation of the alternative complement pathway (8, 21). The K1 capsule is also a poor immunogen, a property attributed to molecular mimicry of the polysialic acid capsule to polysialosylglycopeptides on human fetal neuronal tissue (33).
Biosynthesis and transport of the E. coli K1 capsule are encoded by the 17-kb kps gene cluster that is located at 67 min on the E. coli chromosome (4, 37, 40). The kps cluster is functionally divided into three regions, with central region 2 being unique among capsular types. In E. coli K1, region 2 contains the neu genes, which direct the biosynthesis, activation, and polymerization of NeuNAc (4, 37, 40). Regions 1 and 3 encode proteins that function in the assembly of the capsule and its transport to the bacterial cell surface.
Biosynthesis of the polysialic acid capsule of E. coli K1 requires the intracellular condensation of N-acetylmannosamine (ManNAc) and phosphoenolpyruvate (PEP) to form NeuNAc. This reaction is catalyzed by NeuB (36). Activation of NeuNAc is performed by NeuA, which adds a nucleotide monophosphate to the sugar to form CMP-NeuNAc (35, 42). NeuS is the polysialyltransferase that polymerizes activated CMP-NeuNAc to polysialic acid (31). NeuC (41) and NeuD (9) mutants are complemented by sialic acid and therefore appear to be involved in sialic acid biosynthesis. Reactions 1 and 2 summarize the postulated mechanism of sialic acid synthesis in E. coli K1:
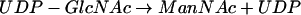 |
(1) |
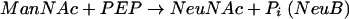 |
(2) |
In mammals, the biosynthesis of sialic acid follows a pathway different from that observed in prokaryotes. These differences potentially can be exploited as targets for chemotherapeutic intervention. Reactions 3 to 6 summarize the postulated mechanism of sialic acid synthesis in eukaryotes:
 |
(3) |
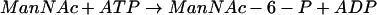 |
(4) |
 |
(5) |
 |
(6) |
The formation of ManNAc from UDP-N-acetylglucosamine (GlcNAc) is the first committed step in sialic acid synthesis. A bifunctional enzyme catalyzing the epimerization of UDP-GlcNAc to ManNAc and its subsequent phosphorylation has been purified to homogeneity from rat liver (14). The enzyme associates with itself both as a homodimer and as a hexamer. The dimer catalyzes only the ManNAc kinase reaction (reaction 2 above), while the hexamer displays both UDP-GlcNAc 2-epimerase and ManNAc kinase activities (reactions 1 and 2) (12). In this report, we show that the product of the neuC gene from region 2 of the kps cluster is the UDP-GlcNAc 2-epimerase that converts UDP-GlcNAc to ManNAc in E. coli K1. This finding is consistent with the observation that the amino acid sequence of NeuC bears homology to that of the UDP-GlcNAc 2-epimerase portion of the bifunctional eukaryotic enzyme described above.
MATERIALS AND METHODS
Bacterial strains, plasmids, bacteriophages, and media.
Descriptions of the bacterial strains and plasmids used in this study are listed in Table 1. The capsule-specific bacteriophage K1F was described previously (39). Bacteriophage P1 vir was used to transduce the nanA4 allele (38). In general, bacterial cultures were grown at 37°C in Luria-Bertani (LB) broth or on LB agar supplemented with appropriate antibiotics. Sucrose media for suicide vector selection were as previously described (11). Horse 46 antiserum agar plates were used to assay K1 capsule precipitin halo formation as described previously (29, 39).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| HB101 | hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 supE44 leu | Laboratory collection |
| EV36 | galP23 rpsL9 (argA+rha+kps+) | 38 |
| RS2444 | Source of nanA4 zgj-791::Tn10 allele | 2 |
| RS2918 | EV36 ΔneuC | This work |
| RS2921 | EV36 ΔneuC nanA4 | This work |
| RS2929 | pSR647 in HB101 | |
| RS2992 | pSR685 in HB101 | This work |
| Plasmids | ||
| pSR647 | neuC in pCYB4 | This work |
| pSR685 | neuC in pTrcHis | This work |
| Bluescript KS(+) | Phagemid cloning vector | Stratagene |
| pCYB4 | Intein fusion plasmid from IMPACT-1 kit | New England Biolabs |
| pDC123 | Cloning vector for GBS | 6 |
| pDC128 | PDC123 carrying sialic acid (neu) genes from GBS capsular poly- saccharide synthesis locus | This work |
| pSR641 | neuC deletion construct plasmid | This work |
| pSR652 | siaA in pCYB4 | This work |
| pSR653 | rfbC in Bluescript KS(+) | This work |
Allelic exchange methodology.
The neuC deletion strain, RS2918, was constructed essentially as described previously (11). Colony PCR with primers that flanked neuC confirmed the deletion, which reduced the gene from 1,195 to 110 bp.
Bacteriophage lysis assays.
Fifty-milliliter cultures of EV36, the K-12-K1 hybrid strain, the ΔneuC strain RS2918, and RS2918 carrying wild-type neuC in trans were grown to stationary phase, diluted 1:50 in LB broth, and grown to mid-log phase at 37°C with appropriate antibiotics. Bacteriophage K1F was added at a multiplicity of infection of 0.5. The cultures were monitored at 600 nm for clearing, indicative of capsule synthesis resulting in cell lysis due to bacteriophage infection and propagation.
For K1F bacteriophage plaque assays, 100-μl stationary-phase cultures of the host strain were mixed with 100 μl of an appropriate dilution of bacteriophage K1F in 3 ml of LB soft agar (LB medium with 7 g of Bacto Agar per liter) kept at 50°C. The mixtures were immediately poured onto the surface of LB agar plates and incubated at 37°C for 4 to 6 h.
Complementation assays.
Transformants that were making a polysialic acid capsule were detected on a minimal agar plate supplemented with the appropriate antibiotics by streaking K1-specific bacteriophage across the diameter of the plate. After the streak had dried, transformants were cross-streaked at a 90° angle to the bacteriophage and incubated at 37°C for 4 to 6 h. If a transformant expressed a capsule, growth stopped at the margin of the bacteriophage streak. Phage-sensitive transformants also were tested for halo formation on horse 46 antiserum agar plates.
DNA manipulation and sequencing.
Restriction endonucleases, DNA polymerases, and DNA ligases were purchased from Gibco-BRL (Gaithersburg, Md.) and used according to the manufacturer's instructions. Enzymatic manipulations of DNA and the preparation of competent cells for transformation were done as previously described (3). DNA for sequence analysis was prepared by PCR followed by column purification with a Wizard PCR Prep DNA purification system (Promega, Madison, Wis.) or by plasmid purification with a Wizard Plus SV Miniprep DNA purification system (Promega) followed by isopropanol precipitation. DNA sequencing was performed at the University of Rochester Core Nucleic Acid Laboratory.
DNA and protein computer analyses.
DNA and protein sequence analyses were done with software from The Genetics Computer Group, Inc., Madison, Wis (10). Searches of DNA and protein databases were done at the National Center for Biotechnology Information site at www.ncbi.nlm.nih.gov. Computer analysis of protein sequences also was performed with the ExPASy Molecular Biology Server at www.expasy.ch, operated by the Swiss Institute of Bioinformatics (Geneva, Switzerland).
Protein purification and quantitation.
A stationary-phase culture of either RS2929 or DH5α(pSR647) was used to inoculate 1.5 liters of LB broth containing 100 μg of ampicillin/ml. Cells were shaken at 37°C until the culture reached an A600 of 0.6 and then were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The temperature was immediately decreased to 22°C, and the cells were shaken for an additional 21 h. Cells were harvested by centrifugation and resuspended in 20 ml of 20 mM HEPES-500 mM NaCl (pH 7.8) containing 1 μg of pepstatin/ml and 1 μg of aprotonin/ml. The cell suspension was lysed in a French pressure cell (20,000 lb/in2) and then centrifuged at 10,000 × g for 15 min at 4°C to remove unbroken cells. The supernatant (20 ml) was loaded onto a 12-ml chitin column (New England Biolabs) equilibrated with cold 20 mM HEPES-500 mM NaCl (pH 7.8). The resin was washed with 120 ml of 20 mM HEPES-500 mM NaCl (pH 7.8) followed by 25 ml of 50 mM dithiothreitol (DTT)-20 mM HEPES-500 mM NaCl (pH 7.8). The column was sealed and incubated for 18 h at 4°C to allow cleavage by the DTT. The protein was eluted with 10 ml of 50 mM DTT-20 mM HEPES-500 mM NaCl (pH 7.8) followed by 50 ml of 20 mM HEPES-500 mM NaCl (pH 7.8). The protein was stored as a 90% ammonium sulfate suspension. As needed, aliquots of this enzyme suspension were centrifuged, and the precipitate was dissolved and dialyzed against 50 mM morpholinepropanesulfonic acid (MOPS)-100 mM NaCl (pH 7.5).
Proteins were quantitated by the method of Bradford (5) with a Bio-Rad (Hercules, Calif.) reagent kit according to the manufacturer's instructions.
Enzyme assays by NMR.
For nuclear magnetic resonance (NMR) experiments, the protein was isolated from the chitin resin as described above with 12 ml of HEPES buffer and exchanged into deuterated sodium phosphate buffer (10 mM, pH 7.5) by four rounds of concentration and dilution with 4-ml centrifugal filters (10-kDa MWCO; Millipore).
The enzyme was diluted to 665 μl and transferred to an NMR tube containing Chelex-100 resin (20 mg, 200/400 mesh, Na+ form, previously rinsed with D2O). The reaction was initiated by the addition of 35 μl of 100 mM UDP-GlcNAc in D2O, and the resulting solution (5 mM UDP-GlcNAc) was incubated at 37°C for 2 days. 1H and proton-decoupled 31P NMR spectra were obtained at timed intervals by using a Bruker 300-MHz spectrometer.
The final solution was stirred with 1 ml of Dowex AG1-X8 resin (formate form, 100/200 mesh; Bio-Rad) for 1 h, filtered through glass wool, and lyophilized to dryness. The sample was dissolved in 700 μl of D2O, and a 1H NMR spectrum was obtained; it was indistinguishable from that of a standard of 2-acetamidoglucal previously synthesized by known methods (18, 23). The sample was lyophilized to dryness again and analyzed by mass spectrometry: 226 (M + Na+).
From a separate preparation of NeuC, two 665-μl samples (0.3 mg/ml) were prepared in deuterated buffer in NMR tubes as described above. To one tube, 0.5 mg of CMP-NeuNAc (1 mM) was added, and then to both tubes, 35 μl of 100 mM UDP-GlcNAc was added. The reaction mixtures were incubated at 37°C with monitoring by 1H NMR.
SDS-PAGE.
Discontinuous sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) of proteins was done according to the method of Laemmli (17).
Sugar epimerase assays.
The reaction mixture for sugar epimerase assays consisted of 7 to 9 mg of dialyzed NeuC protein/ml, 45 mM MOPS, 45 mM MgCl2 (pH 7.5), and 10 μM UDP-[14C]GlcNAc (266 mCi/mmol) in a 45- to 160-μl volume. The reaction mixture was incubated at 37°C for 3 h and then spotted (two 20-μl samples) on borate-impregnated Whatman no. 3 paper (soaked in 1% sodium tetraborate and dried prior to chromatography). The chromatogram was developed for 16 h by descending chromatography with ethyl acetate-2-propanol-pyridine-water (50:22:14:14) solvent (43). The radioactive portions of the paper were visualized with a PhosporImager (ImageQuant). The unlabeled sugar standards were detected by a silver nitrate dipping procedure. The sugar standards were solutions (1 mg/ml) of ManNAc, GlcNAc (Sigma Chemical Co.), and 2-acetamidoglucal.
To test for the formation of ManNAc, the 3-h reaction mixture described above was treated as follows. After the 3-h incubation, the reaction mixture was adjusted to 0.7, 18, or 35 mM pyruvate in 16 mM potassium phosphate (pH 7.2) followed by the addition of 2 U of N-acetylneuraminic acid aldolase (Sigma A-6680). The reaction mixture was incubated at 37°C for an additional 3 h prior to spotting for paper chromatography and development in ethyl acetate-2-propanol-pyridine-water (50:22:14:14) to resolve ManNAc and ethanol-1 M ammonium acetate (pH 7.5) (7.5:3, vol/vol) to resolve NeuNAc.
RESULTS
NeuC is homologous to sugar 2-epimerases.
The nonredundant GenBank translation database was searched with the peptide sequence of NeuC as the query to determine the probable function of this protein. Several known and some putative UDP-GlcNAc 2-epimerases from both prokaryotes and eukaryotes were identified as having significant homology to NeuC (Table 2) based on an analysis with BLAST software (1). Interestingly, the sequence with the highest homology belonged to the capsule gene cluster from Streptococcus agalactiae group B (group B streptococci [GBS]). This gram-positive bacterium synthesizes an unrelated capsule with a branched-chain repeat unit having a side chain of galactose that terminates with a sialic acid residue. The GBS NeuC homologue, NeuIIIC, displays 57% similarity and 45% identity to NeuC. Although NeuIIIC is essential for sialylation of the GBS capsule, the exact function of this protein has not been described.
TABLE 2.
Homologues of NeuCa
| Homologuea | Species | %
|
|
|---|---|---|---|
| Similarity | Identity | ||
| NeuIIIC | Streptococcus agalactiae group B | 57 | 45 |
| Bifunctional enzyme | Rattus norvegicus | 53 | 27 |
| Bifunctional enzyme | Mus musculus | 53 | 27 |
| Bifunctional enzyme | Homo sapiens | 53 | 27 |
| SiaA | Neisseria meningitidis | 52 | 32 |
| NeuVC167C | Campylobacter coli | 47 | 36 |
The GenBank nucleotide sequence accession numbers of the NeuC homologues used in this study are AF163833 (S. agalactiae group B), AF195053 (C. coli), M95053 (N. meningitidis), CAA69204 (R. norvegicus), CAB36908 (M. musculus), and NP_005467 (H. sapiens). The GenBank accession number for NeuC is M84026.
The bifunctional UDP-GlcNAc 2-epimerase-ManNAc kinase enzyme found in human, mouse, and rat liver displays homology to NeuC with 53% similarity and 27% identity over the first 400 amino acids. Epimerase activity is localized to the N-terminal portion of the bifunctional liver enzyme (12, 14).
A Campylobacter coli VC167 gene product, NeuVC167C, also shows significant similarity to NeuC (Table 2). NeuVC167C is involved in modification of C. coli VC167 flagellin with the sialic acid analogue pseudaminic acid (19). SiaA is a GlcNAc-6-P 2-epimerase necessary for sialic acid synthesis in Neisseria meningitidis group B (22). This gram-negative organism synthesizes a polysialic acid capsule that is identical to that of E. coli K1 (15). SiaA displays 52% similarity and 32% identity to NeuC. RfbC, which encodes a UDP-GlcNAc 2-epimerase that converts UDP-GlcNAc to UDP-ManNAc (16), is 42% similar and 23% identical to NeuC. Salmonella rffE (wecB) has been demonstrated to encode a UDP-GlcNAc 2-epimerase. This protein shows 32% similarity and 21% identity to NeuC.
Of the homologues described above, the eukaryotic bifunctional enzyme has been most extensively studied. It has been demonstrated clearly that this epimerase catalyzes the formation of ManNAc directly from UDP-GlcNAc via a 2-acetamidoglucal intermediate with the release of UDP. The sequence homology suggests that the E. coli K1 neuC gene encodes a UDP-GlcNAc epimerase.
The NeuC homologue from GBS complements a strain with a nonpolar chromosomal deletion in neuC.
RS2918 is a derivative of the K-12-K1 hybrid EV36 with a deletion in the neuC gene constructed by allelic exchange methodology. The resulting phenotype is acapsular, as shown by resistance to capsule-specific bacteriophage K1F and the lack of precipitin haloes on antiserum agar. Supplying neuC in trans on plasmid pSR647 restored capsule synthesis to RS2918 (data not shown). Moreover, the addition of exogenous NeuNAc to the medium restored capsule synthesis to RS2921, a nanA (NeuNAc aldolase) derivative of RS2918 (data not shown). The mutation in nanA ensures that the NeuNAc added to the media was targeted for polymer synthesis and not a catabolic pathway. These observations are consistent with those of previous studies (29, 41) and support the notion that NeuC is an essential enzyme in the biosynthesis of sialic acid. Zeitler et al. previously showed that capsular polysaccharide production by a neuC insertion mutant was not complemented by ManNAc (43).
The gene product with the highest identity to NeuC was NeuIIIC from GBS. Plasmid pDC128, which contains the NeuIIIC gene, was used to transform RS2918. The resulting strain regained the ability to synthesize a capsule. We also tested the ability of pSR653 to complement the neuC defect in pRS2918. pSR653 carries the rfbC gene from Salmonella enterica serovar Borreze. RfbC, a UDP-GlcNAc 2-epimerase that converts UDP-GlcNAc to UDP-ManNAc (16), is 23% identical and 42% similar to NeuC. RfbC did not complement the NeuC defect in RS2918.
UDP-GlcNAc 2-epimerase assays of NeuC.
NeuC was overexpressed by plasmid pSR647 as an intein-chitin binding protein fusion and purified by affinity chromatography on chitin resin. The purified enzyme migrated as a single major protein band on SDS-PAGE with an expected molecular weight of 45,000 (Fig. 1).
FIG. 1.
Polyacrylamide gel-purified:NeuC. The mobilities of molecular weight (M.W.) standards (Std.) are shown at the right.
UDP-GlcNAc 2-epimerase assays were performed with purified NeuC. Figure 2, lane 1, shows the radioactive reaction products resulting from the incubation of 10 μg of the purified NeuC fraction with UDP-[14C]GlcNAc. The major product of the reaction migrated as a rapidly moving spot that comigrated with 2-acetamidoglucal, a putative intermediate in the epimerization of UDP-GlcNAc to ManNAc or UDP-ManNAc. In addition, a fainter, more slowly moving product that comigrated with the ManNAc standard was observed. The third, even fainter slowly moving spot near the origin has not been identified. When the enzyme fraction was heated to 95°C for 5 min prior to incubation (Fig. 2, lane 2), no products were observed. The identity of the slowly moving spot that comigrated with the ManNAc standard was confirmed by the following experiment. Purified NeuC was incubated with UDP-[14C]GlcNAc as described above for 3 h and then treated with NeuNAc aldolase and pyruvate. This procedure resulted in the appearance of a spot that comigrated with sialic acid and a decrease in the intensity of the putative ManNAc spot, as expected (data not shown). These results are consistent with the hypothesis that NeuC is a UDP-GlcNAc 2-epimerase.
FIG. 2.
UDP-GlcNAc 2-epimerase activity of NeuC. (A) Autoradiogram. Purified enzyme was incubated with UDP-[14C]GlcNAc for 3 h at 37°C (lane 1). As a control, enzyme was boiled for 5 min prior to the addition of substrate (lane 2). A total of 40 μl of each reaction mixture was spotted at the origin of borate-impregnated paper, and the chromatogram was developed in ethyl acetate-2-propanol-pyridine-H2O (50:22:14:14). (B) Sugar standard. One milligram of each sugar per milliliter was spotted (25 μl), and the chromatogram was developed with silver nitrate.
To determine whether NeuC could use GlcNAc as a substrate, 10 μg of the purified NeuC fraction was incubated with 9 nmol of [14C]GlcNAc. An identical incubation was performed with 10 μg of bovine serum albumin as a negative control. No epimerization from GlcNAc to ManNAc was detected (data not shown). This finding indicated that GlcNAc is not a substrate for NeuC. To determine whether GlcNAc-6-P is a substrate for NeuC, a 5 mM sample was incubated with the enzyme and monitored by 1H NMR spectroscopy (see assay below). The absence of any reaction under these conditions indicated that GlcNAc-6-P is not a substrate for NeuC.
The enzymatic reaction was examined by 1H and 31P NMR spectroscopy to provide further identification of the reaction products. A sample of NeuC was purified by affinity chromatography on chitin resin, followed by exchange into phosphate buffer prepared in D2O. UDP-GlcNAc was added, and 1H and 31P NMR spectra (Fig. 3A and 4A, respectively) were obtained immediately. After incubation at 37°C for 43.5 h, a new singlet at 6.59 ppm in the 1H NMR spectrum was observed (Fig. 3B). This finding is consistent with the formation of 2-acetamidoglucal (Fig. 3C), whereby the anomeric proton of UDP-GlcNAc at 5.40 ppm has been converted to a vinylic proton. Integration of the H-1 signals indicates that 8% conversion occurred over the course of 15 h and that 19% conversion was achieved by 43.5 h. Repeated measurements confirmed that the rate of conversion was dependent on the enzyme concentration, and control experiments lacking enzyme showed no detectable formation of 2-acetamidoglucal under otherwise identical conditions. No signals attributable to ManNAc (5.02 ppm for H-1 of the α-anomer and 4.92 ppm for H-1 of the β-anomer [7]) were observed in the spectrum, presumably due to the inherent insensitivity of the technique (<1% product would not be detected by this method). Additionally, the spectrum revealed a new doublet at 7.87 ppm, adjacent to the doublet from H-5 of the uracil ring in the substrate, indicative of the formation of UDP. The 31P NMR spectrum (Fig. 4B) further supports these data, as a pair of doublets appeared at −6.26 and −9.75 ppm, downfield from those of UDP-GlcNAc. A spectrum for UDP was obtained for comparison (Fig. 4C).
FIG. 3.
(A and B) 1H NMR spectra of the incubation of UDP-GlcNAc with NeuC after 3 min (A) and 43.5 h (B). t, time. (C) 1H NMR spectrum of an authentic sample of 2-acetamidoglucal. The position of the anomeric proton is indicated by the arrow.
FIG. 4.
(A and B) 31P NMR spectra of the incubation of UDP-GlcNAc with NeuC after 6 min (A) and 43.5 h (B). t, time. (C) 31P NMR spectrum of an authentic sample of UDP.
To further confirm that 2-acetamidoglucal had been formed, the enzyme solution was treated with an anion-exchange resin to remove all phosphate-containing species. The 1H NMR and mass spectra of the resulting material were in agreement with those of authentic 2-acetamidoglucal (18, 23).
It has been shown that CMP-NeuNAc is a feedback inhibitor of the mammalian UDP-GlcNAc 2-epimerase (14). Therefore, CMP-NeuNAc was tested as a possible regulator of NeuC. A sample of NeuC in deuterated buffer was incubated with 5 mM UDP-GlcNAc in the presence of 1 mM CMP-NeuNAc, and the reaction was monitored by NMR. However, no difference in the products or extent of the reaction was detected relative to those of a control lacking CMP-NeuNAc (data not shown). In a similar experiment, the enzyme was assayed in the presence of either 5 mM NAD+ or 5 mM NADP+, and no detectable changes in activity were observed.
DISCUSSION
In this report, we present evidence that NeuC catalyzes the epimerization of UDP-GlcNAc to ManNAc, the first committed step in sialic acid biosynthesis in E. coli K1. These data are in good agreement with both the protein homologies and the indirect results reported by other investigators (29, 41). Two radiolabeled reaction products that comigrated in paper chromatography with ManNAc and 2-acetamidoglucal resulted from the in vitro incubation of purified NeuC protein with UDP-[14C]GlcNAc. However, no products were observed when the enzyme fraction was boiled or when an excess of unlabeled UDP-GlcNAc was added (Fig. 2). This result indicated that neither product was the result of nonenzymatic hydrolysis or epimerization of the substrate. In their studies of UDP-GlcNAc 2-epimerase purified from rat liver, Sommar and Ellis (30) proposed a reaction mechanism that could explain these data (Fig. 5). They posited that the enzymatic mechanism was an ordered one in which the first product released was UDP, followed by the irreversible formation of ManNAc. This mechanism included a 2-acetamidoglucal intermediate, which was enzyme bound. This mechanism does not require a cofactor and is consistent with the requirement for a nucleotide sugar in the formation of ManNAc. It is also consistent with the observation that purified NeuC did not catalyze the epimerization of GlcNAc to ManNAc.
FIG. 5.
Proposed mechanism of the reaction catalyzed by the mammalian UDP-GlcNAc 2-epimerase.
The proposed role for NeuC as a UDP-GlcNAc 2-epimerase is supported by the observed sequence homology between this enzyme and the well-characterized mammalian and E. coli (rffE) UDP-GlcNAc 2-epimerases (27 and 21% identities, respectively). This enzyme clearly is required for sialic acid biosynthesis. It is therefore expected that this enzyme converts UDP-GlcNAc to ManNAc and UDP. Both the chromatographic and the NMR spectroscopic assays indicate, however, that the primary reaction observed is the conversion of UDP-GlcNAc to 2-acetamidoglucal and UDP. The chromatographic assay indicated that a very low level of ManNAc is also formed, but this was an extremely slow process. Thus, two scenarios emerge: either a second enzyme, such as a glycosidase (glycosidases are known to hydrate glycals [18]), is required to complete the conversion to ManNAc, or the purified enzyme is missing an important regulator or coenzyme that is required to complete the catalytic cycle. The second scenario is consistent with the observation that even the formation of 2-acetamidoglucal seems to be quite slow. One attempt was made to determine whether CMP-N-acetylneuraminic acid is an allosteric activator required for full activity; however, no effect was observed.
The formation of 2-acetamidoglucal nevertheless strengthens the link between NeuC and other UDP-GlcNAc 2-epimerases (27). The E. coli UDP-GlcNAc 2-epimerase, RffE, is a true epimerase that interconverts UDP-GlcNAc and UDP-ManNAc. Several lines of evidence support a mechanism involving the anti-elimination of UDP to form 2-acetamidoglucal, followed by the syn-addition of UDP to form a product (20). In fact, the enzyme is known to release 2-acetamidoglucal and UDP into solution once every 1,000 turnovers.
The mammalian UDP-GlcNAc 2-epimerase converts UDP-GlcNAc to UDP and free ManNAc (this action essentially is irreversible and technically is not epimerization) and plays a key role in sialic acid biosynthesis. This enzyme also is thought to use a mechanism involving the anti-elimination of UDP to give 2-acetamidoglucal, followed by the syn-addition of water to give ManNAc (7, 30) (Fig. 5). It has been shown that this enzyme will accept the intermediate 2-acetamidoglucal from solution and hydrate it to form ManNAc. Given that NeuC also catalyzes the anti-elimination of UDP to form 2-acetamidoglucal, it seems reasonable to assume that it also functions as a UDP-GlcNAc 2-epimerase in vivo. During the in vitro studies reported in this article, it is possible that a key protein or regulatory molecule that is required for NeuC to complete the reaction was lacking. Therefore, we simply might have been seeing the products of a “crippled” enzyme that was unable to complete its normal reaction under the specific conditions of the assay.
While the previous information argues strongly for the assignment of NeuC as a UDP-GlcNAc 2-epimerase, it is conceivable that NeuC actually catalyzes a different reaction in vivo. One possibility is that the true substrate is UDP-ManNAc and that the role of NeuC simply is to catalyze hydrolysis of the glycosyl-UDP bond. This possibility seems unlikely, however, since 2-acetamidoglucal and UDP would be the expected intermediates in this process and the former should be converted readily to ManNAc. An alternate possibility is that NeuC is actually a glycosyl transferase that utilizes UDP-GlcNAc as a substrate. This possibility is not unreasonable, since the UDP-GlcNAc 2-epimerases share structural similarities with a family of glycosyltransferases (26). In the absence of an acceptor molecule, the glycal could be formed as an unnatural product. This possibility also seems unlikely, since one would not expect to observe the formation of any ManNAc in this process, and it does not help to explain the role of NeuC in sialic acid biosynthesis.
Surface-displayed sialic acid is an important virulence determinant in a number of bacterial pathogens besides E. coli K1. These include the N. meningitidis polysialic acid capsule, terminal sialic acid residues on the S. agalactiae capsule, and the sialylated flagella of C. coli. The synthesis of sialic acid differs in prokaryotes and eukaryotes.
GBS synthesize a branched-chain polysaccharide capsule, and the only similarity with the E. coli K1 capsule is a single terminal sialic acid residue. The NeuIIIC gene of GBS, however, complements ΔneuC. It has also been shown that the NeuIIIA gene (previously designated cpsF) of GBS can complement a mutation in the E. coli K1 neuA gene, which encodes the CMP-N-acetylneuraminic acid synthetase (13). Indeed, a plasmid containing the four GBS genes involved in sialic acid synthesis, pDC128, successfully complemented strains with mutations in neuD, neuB, neuC, or neuA, the E. coli K1 region 2 sialic acid synthesis genes (D. Daines, unpublished data). This result indicates that the sialic acid biosynthetic pathways for incorporation into capsular polysaccharides are probably identical in these two pathogens.
The epimerases encoded by rffE and rfbC catalyze the conversion of UDP-GlcNAc to UDP-ManNAc. That rfbC does not complement the neuC deletion is not surprising, since NeuC cleaves UDP-GlcNAc during catalysis. During the preparation of this article, Ringenberg et al. reported (25) that rffE is not necessary for polysialic acid synthesis in E. coli K1. These authors also reported that ManNAc-6-phosphate is not involved as an intermediate in the formation of sialic acid. The observations of these authors support our suggestion that NeuC converts UDP-GlcNAc to ManNAc and fit well with the observed homology to the epimerase domain of the mammalian UDP-GlcNAc 2-epimerase.
Acknowledgments
We thank Virginia Clark for providing the N. meningitidis group B strain, Chris Whitfield for providing the rfbC gene.
This work was supported by NIH grants AI39615 to R.P.S. and AI25152 and AI22498 to C.E.R. D.A.D. was supported by Molecular Pathogenesis of Bacteria and Viruses training grant AI07362 from the Public Health Service. This work was also supported by NSERC grant AI07362 to A.S.M. and M.E.T.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annunziato, P. W., L. F. Wright, W. F. Vann, and R. P. Silver. 1995. Nucleotide sequence and genetic analysis of the neuD and neuB genes in region 2 of the polysialic acid gene cluster of Escherichia coli K1. J. Bacteriol. 177:312-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1988. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Bliss, J. M., and R. P. Silver. 1996. Coating the surface: a model for expression of capsular polysialic acid in Escherichia coli K1. Mol. Microbiol. 21:221-231. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Chaffin, D. O., and C. E. Rubens. 1998. Blue/white screening of recombinant plasmids in gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219:91-99. [DOI] [PubMed] [Google Scholar]
- 7.Chou, W. K., S. Hinderlich, W. Reutter, and M. E. Tanner. 2003. Sialic acid biosynthesis: stereochemistry and mechanism of the reaction catalyzed by the mammalian UDP-N-acetylglucosamine 2-epimerase. J. Am. Chem. Soc. 125:2455-2461. [DOI] [PubMed] [Google Scholar]
- 8.Cross, A. S. 1990. The biologic significance of bacterial encapsulation. Curr. Top. Microbiol. Immunol. 150:87-95. [DOI] [PubMed] [Google Scholar]
- 9.Daines, D. A., L. F. Wright, D. O. Chaffin, C. E. Rubens, and R. P. Silver. 2000. NeuD plays a role in the synthesis of sialic acid in Escherichia coli K1. FEMS Microbiol. Lett. 189:281-284. [DOI] [PubMed] [Google Scholar]
- 10.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Effertz, K., S. Hinderlich, and W. Reutter. 1999. Selective loss of either the epimerase or kinase activity of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase due to site-directed mutagenesis based on sequence alignments. J. Biol. Chem. 274:28771-28778. [DOI] [PubMed] [Google Scholar]
- 13.Haft, R. F., M. R. Wessels, M. F. Mebane, N. Conaty, and C. E. Rubens. 1996. Characterization of cpsF and its product CMP-N-acetylneuraminic acid synthetase, a group B streptococcal enzyme that can function in K1 capsular polysaccharide biosynthesis in Escherichia coli. Mol. Microbiol. 19:555-563. [DOI] [PubMed] [Google Scholar]
- 14.Hinderlich, S., R. Stasche, R. Zeitler, and W. Reutter. 1997. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis in rat liver. J. Biol. Chem. 272:24313-24318. [DOI] [PubMed] [Google Scholar]
- 15.Kasper, D. L., J. L. Winkelhake, D. W. Zollinger, B. L. Brandt, and M. S. Artenstein. 1973. Immunochemical similarity between polysaccharide antigens of Escherichia coli O7:K1L:NM and group B Neisseria meningitidis. J. Immunol. 110:262-268. [PubMed] [Google Scholar]
- 16.Keenleyside, W. J., and C. Whitfield. 1996. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J. Biol. Chem. 271:28581-28592. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lai, E. C. K., and S. G. Withers. 1994. Stereochemistry and kinetics of the hydration of 2-acetamido-d-glucal by β-N-acetylhexosaminidases. Biochemistry 33:14743-14749. [DOI] [PubMed] [Google Scholar]
- 19.Logan, S. M., J. F. Kelly, P. Thibault, C. P. Ewing, and P. Guerry. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46:587-597. [DOI] [PubMed] [Google Scholar]
- 20.Morgan, P. M., R. F. Sala, and M. E. Tanner. 1997. Eliminations catalyzed by UDP-N-acetylglucosamine 2-epimerase. J. Am. Chem. Soc. 119:10269-10277. [Google Scholar]
- 21.Moxon, E. R., and J. S. Kroll. 1990. The role of bacterial polysaccharide capsules as virulence factors. Curr. Top. Microbiol. Immunol. 150:65-85. [DOI] [PubMed] [Google Scholar]
- 22.Petersen, M., Fessner, W.-D., M. Frosch, and E. Luneberg. 2000. The siaA gene involved in capsule polysaccharide biosynthesis of Neisseria meningitidis B codes for N-acylglucosamine-6-phosphate 2-epimerase activity. FEMS Microbiol. Lett. 184:161-164. [DOI] [PubMed] [Google Scholar]
- 23.Pravdic, N., I. Franjic-Mihalic, and B. Danilov. 1975. An improved synthesis of the 2-acetamido-d-glucal derivative 3,4,6-tri-O-acetyl-2-(N-acetylacetamido)-1,5-anhydro-2-deoxy-d-arabino-hex-1-enitol. Carbohydr. Res. 45:302-306. [Google Scholar]
- 24.Rick, P. D., and R. P. Silver. 1996. Enterobacterial common antigens and capsular polysaccharides, p. 104-122. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 25.Ringenberg, M. A., S. M. Steenbergen, and E. R. Vimr. 2003. The first committed step in the biosynthesis of sialic acid by Escherichia coli K1 does not involve a phosphorylated N-acetylmannosamine intermediate. Mol. Microbiol. 50:961-975. [DOI] [PubMed] [Google Scholar]
- 26.Robbins, J. B., George, J. H. McCracken, E. C. Gotschlich, F. Orskov, I. Orskov, and L. A. Hanson. 1974. Escherichia coli K1 capsular polysaccharide associated with neonatal meningitis. N. Engl. J. Med. 290:1216-1220. [DOI] [PubMed] [Google Scholar]
- 27.Samuel, J., and M. E. Tanner. 2002. Mechanistic aspects of carbohydrate epimerization. Nat. Prod. Rep. 19:261-277. [DOI] [PubMed] [Google Scholar]
- 28.Silver, R. P., and E. R. Vimr. 1990. Polysialic acid capsule of Escherichia coli K1, p. 39-60. In V. L. C. B. H. Iglewski (ed.), Molecular basis of microbial pathogenesis. Academic Press, Inc., San Diego, Calif.
- 29.Silver, R. P., W. F. Vann, and W. Aaronson. 1984. Genetic and molecular analyses of Escherichia coli K1 antigen genes. J. Bacteriol. 157:568-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommar, K. M., and D. B. Ellis. 1972. Uridine diphosphate N-acetyl-d-glucosamine-2-epimerase from rat liver. II. Studies on the mechanism of action. Biochim. Biophys. Acta 268:590-595. [DOI] [PubMed] [Google Scholar]
- 31.Steenbergen, S. M., T. J. Wrona, and E. R. Vimr. 1992. Functional analysis of the sialyltransferase complexes in Escherichia coli K1 and K92. J. Bacteriol. 174:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sussman, M. 1997. Escherichia coli and human disease, p. 3-48. In M. Sussman (ed.), Escherichia coli: mechanisms of virulence. Cambridge University Press, Cambridge, England.
- 33.Troy, F. A. 1992. Polysialylation: from bacteria to brains. Glycobiology 2:5-23. [DOI] [PubMed] [Google Scholar]
- 34.Unhanand, M., M. M. Mustafa, J. G. H. McCracken, and J. D. Nelsen. 1993. Gram-negative enteric bacillary meningitis: a twenty-one-year experience. J. Pediatr. 122:15-21. [DOI] [PubMed] [Google Scholar]
- 35.Vann, W. F., R. P. Silver, C. Abeijon, K. Chang, W. Aaronson, A. Sutton, C. W. Finn, W. Lindner, and M. Kotsatos. 1987. Purification, properties, and genetic location of Escherichia coli cytidine 5′-monophosphate N-acetylneuraminic acid synthetase. J. Biol. Chem. 262:17556-17562. [PubMed] [Google Scholar]
- 36.Vann, W. F., J. J. Tavarez, J. Crowley, E. Vimr, and R. P. Silver. 1997. Purification and characterization of the Escherichia coli K1 neuB gene product N-acetylneuraminic acid synthetase. Glycobiology 7:697-701. [DOI] [PubMed] [Google Scholar]
- 37.Vimr, E., Steenbergen, S., and M. Cieslewicz. 1995. Biosynthesis of the polysialic acid capsule in Escherichia coli K1. J. Ind. Microbiol. 15:352-360. [DOI] [PubMed] [Google Scholar]
- 38.Vimr, E. R., and F. A. Troy. 1985. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J. Bacteriol. 164:845-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vimr, E. R., R. D. McCoy, H. F. Vollger, N. C. Wilkison, and F. A. Troy. 1984. Use of prokaryotic-derived probes to identify poly(sialic acid) in neonatal neural membranes. Proc. Natl. Acad. Sci. USA 81:1971-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 41.Zapata, G., J. M. Crowley, and W. F. Vann. 1992. Sequence and expression of the Escherichia coli K1 neuC gene product. J. Bacteriol. 174:315-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zapata, G., W. F. Vann, W. Aaronson, M. S. Lewis, and M. Moos. 1989. Sequence of the cloned Escherichia coli K1 CMP-N-acetylneuraminic acid synthetase gene. J. Biol. Chem. 264:14769-14774. [PubMed] [Google Scholar]
- 43.Zeitler, R., B. J., C. Bauer, and W. Reutter. 1992. Inhibition of the biosynthesis of N-acetylneuraminic acid by metal ions and selenium in vitro. Biometals 5:103-109. [DOI] [PubMed] [Google Scholar]







