Abstract
FtsE and FtsX have homology to the ABC transporter superfamily of proteins and appear to be widely conserved among bacteria. Early work implicated FtsEX in cell division in Escherichia coli, but this was subsequently challenged, in part because the division defects in ftsEX mutants are often salt remedial. Strain RG60 has an ftsE::kan null mutation that is polar onto ftsX. RG60 is mildly filamentous when grown in standard Luria-Bertani medium (LB), which contains 1% NaCl, but upon shift to LB with no NaCl growth and division stop. We found that FtsN localizes to potential division sites, albeit poorly, in RG60 grown in LB with 1% NaCl. We also found that in wild-type E. coli both FtsE and FtsX localize to the division site. Localization of FtsX was studied in detail and appeared to require FtsZ, FtsA, and ZipA, but not the downstream division proteins FtsK, FtsQ, FtsL, and FtsI. Consistent with this, in media lacking salt, FtsA and ZipA localized independently of FtsEX, but the downstream proteins did not. Finally, in the absence of salt, cells depleted of FtsEX stopped dividing before any change in growth rate (mass increase) was apparent. We conclude that FtsEX participates directly in the process of cell division and is important for assembly or stability of the septal ring, especially in salt-free media.
In Escherichia coli, the division septum forms via the coordinated inward growth of all three layers of the cell envelope—the cytoplasmic membrane, the peptidoglycan wall, and the outer membrane. To date, about a dozen E. coli genes are known to be specifically required for septation (3, 11). These genes share two important properties: (i) loss of function mutations result in the formation of long, aseptate filaments with regularly spaced nucleoids (the fts, or filamentation temperature-sensitive phenotype), and (ii) the proteins encoded by these genes localize to the division site. Because cell division genes are generally essential and because lesions in many housekeeping genes can affect cell division indirectly, there have not been any exhaustive screens for division mutants. Thus, it seems likely that more division genes remain to be described.
A number of mutant hunts, starting with the pioneering work of Hirota and coworkers in the 1960s, suggested that there is an important cell division gene located at about 76 min on the E. coli chromosome (30). This locus was originally designated ftsE. One interesting property of ftsE mutants is that many are salt remedial, meaning that viability is restored by inclusion of NaCl in the growth medium. The amount of salt required for rescue is strain dependent, but generally in the range of 0.5%. Studies by Salmond and colleagues in the 1980s revealed that “ftsE” comprised two genes, which were then designated ftsE and ftsX (13). Moreover, the sequence of these genes revealed clear homology to ABC transporters; FtsE is the ATP-binding cassette (ABC) component, while FtsX is the membrane component. ABC transporters use energy from ATP to transport a wide variety of substrates either into or out of cells (or subcellular compartments). These observations led to the proposal that FtsEX transports an ion needed for division but not for growth per se (13).
Subsequently, Woldringh and colleagues questioned whetherftsE is really a division gene, after studying one allele and finding that this mutant produced filaments in broth but not minimal medium (34). Their thinking was influenced by having just completed a study of ftsB, which only filamented at high growth rates and turned out to be an allele of nrd, a gene needed for synthesis of DNA precursors (35). The view that ftsEX affects cell division indirectly seems to have gained ascendancy, as most of the review articles on bacterial cell division published in the last 10 years make no mention of ftsE or ftsX (e.g., references 11, 22, and 31), and recent work in E. coli has explored potential connections to membrane protein insertion (10, 37). During this same time period, however, mutants of ftsE and/or ftsX have been reported in Flavobacterium, Neisseria, and Aeromonas (2, 20, 24). Interestingly, these mutants are viable but have morphological defects suggestive of impaired division.
Here we report on experiments intended to determine whether FtsE and FtsX participate directly in cell division in E. coli. Our findings establish that FtsE and FtsX are bona fide division proteins.
MATERIALS AND METHODS
Strains.
Strains used in this study are listed in Table 1. RG60 has been described previously (10). Phage P1 grown on RG60 was used to transduce MG1655/pDSW610 to Kanr to create EC1335. EC1335 was transduced to Ampr with P1 phage grown on EC447, EC436, EC535, and EC442 (8, 23, 38) to create EC1363, EC1366, EC1391, and EC1392, respectively. EC1335 was transformed with pCH205 (18) to create EC1386. EC1063 and EC1065 are derivatives of MG1655 and were constructed from pDSW513 and pDSW512 using λInCh1 pSX102(Ampr) (5). EC1116 was constructed by integrating pDSW533 into attφ80 of MG1655 (16). EC1116 was transduced to Tetr with P1 grown on MM61 [ftsA12(Ts) leu::Tn10] or DRC14 [ftsZ84(Ts) leu::Tn10] to create EC1152 and EC1158, respectively. EC1159 was constructed by transducing EC1116/pDSW406 to Kanr with P1 grown on EC912 (23). EC1179, EC1180, and EC1181 were constructed by integrating pDSW533 into attφ80 of JOE170, JMG265, and EC549 (8, 12, 38). EC1295 was constructed by transducing JOE563 (7) to Kanr with P1 grown on EC1063. EC1340 was constructed by transducing PS223 (28) to Kanr with P1 grown on EC1065. EC1384 was constructed by transforming DHB4 (5) with pDSW609.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant features | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | Wild type | Lab collection |
| DHB4 | Wild type with F′ lacIq | 5 |
| EC549 | ftsI::TnphoA (Kanr)/pBAD33-ftsI | 38 |
| JOE170 | ftsQ::TnphoA (Kanr)/pBAD33-ftsQ | 8 |
| JOE563 | ftsK::cat/pBAD42-ftsK | 7 |
| JMG265 | ftsL::TnphoA (Kanr)/pBAD33-ftsL | 12 |
| PS223 | W3110 zipA1(Ts) | 28 |
| RG60 | MG1655 ftsE::kan | 10 |
| EC1063 | MG1655 Δ(attL-lom)::(kan lacIq P204-ftsX-gfp) | This study |
| EC1065 | MG1655 Δ(attL-lom)::(kan lacIq P206-gfp-ftsX) | This study |
| EC1116 | MG1655 attφ80::pDSW533 (P206-gfp-ftsX) | This study |
| EC1152 | EC1116 ftsA12(Ts) leu::Tn10 | This study |
| EC1158 | EC1116 ftsZ84(Ts) | This study |
| EC1159 | EC1116 ftsW::kan/pBAD33-ftsW | This study |
| EC1179 | JOE170 attφ80::pDSW533 (P206-gfp-ftsX) | This study |
| EC1180 | JMG265 attφ80::pDSW533 (P206-gfp-ftsX) | This study |
| EC1181 | EC549 attφ80::pDSW533 (P206-gfp-ftsX) | This study |
| EC1295 | JOE563 Δ(attL-lom)::(kan P204-ftsX-gfp) | This study |
| EC1335 | MG1655 ftsE::kan/pBAD33-ftsEX | This study |
| EC1340 | PS223 attφ80::pDSW533 (P206-gfp-ftsX) | This study |
| EC1363 | EC1335 Δ(attL-lom)::(bla lacIq P206-ftsA-gfp) | This study |
| EC1366 | EC1335 Δ(attL-lom)::(bla lacIq P204-gfp-ftsI) | This study |
| EC1386 | EC1335/pCH205 [Plac-ftsK(1-266)-gfp] | This study and ref. 18 |
| EC1391 | EC1335 Δ(attL-lom)::(bla lacIq P206-zipA-gfp) | This study |
| EC1392 | EC1335 Δ(attL-lom)::(bla lacIq P206-gfp-ftsQ) | This study |
| Plasmids | ||
| pBAD33 | Arabinose regulation (PBAD), p15A ori, Camr | 15 |
| pDSW206 | P206 promoter, ColE1 ori, Ampr | 38 |
| pDSW209 | gfp-fusion vector, P206 promoter, ColE1 ori, Ampr | 38 |
| pDSW210 | gfp-fusion vector, P206 promoter, ColE1 ori, Ampr | 38 |
| pDSW533 | pJC69-P206-gfp-ftsX (lacIq, oriRR6Kγ, attPφ80, Spcr) | This study and ref. 7 |
| pDSW609 | pTH18kr-ftsE-3xHA (Plac, Kanr, pSC101 ori) | This study and ref. 19 |
| pDSW610 | pBAD33-ftsEX | This study |
| pDSW621 | pBAD33-ftsE | This study |
| pDSW622 | pBAD33-ftsX | This study |
| pDSW636 | pDSW209-ftsX | This study |
| pDSW637 | pDSW210-ftsX | This study |
| pDSW638 | pDSW206-ftsE-3xHA | This study |
Plasmids.
Plasmid pDSW512 (P206-gfp-ftsX) was constructed by amplifying ftsX from the chromosome of MG1655 using primers P481 (CGAGAATTCAACAACAACGTCACTTGCATGGAGGCGTGG) and P482 (TGCTCTAGATATTCAGGCGTAAAGTGGCG). The 1,111-bp product was cut with EcoRI and XbaI (sites underlined) and ligated into the same sites of pDSW286, a Kanr derivative of pDSW209 (38). pDSW513 (P204-ftsX-gfp) was constructed similarly using primers P483 (CAGGAATTCGTCACTTGCATGGAGGCGTGG) and P484 (CTGCTGCAGGTTGTTGTTTTCAGGCGTAAAGTGGCGTAA), with the product being cloned into the EcoRI-PstI sites of pDSW256, a Kanr derivative of pDSW208 (38). To construct pDSW533 (P206-gfp-ftsX, Spcr, attPφ80), pDSW512 was digested with MunI and XbaI, and the 1,252-bp fragment carrying ftsX was isolated and ligated into the same sites of pJC118 (7). Plasmid pDSW609 was constructed in several steps. First, the triple hemagglutinin (HA) tag was PCR amplified from pMYP-3xHA (32) using primers P578 (catggaggcgtgggccatgaaaacaacaacTCTAGATACCCATACGATGTTCCTGAC) and P577 (CTGAAGCTTACTaAGCAGCGTAATCTGGAACGTC). The upstream primer has 20 bases homologous to the 3′ end of ftsE (lowercase letters), omitting the stop codon. The resulting 144-bp product was isolated and used as a primer in a second PCR together with P579 (CACGAATTCATAACACTTTTTGCCCGAGAGGATTAAC), which anneals to the 5′ end of ftsE. This reaction produced a 742-bp ftsE-3xHA fusion product that was digested with EcoRI and HindIII (sites underlined in P579 and P577) and ligated into the same sites of pTH18kr (19). Plasmid pDSW610 (pBAD33-ftsEX) was constructed in two steps. First, ftsEX was amplified from the chromosome of MG1655 with primers P477 (CAGCCATGGTTCGCTTTGAACATGTCAGC) and P488 (GTCAAGCTTATTCAGGCGTAAAGTGGCGT). The 1,734-bp product was cut with NcoI and HindIII (sites underlined) and ligated into the same sites of pBAD24 (15) to create pDSW519. Then, the 1,799-bp BamHI-HindIII fragment carrying ftsEX from pDSW519 was moved into the same sites of pBAD33 (15) to create pDSW610. The ftsX gene was deleted from pDSW610 by digestion with PshAI and HindIII, followed by treatment with T4 DNA polymerase, and then ligation to create pDSW620 (pBAD33-ftsE). Similarly, ftsE was deleted by digesting pDSW610 with PciI and SphI, treatment with T4 DNA polymerase, and ligation to create pDSW621 (pBAD33-ftsX). Plasmid pDSW636 (P206-gfp-ftsX) was constructed by ligating the 1,098-bp EcoRI-XbaI fragment carrying ftsX from pDSW512 into pDSW209 (38). The 1,095-bp EcoRI-PstI fragment carrying ftsX from pDSW513 was ligated into the same sites of pDSW210 (38) to create pDSW637 (P206-ftsX-gfp). Plasmid pDSW638 (P206-ftsE-3xHA) was constructed by cloning the 790-bp EcoRI-HindIII fragment carrying ftsE with three tandem repeats of the HA epitope tag into the same sites of pDSW206.
Media, chemicals, and molecular biological procedures.
Luria-Bertani medium (LB) consisted of 10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl (1% NaCl) and, for plates, 15 g of agar per liter. LB always contained 1% NaCl except where stated that it was omitted. Antibiotics were used at the following concentrations: 40 μg of kanamycin/ml, 30 μg of chloramphenicol/ml, 50 μg of spectinomycin/ml. l-Arabinose and d-glucose were used at 0.2%, unless otherwise indicated, to modulate expression from the araBAD promoter PBAD. Isopropyl-β-d-galactopyranoside (IPTG) was added at the following concentrations: 5 μM (gfp-ftsI), 1 mM (zipA-gfp), 100 μM (ftsA-gfp), 50 μM (ftsX-gfp), 100 μM (gfp-ftsX), 40 μM (ftsK (1-266)-gfp), or 10 μM (gfp-ftsQ). Enzymes used to manipulate DNA were from New England Biolabs (Beverly, Mass.). Oligonucleotides were from Integrated DNA Technologies (Coralville, Iowa). DNA sequencing was performed by the DNA Core Facility of the University of Iowa. All constructs made by PCR were sequenced to verify their integrity.
Depletion of FtsEX.
A culture of EC1335 (ftsE::kan/pBAD33-ftsEX) was grown overnight at 30°C in 5 ml of LB containing kanamycin, chloramphenicol, and arabinose. The next morning, this culture was diluted 1:20 into LB with no NaCl but containing kanamycin, chloramphenicol, 0.02% arabinose, and 0.2% glucose. This culture was grown at 30°C for 2 h to an optical density at 600 nm (OD600) of ∼0.5. Cells were washed to remove sugars by pelleting 1 ml of culture in a microcentrifuge and resuspending in 1 ml of LB with no NaCl. The washed cells were diluted 1:150 into a flask of LB with no NaCl but containing chloramphenicol and either arabinose or glucose. Growth and cell morphology were monitored periodically by the OD600 and with microscopy, respectively.
Localization of GFP fusions to FtsX.
Strains EC1063 (P204-ftsX-gfp) and EC1065 (P206-gfp-ftsX) are MG1655 derivatives that harbor fusions of gfp to ftsX integrated into the chromosome in single copy at the λ attachment site. These strains were grown overnight at 30°C in LB-kanamycin. The next morning cultures were diluted 1:1,000 into LB without antibiotic but containing IPTG to induce expression of the gfp fusion, and cultures were grown to an OD600 of ∼0.3 and then fixed in the growth medium with cross-linking agents and processed for fluorescence microscopy as described previously (38). Dependence of green fluorescent protein (GFP)-FtsX localization on other division proteins was determined in strains that harbored conditional alleles of these proteins. Growth of such strains under permissive and nonpermissive conditions has been described elsewhere (38). Briefly, cultures were grown in LB with IPTG to induce the gfp fusion and antibiotics to maintain any plasmids until early exponential phase, at which time cultures were shifted to the nonpermissive condition. Cells were fixed when they appeared filamentous. Typically, this was 1 h after shift to 42°C for Ts mutants and 4 h after dilution into glucose-containing medium for arabinose-dependent depletion strains.
Localization of GFP-Fts fusions in an FtsEX depletion background.
Fusions of gfp to various division genes were introduced into the FtsEX depletion strain EC1335 by transduction or transformation. Depletion of FtsEX was done in LB with no NaCl as described above, except that the medium contained IPTG to induce the respective gfp fusion. Cells were fixed when the glucose-grown culture became filamentous and were examined by fluorescence microscopy.
Localization of FtsE-3xHA.
A culture of DHB4/pDSW609 (Plac-ftsE-3xHA) was grown overnight at 30°C in LB with kanamycin. This culture was diluted 1:200 into LB with 100 μM IPTG and grown at 30°C to an OD600 of ∼0.5. Then, cells were fixed and processed for immunofluorescence microscopy as described previously (33). FtsE-3xHA was detected with anti-HA monoclonal antibody (HA.11; BabCo, Berkeley, Calif.) diluted 1:200 and incubated overnight, followed by goat anti-mouse antibody conjugated to Alexa488 (Molecular Probes, Eugene, Oreg.) diluted 1:200 and incubated for 2 h.
Localization of FtsN by immunofluorescence microscopy.
Wild-type MG1655 or ftsE::kan mutant RG60 cells grown at 30°C in LB with 1% NaCl were fixed with cross-linking agents and processed for immunofluorescence as described previously (29). Purified anti-FtsN serum (39) was used at a dilution of 1:500 overnight at 4°C. The secondary antibody was goat anti-rabbit conjugated to Texas Red (Molecular Probes) at 1:400 for 2 h.
RESULTS
Localization of FtsN in RG60.
Strain RG60 has been described elsewhere (10). It is an MG1655 derivative with an ftsE::kan insertion, now designated ftsE400::kan, that is predicted to be polar onto ftsX. RG60 was reported to be filamentous and required at least 0.5% NaCl for viability. Other electrolytes found to rescue RG60 on plates included NaH2PO4, Na2SO4, KCl, NH4Cl, and CaCl2, but not K2SO4, MgSO4, or MnCl2. Osmolytes such as sucrose, glycine betaine, and proline also failed to rescue RG60.
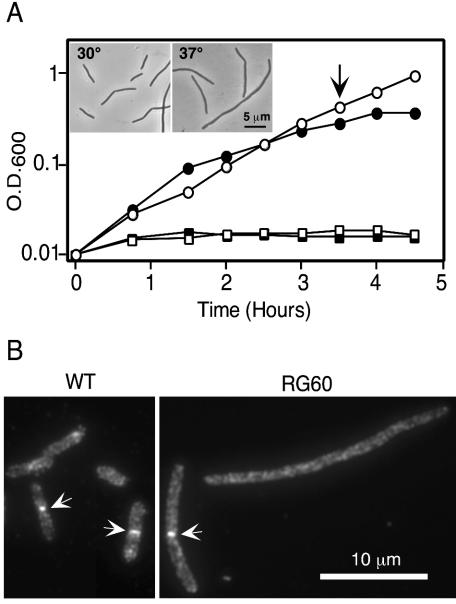
We found that RG60 grew somewhat slowly and was mildly filamentous in standard LB broth, which contains 1% NaCl (Fig. 1A). These defects were more pronounced at elevated temperatures. Upon shift to LB broth that lacked NaCl, growth and division essentially stopped.
FIG. 1.
(A) Effect of salt and temperature on growth of RG60. RG60 (ftsE400::kan) growing in LB with 1% NaCl at 30°C was subcultured into LB with 1% (circles) or no (squares) NaCl at 30°C (open symbols) or 37°C (closed symbols). The inset shows a phase-contrast micrograph of cells harvested at the time indicated by the arrow from the cultures growing with salt. (B) Localization of FtsN. Cells of wild type (MG1655) or RG60 in exponential growth in LB with 1% NaCl at 30°C were fixed, and FtsN was visualized by immunofluorescence microscopy. Arrows point to septal localization of FtsN.
We attempted to transduce various gfp-fts fusion genes into RG60 so that we could determine whether the division block in this strain occurs before, during, or after assembly of the septal ring. These efforts were generally unsuccessful, although we could move the fusions into the parental strain, MG1655 (data not shown). Other approaches, such as transducing the ftsE400::kan allele into the gfp fusion strains or transforming RG60 with plasmids that express the fusions, also failed. In some cases we simply never recovered transductants or transformants, while in others we could recover them and confirm by PCR that they were correct, but they had a small colony phenotype and lysed when grown in broth, even with high salt. These findings suggest that the gfp-fts fusion genes are toxic when combined with the ftsE::kan mutation, which is somewhat surprising because they are not very toxic in a wild-type background and some of them, like gfp-ftsI, complement (38).
Ultimately, we were able to use immunofluorescence microscopy to show that FtsN, a late recruit to the division site (1), can localize in RG60 grown under permissive conditions (1% NaCl, 30°C). About 60% of the RG60 cells exhibited FtsN localization, compared to about 35% of the cells of a wild-type control strain, MG1655 (Fig. 1B). However, the RG60 cells are long enough that close to 100% should exhibit FtsN localization if the ftsE::kan mutation has no effect on assembly or stability of the septal ring. Thus, FtsE improves, but is not required for, assembly or stability of the septal ring. This finding is consistent with RG60 having a leaky division defect.
Depleting cells of FtsEX blocks cell division.
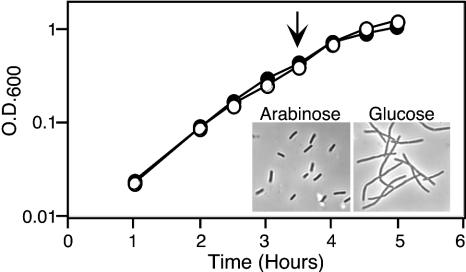
It is not obvious why an ABC transporter would be needed specifically for cell division, and strain RG60 grows poorly even in the presence of salt. These considerations suggested that the division defect in RG60 might be a secondary consequence of a metabolic defect that renders the cells generally unhealthy. We therefore constructed a strain, EC1335, in which FtsEX expression is under control of an arabinose-dependent promoter, PBAD (15). In the presence of glucose, which prevents ftsEX expression, EC1335 formed colonies on LB plates that contained 1% NaCl, but not on LB plates that lacked NaCl. Colony formation in the absence of NaCl was rescued by arabinose. To investigate the relationship between cell division and overall health, EC1335 was grown in LB broth with no salt but containing 0.2% arabinose or 0.2% glucose. Both cultures grew at the same rate as judged by OD600, but the glucose-grown cells stopped dividing after about 2 h and were clearly filamentous after about 3.5 h (Fig. 2). We conclude that the division defect is a primary defect rather than a secondary consequence of the cells becoming unhealthy.
FIG. 2.
Effect of FtsEX depletion on growth and division. EC1335 (ftsE::kan/pBAD33-ftsEX) was grown in LB with no NaCl but containing either arabinose (closed symbols) or glucose (open symbols) to modulate expression of the plasmid-borne ftsEX genes. Samples were removed periodically to monitor growth by OD600 or cell morphology. The inset shows a phase-contrast micrograph of cells harvested at the time indicated by the arrow.
FtsE and FtsX localize to the division site.
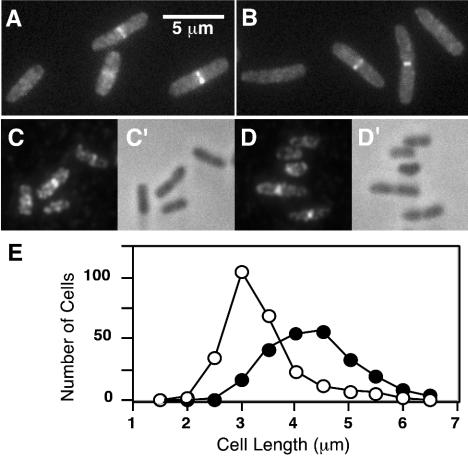
To determine whether FtsE and FtsX localize to the division site, we fused ftsE and ftsX to gfpmut2 (9), which encodes a bright variant of GFP. The fusion genes were integrated into the chromosome at the λ attachment site by selecting for a tightly linked kanamycin resistance marker (5). Expression of the gfp fusions was under control of an IPTG-inducible promoter. Strains were grown in LB containing IPTG to an OD600 of ∼0.3, fixed with cross-linking agents, and examined by fluorescence microscopy. With FtsX-GFP and GFP-FtsX, about half of the cells had a bright band of fluorescence at the midcell (Fig. 3A and B). More precisely, the fraction of cells exhibiting septal localization was about 40% in an MG1655 background and about 60% in an MC4100 background. No other sites of localization were apparent. Convincing localization of GFP-FtsE and FtsE-GFP was not observed, perhaps because GFP interferes with the proper function of this protein. We therefore constructed a low-copy plasmid that expressed ftsE with three tandem copies of an HA epitope tag at the C terminus (32). Immunofluorescence microscopy revealed septal localization of the FtsE-3xHA fusion in about 50% of the cells (Fig. 3C and D). Similar fluorescent bands were not observed unless production of FtsE-3xHA was induced with IPTG, verifying the specificity of the antibodies used to detect the tagged protein. Cells with FtsE and FtsX at the midcell were on average longer than those without (Fig. 3E), indicating that these proteins are recruited to the division site during the later stages of cell growth and remain there until division is complete.
FIG. 3.
Localization of FtsE and FtsX to the division site. (A to D) Cells in exponential growth in LB with NaCl were fixed and examined by fluorescence microscopy directly (A and B), by indirect immunofluorescence microscopy (C and D), or by phase-contrast microscopy (C′ and D′). Strains shown are EC1063 (P204-ftsX-gfp) (A); EC1065 (P206-gfp-ftsX) (B); DHB4/pDSW609 (Plac-ftsE-3xHA) (C and D). (E) Relationship between cell length (age) and septal localization of FtsX. 509 cells of EC1063 were measured and scored for the presence (closed symbols) or absence (open symbols) of a fluorescent band at the midcell.
Complementation tests were used to determine whether the fusion genes used in the localization studies encoded functional division proteins. This was done by cloning the fusion genes into plasmids that confer Ampr and express the fusions under control of a weak IPTG-inducible promoter. Plasmids that express gfp-ftsX or ftsX-gfp rescued growth of RG60 on LB plates lacking NaCl when cotransformed with pBAD33-ftsE (Table 2). Likewise, a plasmid that expressed ftsE-3xHA rescued RG60 when cotransformed with pBAD33-ftsX. We conclude that our fusions to ftsE and ftsX function in cell division. Moreover, these results confirmed the expected polarity of ftsE400::kan onto ftsX, since expression of ftsE alone was not sufficient to rescue RG60.
TABLE 2.
Complementation of the ftsE400::kan allele in RG60a
| Plasmid(s) | Arabinose | Glucose |
|---|---|---|
| pBAD33 | − | − |
| pBAD33-ftsE | − | − |
| pBAD33-ftsX | − | − |
| pBAD33-ftsEX | + | − |
| pBAD33-ftsE + pDSW209 (vector) | − | − |
| pBAD33-ftsE + pDSW636 (gfp-ftsX) | + | − |
| pBAD33-ftsE + pDSW210 (vector) | − | − |
| pBAD33-ftsE + pDSW637 (ftsX-gfp) | + | − |
| pBAD33-ftsX + pDSW206 (vector) | − | − |
| pBAD33-ftsE + pDSW638 (ftsE-3xHA) | + | − |
Complementation was determined by colony formation when streaked onto LB plates with no NaCl but containing arabinose or glucose to modulate expression of genes under control of PBAD. IPTG was not necessary for complementation.
Position of FtsX in the recruitment pathway.
Numerous studies of Fts protein localization in E. coli have revealed a set of dependencies that are generally interpreted to reflect the order of assembly of these proteins into a multiprotein complex, sometimes called the septal ring (reviewed in references 11, 22, and 31). The first protein to localize is FtsZ, which polymerizes into a ring on the inner surface of the cytoplasmic membrane. FtsA, ZipA, and ZapA (14) bind directly to FtsZ and localize next. Then come FtsK, FtsQ, FtsL/YgbQ (probably as a complex), FtsW, FtsI, FtsN, and AmiC. To determine where FtsX fits into this hierarchy, we examined the effects of inactivation of various fts genes on septal localization of FtsX fused to GFP. We also did a complementary set of experiments that investigated which Fts proteins do and do not localize properly upon depletion of FtsEX.
We introduced our fusions into strains that had conditional alleles of ftsZ, ftsA, zipA, ftsK, ftsQ, ftsL, or ftsI. Because several of these conditional mutants have kanamycin resistance elements inserted into the gene of interest, we subcloned our original gfp-ftsX fusion, which was linked to a kanamycin marker, into a plasmid that confers spectinomycin resistance (7). This plasmid, pDSW533, was then integrated into the chromosome of MG1655 at the attachment site for phage φ80 as described elsewhere (16) to create strain EC1116. About half the cells of EC1116 displayed localization of GFP-FtsX to the septal ring (Table 3), as was observed for the same fusion integrated at the phage λ attachment site in EC1065 (Fig. 3B).
TABLE 3.
Localization of GFP-FtsX or FtsX-GFP in fts mutant backgroundsa
| Mutant | Growth condition | No. of cells scored | Avg cell length (μm) | Total no. of FtsX rings | % Cells with a ring(s) | Spacing of ringsb |
|---|---|---|---|---|---|---|
| Wild type | 30°C | 254 | 4.5 | 103 | 44 | 10 |
| 42°C | 225 | 7.2 | 91 | 40 | 18 | |
| Temperature-sensitive mutants | ||||||
| ftsZ(Ts) | 30°C | 135 | 13 | 36 | 27 | 15 |
| 42°C | 101 | 34 | 0 | 0 | >3,400 | |
| ftsA(Ts) | 30°C | 218 | 6.3 | 121 | 56 | 11 |
| 42°C | 112 | 36 | 95c | 60 | 42 | |
| zipA(Ts) | 30°C | 174 | 4.0 | 53 | 30 | 13 |
| 42°C | 105 | 33 | 4 | 3.8 | 880 | |
| Depletion strains | ||||||
| FtsK | Arabinose | 208 | 3.0 | 112 | 54 | 5.7 |
| Glucose | 145 | 18 | 261 | 98 | 10 | |
| FtsQ | Arabinose | 187 | 6.9 | 123 | 66 | 11 |
| Glucose | 132 | 28 | 231 | 91 | 18 | |
| FtsL | Arabinose | 220 | 8.1 | 157 | 71 | 11 |
| Glucose | 72 | 38 | 152 | 83 | 18 | |
| FtsW | Arabinose | 265 | 4.5 | 113 | 43 | 11 |
| Glucose | 61 | 29 | 104 | 89 | 17 | |
| FtsI | Arabinose | 212 | 4.5 | 111 | 52 | 8.6 |
| Glucose | 111 | 22 | 230 | 92 | 10 |
Strains used were EC1116, EC1158, EC1152, EC1340, EC1295, EC1179, EC1180, EC1159, and EC1181.
Units are micrometer per ring. The spacing is a measure of the frequency of rings per unit cell mass and was calculated by dividing the total number of rings (column 5) into the total length of cells or filaments scored (column 3 multiplied by column 4).
These bands were faint, and so the numbers given understate the degree of dependence on FtsA.
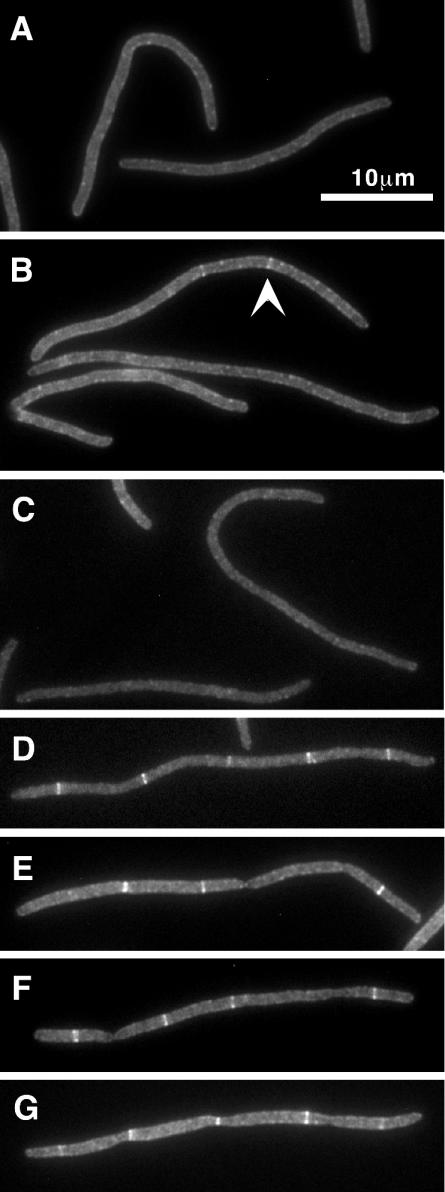
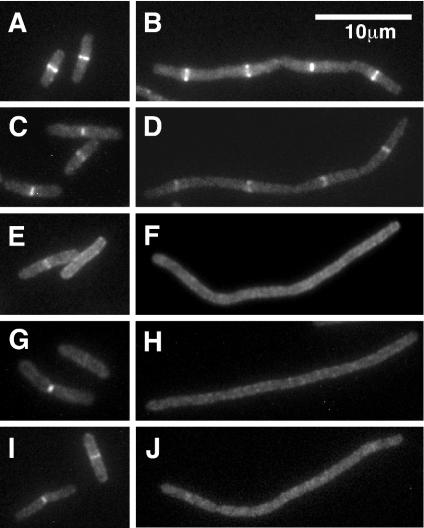
Localization of GFP-FtsX was then assayed in filaments that formed upon inactivation or depletion of the indicated essential division proteins. These results are summarized in Fig. 4 and Table 3. GFP-FtsX did not localize in filaments that developed when FtsZ ring formation was inhibited by shift of an ftsZ(Ts) mutant to the nonpermissive temperature. Similar results were obtained when FtsZ ring assembly was blocked by induction of the FtsZ-binding protein sulA (25, 36) from a pBAD plasmid (38) in cells maintained at 30°C (data not shown). Inactivation of the zipA1(Ts) allele, or depletion of ZipA (17) (data not shown), also greatly reduced localization of GFP-FtsX. Similarly, GFP-FtsX localized poorly in an ftsA12(Ts) background at the nonpermissive temperature. Although some GFP-FtsX localization was seen in the ftsA12(Ts) filaments, these bands were invariably faint (e.g., Fig. 4B, top filament). Whether this residual localization reflects leakiness of the Ts phenotype or a small degree of FtsA independence is not known. In contrast, most filaments formed by depleting cells of FtsK contained multiple fluorescent bands that were as bright as those seen in dividing cells. Likewise, GFP-FtsX localized well in filaments depleted of FtsQ, FtsL, or FtsI. Taken together, these results imply that FtsX localizes after FtsZ, FtsA, and ZipA, but before FtsK and subsequent proteins.
FIG. 4.
Effect of fts mutations on localization of FtsX to potential division sites in filamentous E. coli cells. Strains induced to express GFP-FtsX or FtsX-GFP were grown under nonpermissive conditions until they became filamentous, at which time they were fixed and examined by fluorescence microscopy to visualize GFP. Relevant division mutations are as follows: ftsZ84(Ts) in EC1158 (A), ftsA12(Ts) in EC1152 (B), and zipA1(Ts) in EC1340 (C); and FtsK depletion in EC1295 (D), FtsQ depletion in EC1179 (E), FtsW depletion in EC1159 (F), and FtsI depletion in EC1181 (G). The arrowhead in panel B points to a faint band sometimes observed in ftsA(Ts) filaments. 4′,6′-Diamidino-2-phenylindole staining was done to verify proper nucleoid segregation (data not shown).
To determine which Fts proteins are dependent upon FtsEX for septal localization, we constructed a set of FtsEX depletion strains that harbored gfp fusions to various division genes. The results are summarized in Fig. 5 and Table 4. Depletion of FtsEX had only a minimal effect on localization of FtsA and ZipA. In contrast, localization of FtsK, FtsQ, and FtsI was markedly reduced in FtsEX depletion filaments, and the occasional fluorescent bands observed were quite faint. Sporadic localization of small amounts of FtsK, FtsQ, and FtsI might reflect partial assembly of the septal ring in the absence of FtsEX. Alternatively, there might be some residual FtsEX in the filaments. In either case, these findings imply that FtsEX localizes after FtsZ, FtsA, and ZipA, but before FtsK and other downstream proteins, and are consistent with the behavior of the GFP-FtsX protein in fts mutant backgrounds (see above).
FIG. 5.
Localization of various division proteins after depletion of FtsEX. The strains used express ftsEX under control of an arabinose-dependent promoter and harbor gfp fusions to different division genes. These strains were grown in parallel in media containing arabinose (short cells) or glucose (filaments) and then fixed and examined by fluorescence microscopy to visualize GFP. (A and B) FtsA-GFP in EC1363; (C and D) ZipA-GFP in EC1391; (E and F) FtsK (1-266)-GFP in EC1386; (G and H) GFP-FtsQ in EC1392; (I and J) GFP-FtsI in EC1366. 4′,6′-Diamidino-2-phenylindole staining was done to verify nucleoid segregation (data not shown).
TABLE 4.
Localization of Fts proteins in FtsEX depletion backgrounda
| GFP fusion | Sugar | No. of cells scored | Avg cell length (μm) | % of cells with the indicated no. of rings
|
Spacing of ringsb | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | |||||
| FtsA-GFP | Arabinose | 164 | 4.8 | 7 | 85 | 8 | 0 | 0 | 0 | 3.6 |
| Glucose | 93 | 35 | 0 | 7 | 23 | 13 | 23 | 34 | 7.3 | |
| ZipA-GFP | Arabinose | 224 | 4.1 | 20 | 79 | 1 | 0 | 0 | 0 | 5.0 |
| Glucose | 112 | 28 | 4 | 10 | 35 | 19 | 19 | 13 | 9.5 | |
| FtsK(1-266)-GFP | Arabinose | 243 | 6.3 | 49 | 51 | 0 | 0 | 0 | 0 | 12 |
| Glucose | 101 | 21 | 67 | 29c | 4c | 0 | 0 | 0 | 55 | |
| GFP-FtsQ | Arabinose | 217 | 7.2 | 35 | 65 | 0 | 0 | 0 | 0 | 11 |
| Glucose | 131 | 24 | 69 | 27c | 4c | 0 | 0 | 0 | 67 | |
| GFP-FtsI | Arabinose | 108 | 5.0 | 63 | 37 | 0 | 0 | 0 | 0 | 14 |
| Glucose | 66 | 23 | 68 | 32c | 0 | 0 | 0 | 0 | 72 | |
Strains used were EC1363, EC1391, EC1386, EC1392, and EC1366.
See Table 3, footnote b.
These bands were faint, and so the numbers given understate the degree of dependence on FtsEX.
The above experiments concerning localization of various Fts proteins in cells depleted of FtsEX were done in media without salt. Depletion of FtsEX in LB with 1% NaCl caused the cells to form short filaments (∼10 μm), about 40% of which exhibited localization of GFP-FtsI (data not shown). Because we lack antibodies against FtsEX, we do not know whether the better localization of GFP-FtsI was due to salt rescue of localization (as expected) or less effective depletion of FtsEX.
DISCUSSION
We have shown that FtsE and FtsX localize to the septal ring, which implies that FtsEX participates directly in the division process. In a culture growing with a doubling time of about 30 min, about half of the cells exhibited septal localization of FtsE and FtsX. These cells were the longer (older) ones in the population and included cells with deep constrictions. Apparently, FtsEX is not present at the division site in newborn cells but gets recruited to that site during the later stages of cell growth and remains there until division is complete. This pattern of timed localization to the septal ring is similar to what has been reported for many other Fts proteins (e.g., references 8, 12, and 38). An additional line of evidence for direct participation in cell division is that during depletion of FtsEX, cells stop dividing before any change in growth rate (mass increase) is apparent. Thus, filamentation does not appear to be a secondary consequence of some metabolic defect or ion imbalance only tangentially related to septum assembly. This is not a trivial observation, as there are numerous examples of mutations that exert indirect effects on cell division, such as mutations in nrdB, dnaK, secA, and ffh (6, 26, 27, 35).
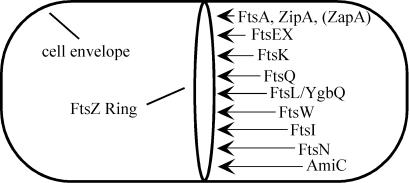
Previous studies from several laboratories have produced a model for the order of assembly of proteins into the septal ring in E. coli (for a recent review, see reference 11). A version of this model, revised to incorporate our new findings, is presented in Fig. 6. We infer that FtsEX localizes after FtsZ, FtsA, and ZipA and is important for recruitment of FtsK and all subsequent division proteins. Although we only demonstrated this directly for FtsK, FtsQ, and FtsI, previous work has established that septal localization of FtsL, YgbQ, FtsW, FtsN, and AmiC (3) requires prior localization of upstream proteins.
FIG. 6.
Model for recruitment of proteins to the septal ring. The first event is polymerization of FtsZ into the Z-ring. FtsA, ZipA, and ZapA bind directly to FtsZ and localize next or concomitantly with Z-ring assembly. Once either FtsA or ZipA has joined the septal ring, the remaining proteins localize in the order indicated. Whether any E. coli proteins are dependent upon ZapA is not yet known.
The ability of salt to rescue division in an FtsEX null mutant is enigmatic if FtsEX is needed for proper assembly or stability of the septal ring. Presumably, the downstream essential division proteins, FtsK through FtsN, all of which are required for division even on media containing salt, have some ability to localize in the absence of FtsEX. Consistent with this, RG60 is sensitive to β-lactams that target FtsI (D. Weiss, unpublished data) and we observed localization of FtsN, which is a late recruit to the division site (1), when RG60 was grown under permissive conditions. It is tempting to speculate that the ability of downstream proteins to localize independently of FtsEX is why we observed some residual localization of FtsK, FtsQ, and FtsI in filaments depleted of FtsEX in LB with no salt, but we cannot exclude the less interesting possibility that there is residual FtsEX in these filaments. Our depletion strains express ftsEX from a multicopy plasmid, so we sought to reduce the potential for leaky expression by placing a single copy of PBAD-ftsEX on the chromosome. Unfortunately, this configuration resulted in very poor complementation, presumably owing to too little ftsEX expression in the presence of arabinose.
To explain the salt-remedial nature of ftsEX null mutants, we propose that ionic conditions affect the folding, assembly, and/or function of one or more of the downstream division proteins, FtsK through FtsN. We further suggest that there is a synergistic effect when combined with loss of FtsEX such that the septal ring fails to assemble or function properly if both salt and FtsEX are lacking, but salt can rescue the ring in the absence of FtsEX, albeit poorly (recall that RG60 is filamentous even in the presence of salt).
An important question is whether FtsEX has any role in cell division beyond serving as an assembly or stability factor. In particular, one wonders whether FtsEX really is an ABC transporter and, if so, what it transports. Sequence comparisons indicate that FtsEX groups with importers rather than exporters (4). If FtsEX were an importer, it would be expected to function in conjunction with a periplasmic binding protein, although none has been associated with FtsEX as of yet. It has been speculated that FtsEX imports an ion (13), in part because of the salt-remedial nature of the defect in ftsEX mutants, but this notion is difficult to reconcile with the lack of specificity with respect to which salts rescue an ftsEX mutant (10). Moreover, FtsX does not appear to have any charged amino acids in its transmembrane domains, so it is difficult to envision how FtsX would accommodate an ion (R. Arends and D. Weiss, unpublished data). Finally, preliminary transcriptional profiling of an ftsEX null mutant has revealed a number of genes whose expression is altered, and none of these appears to be related to ion transport, ion homeostasis, or osmotic regulation (R. Arends and D. Weiss, unpublished data).
Not all ABC systems that group with importers actually import, or even transport, a substrate. Two interesting examples are the MacAB system and the LolCDE system, both of which are phylogenetically close to FtsEX (4). MacAB is an exporter that confers resistance to macrolides (21), while the LolCDE system is not a transporter at all—it is involved in release of lipoproteins from the cytoplasmic membrane (40). These observations make it worth considering functions for FtsEX that are unrelated to import. It has been suggested that FtsEX might be needed for insertion of a division protein into the cytoplasmic membrane (4, 36). An altogether different possibility is that FtsX serves as a membrane anchor, while FtsE uses ATP hydrolysis to promote constriction of the septal ring.
Acknowledgments
We thank Nienke Buddelmaijer, Joe Chen, Cynthia Hale, and Joe Lutkenhaus for strains and plasmids and Elie Dassa for information on ABC transporters, especially for calling our attention to MacAB and LolCDE.
These studies were supported by grants from the National Institutes of Health to D.S.W. (GM59893) and G.J.P. (GM50836). The DNA facility is supported by the Diabetes and Endocrinology Research Center with National Institutes of Health grant DK25295 and by the School of Medicine.
REFERENCES
- 1.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25:303-309. [DOI] [PubMed] [Google Scholar]
- 2.Bernatchez, S., F. M. Francis, H. Salimnia, T. J. Beveridge, H. Li, and J. A. Dillon. 2000. Genomic, transcriptional and phenotypic analysis of ftsE and ftsX of Neisseria gonorrhoeae. DNA Res. 7:75-81. [DOI] [PubMed] [Google Scholar]
- 3.Bernhardt, T. G., and P. A. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouige, P., D. Laurent, L. Piloyan, and E. Dassa. 2002. Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr. Protein Peptide Sci. 3:541-559. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukau, B., and G. C. Walker. 1989. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J. Bacteriol. 171:2337-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. C., D. S. Weiss, J. M. Ghigo, and J. Beckwith. 1999. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol. 181:521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 10.de Leeuw, E., B. Graham, G. J. Phillips, C. M. ten Hagen-Jongman, B. Oudega, and J. Luirink. 1999. Molecular characterization of Escherichia coli FtsE and FtsX. Mol. Microbiol. 31:983-993. [DOI] [PubMed] [Google Scholar]
- 11.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghigo, J. M., D. S. Weiss, J. C. Chen, J. C. Yarrow, and J. Beckwith. 1999. Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol. 31:725-737. [DOI] [PubMed] [Google Scholar]
- 13.Gill, D. R., G. F. Hatfull, and G. P. Salmond. 1986. A new cell division operon in Escherichia coli. Mol. Gen. Genet. 205:134-145. [DOI] [PubMed] [Google Scholar]
- 14.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale, C. A., and P. A. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale, C. A., and P. A. de Boer. 2002. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 184:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto-Gotoh, T., M. Yamaguchi, K. Yasojima, A. Tsujimura, Y. Wakabayashi, and Y. Watanabe. 2000. A set of temperature sensitive-replication/-segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene 241:185-191. [DOI] [PubMed] [Google Scholar]
- 20.Kempf, M. J., and M. J. McBride. 2000. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J. Bacteriol. 182:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi, N., K. Nishino, and A. Yamaguchi. 2001. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 23.Mercer, K. L., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merino, S., M. Altarriba, R. Gavin, L. Izquierdo, and J. M. Tomas. 2001. The cell division genes (ftsE and X) of Aeromonas hydrophila and their relationship with opsonophagocytosis. FEMS Microbiol. Lett. 198:183-188. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee, A., C. Cao, and J. Lutkenhaus. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver, D. B., and J. Beckwith. 1981. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 25:765-772. [DOI] [PubMed] [Google Scholar]
- 27.Phillips, G. J., and T. J. Silhavy. 1992. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359:744-746. [DOI] [PubMed] [Google Scholar]
- 28.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pogliano, J., K. Pogliano, D. S. Weiss, R. Losick, and J. Beckwith. 1997. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. USA 94:559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard, M., and Y. Hirota. 1973. Process of cellular division in Escherichia coli: physiological study on thermosensitive mutants defective in cell division. J. Bacteriol. 116:314-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothfield, L., S. Justice, and J. Garcia-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 32.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 33.Sun, Q., X. C. Yu, and W. Margolin. 1998. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol. Microbiol. 29:491-503. [DOI] [PubMed] [Google Scholar]
- 34.Taschner, P. E., P. G. Huls, E. Pas, and C. L. Woldringh. 1988. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J. Bacteriol. 170:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taschner, P. E., J. G. Verest, and C. L. Woldringh. 1987. Genetic and morphological characterization of ftsB and nrdB mutants of Escherichia coli. J. Bacteriol. 169:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trusca, D., S. Scott, C. Thompson, and D. Bramhill. 1998. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J. Bacteriol. 180:3946-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ukai, H., H. Matsuzawa, K. Ito, M. Yamada, and A. Nishimura. 1998. ftsE(Ts) affects translocation of K+-pump proteins into the cytoplasmic membrane of Escherichia coli. J. Bacteriol. 180:3663-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wissel, M. C., and D. S. Weiss. 2003. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J. Bacteriol. 186:490-502. [DOI] [PMC free article] [PubMed]
- 40.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212-218. [DOI] [PubMed] [Google Scholar]