Abstract
Background/Aims
We aim to quantify the effect of non-differential genotyping errors on the power of rare variant tests and identify those situations when genotyping errors are most harmful.
Methods
We simulated genotype and phenotype data for a range of sample sizes, minor allele frequencies, disease relative risks and numbers of rare variants. Genotype errors were then simulated using five different error models covering a wide range of error rates.
Results
Even at very low error rates, misclassifying a common homozygote as a heterozygote translates into a substantial loss of power, a result that is exacerbated even further as the minor allele frequency decreases. While the power loss from heterozygote to common homozygote errors tends to be smaller for a given error rate, in practice heterozygote to homozygote errors are more frequent and, thus, will have measurable impact on power.
Conclusion
Error rates from genotype-calling technology for next-generation sequencing data suggest that substantial power loss may be seen when applying current rare variant tests of association to called genotypes.
Key Words: Sequencing data, Power, Case-control, Misclassification
Background
Given the inability of common variants alone to sufficiently explain common disease heritability and the advent of next-generation sequencing (NGS) technology, the attention of genome-wide association (GWA) studies has turned toward the common disease-rare variant hypothesis, which suggests that the primary contributors to common disease susceptibility may be rare genetic variations, typically presumed to be single-nucleotide variants (SNVs) [1,2]. However, the most widely used GWA testing methods are not viable for the analysis of rare SNVs (e.g. those occurring with minor allele frequency (MAF) less than 5%), as single variant testing methods are generally underpowered for detecting signals which occur so infrequently.
For the analysis of rare variant data, a new class of methods has been proposed which attempts to test aggregated sets of SNVs, such as genes or sets derived from metabolic pathways and other biologically relevant sets [3,4,5,6,7,8,9,10,11,12,13]. By aggregating SNVs to sets, the goal is to magnify the strength of the signal so that reasonable power for association tests can be obtained using reasonable sample sizes. While this new class of rare variant tests offers promise for the analysis of rare variants, little is known about these methods outside of the idealized simulated data environments where they have been developed. One realistic consideration that has been largely ignored in the development of rare variant tests is that of genotyping errors, which are being reported at high levels in NGS data [14,15,16,17]; higher than those observed in the early days of SNP microarray technology [18].
Current genotyping algorithms follow a series of three steps to determine genotypes, with errors possible at any of the three steps [16]. In the first step, short reads are genotyped. Errors in determining genotypes at this stage have been documented to follow an auto-regressive process dependent on the true allele [19], with an error of approximately 0.5% per base. The second step of the process aligns these short reads to a reference genome, a process generally accepted to have a very low error rate [19]. Lastly, a Bayesian prior is used when determining genotypes that utilizes known information about the population MAFs at each variant site. The prior is then updated with the observed sequences, yielding a posterior probability that an individual is of a particular genotype. When there are many reads (high coverage/depth, e.g. 30x) it is easy to overcome the prior and call a rare variant. But at lower sequencing depth, this becomes harder to do. Specifically, in low-depth sequence data, some genotype callers (e.g. individual-based callers) underestimate the total amount of rare novel variation present in the sample, while others (e.g. population-based and linkage disequilibrium-aware callers) improve genotype calling for low frequency variants, but perform even more poorly at identifying singletons and doubletons.
The end result of the genotype calling process from rare variants is a high heterozygote to homozygote error rate, and a lower homozygote to heterozygote error rate for rarely seen variants in the same (e.g. singletons, doubletons). As the number of observed alleles in the sample increases, the likelihood that a common homozygote is called a heterozygote increases. One exception is when the reference genome has an allele that is not common in the sample being sequenced, in which case individual-based genotype callers have a difficult time identifying someone as possessing the common homozygote. Potential genotyping errors at all stages of the production of NGS data suggest the need for research into the effect of genotyping errors on downstream analysis of NGS data: including the use of rare variant tests of association [20].
The effects of genotyping errors on common variant tests of association (single-marker methods) have been well explored [21,22,23,24,25,26,27]. Extensions of the results for measured common variants have also been extended to tests conducted with imputed common variants [28,29,30]. Specifically, differential genotyping errors, which occur with different probability in cases and controls, can inflate the type I error rate of common variant tests [22,23,31,32], while ‘non-differential’ genotyping errors, which occur with equal frequency in cases and controls, maintain type I error but decrease statistical power [25,26,27].
In particular, research into the effects of errors on common variant tests have found that errors that misclassify a major allele as a minor allele are most detrimental and that the minimum sample size necessary to maintain power and significance level in the presence of this type of error increases without bound as the MAF approaches 0 [24,25,26]. This last point is particularly concerning for the analysis of rare variant data.
Recent research suggested that differential genotyping error resulting from different sequencing depths for cases and controls can bias rare variant tests and increase type I error [33]. But to date, little effort has been put into exploring the potential effects of non-differential genotyping errors on the power of rare variant tests.
In this paper, we use simulation to evaluate the impact of non-differential genotyping errors on the power of four commonly considered rare variant tests of genetic association (CMC [3], WS [4], PR [5] and CMAT [6]). We start by evaluating the impact on type I error in the tests. Then, we evaluate the effects of genotyping error on power. Specifically, we contrast two types of genotyping error: misclassifying the common homozygote as the heterozygote and misclassifying the heterozygote as the common homozygote, with the goal of identifying situations where genotyping errors are particularly harmful. Our simulation analysis considers genotyping error rates for rare variant tests spanning those reported in recent publications.
Methods
Simulation of Genotypes and Phenotypes
To investigate the impact of genotyping errors on rare variant tests, we first simulated genotype data according to an additive disease model. Simulations to assess power and type I error covered all 24 possible combinations of the following four parameters: disease relative risk (γ = 1.00, 1.25 or 2.00), sample size (n = 500 or 2,000, equally split between cases and controls), number of SNVs in the set (e.g. gene) (M = 8 or 64) and SNV MAF of rare/common SNVs in the set (0.1%/1.0% or 0.5%/5.0%). In each set of SNVs, one quarter of the SNVs in the set are more common (having a MAF of 1 or 5%), and three quarters of the SNVs in the set are rarer (having a MAF of 0.1 or 0.5%).
Genotypes were simulated based on the specific number of SNVs and the MAF distribution of the SNVs, while assuming independence of variants and Hardy-Weinberg equilibrium at each variant site. Simulation of phenotypes followed a model similar to that of Li and Leal for CMC [3]. In our simulation, we set the wild-type penetrance f0 to 0.01 for all individuals with no rare alleles at any of the M variant sites within the set (e.g. gene). In accordance with an additive disease model, individuals with rare alleles had a total disease risk equal to f0 (1 + Σ(gi − 1)), gi = 1 if variant and gi = 0 otherwise. In our simulations, we let γ1i be the same for all i variants in the set. The phenotypes for each individual were simulated using a Bernoulli random variable in R [34].
Simulating Genotyping Errors
Following the simulation of genotypes and phenotypes, errors were added to the genotypes. Let ∊01 denote the probability of misclassifying a common homozygote as heterozygote, and let ∊10 denote the probability of misclassifying a heterozygote as common homozygote. Genotyping errors involving the less common homozygote were not considered here because of their extremely low observed frequency in samples and differences in how rare variant tests handle less common homozygotes, which would limit our ability to compare methods.
Three different error models were considered in the primary simulation study: (model 1) ∊01 = p, ∊10 = 0, (model 2) ∊01 = 0, ∊10 = p and (model 3) ∊01 = ∊10 = p. We considered 10 values of p: 0.000, 0.001, 0.005, 0.010, 0.015, 0.020, 0.025, 0.030, 0.040 and 0.050. Genotyping errors were simulated without regard to an individual's disease status to illustrate non-differential genotyping error. We also extended our simulation study to reflect the fact that the use of a prior distribution means that genotype calling algorithms make it difficult to detect the rarest variants, and so in practice ∊10 may be much greater than ∊01. In an extended simulation we considered the following two models: model 4: (0, 0), (0, 0.01), (0, 0.05), (0, 0.10), (0, 0.15), (0, 0.20), (0, 0.25), (0, 0.30), (0, 0.40) and (0, 0.50), model 5: (0, 0), (0.001, 0.01), (0.005, 0.05), (0.01, 0.10), (0.015, 0.15), (0.02, 0.20), (0.03, 0.30), (0.04, 0.40) and (0.05, 0.50), where ordered pairs of values represent (∊01, ∊10).
Rare Variant Tests Used to Analyze Data
CMC Method
The CMC method aggregates the genotype data from multiple variant sites within a set of interest (e.g. all rare variants within a gene) to a dichotomous variable for each subject (1 if the subject has any minor alleles in the set and 0 otherwise). The CMC approach collapses variants within a priori defined subsets and then performs a multivariate test, treating each collapsed subset as a SNV. In our case, we collapse all rare variants with MAF strictly less than 1% into a single group, and then asymptotically evaluate Hotelling's T2 on the groups (one group of SNVs with MAF less than 1%, in addition to each SNV with MAF 1% or greater treated as its own group).
WS Method
The novelty of the WS method is that it uses a weighting scheme to put more emphasis on rarer SNVs. For each variant site, a weight is calculated proportional to an estimate of the standard error of the total number of minor alleles in controls. For each subject, a score is summed over all variant sites, equal to the number of minor alleles at each site divided by the site's weight. The subjects are ranked by score from greatest to least, and the test statistic is defined as the sum of the rankings of all cases. To obtain a p value for the test, case/control status is permuted among the individuals 1,000 times to obtain an empirical distribution under the null hypothesis.
PR Method
The PR method models subjects’ case/control statuses as binary responses to a univariate logistic regression for which the lone covariate is the proportion of rare variant sites at which a subject has at least one minor allele. Formally, for each subject and rare variant site, a variable is defined as 1 if the subject has any minor alleles at the site and 0 otherwise. For each subject, the covariate is defined as the average of these dichotomous variables across all rare variant sites. A hypothesis test of whether the regression coefficient for the covariate is significantly different from zero is conducted using the asymptotic distribution of the likelihood ratio test statistic.
CMAT Method
CMAT is similar in spirit to a 2×2 χ2 test. The four values constituting the 2×2 table are the total number of minor alleles among all controls, the total number of major alleles among all controls, the total number of minor alleles among all cases and the total number of major alleles among all cases. Rather than using an asymptotic χ2 distribution to obtain the corresponding p value to the test statistic, the significance of the test is determined using phenotype permutations to avoid assumptions such as linkage equilibrium.
Estimating Power and Type I Error
For all cases where the relative risk is set to 1 (no genotype-phenotype association), 3,000 random samples were generated for each combination of simulation model parameters (γ1i = 1, n, M, MAF, ∊01 and ∊10). For situations where the relative risk is greater than 1, 5,000 random samples were generated for each combination of the simulation model parameters. Type I error and power were estimated as the proportion of the simulated samples with p values less than a significance level of 0.05.
Explaining the Impact of Genotyping Error on Power
We used regression models to further explore the effects of simulation parameters (error level, sample size, MAF and relative risk) on power loss. Specifically, we created a regression model predicting the change (y) in power due to a 0.5% increase in error rates. Explanatory variables in the model included power (x1) before the 0.5% increase in error, error rate (x2) before increase, sample size (x3), MAF (x4) and relative risk (x5).
y = β0 + β1x1 + β2x2 +β3x3 +β4x4 +β5x5 + ∊: ∊ ∼ N(0, σ2)
Significant explanatory variables in this model indicate variables which impact power loss differentially for a fixed error increase. In order to evaluate change in power at 0.5% increments of change in error, only six levels of error are considered: 0, 0.5, 1.0, 1.5, 2.0, and 2.5%. Thus our regression spans 96 data points (16 simulation configurations × 6 error levels).
Results
Type I Error Rate
For each of the 80 simulation settings used to assess type I error rate (all combinations of n, M and MAF for the 10 error levels in error model 3), the four rare variant test methods maintained a 5% type I error rate. The average type I error rate across the 80 simulation settings was 5 (WS), 4.7 (CMC and PR) and 4.3% (CMAT). Linear models (details not given), regressing the type I error rate on the simulation parameters (sample size, number of SNVs, SNV MAF and error rate), suggest that a small sample size makes CMC more conservative; while a small sample size, a small number of SNVs and a small SNV MAF each cause both PR and CMAT to be more conservative. The type I error rate of WS was unaffected by any of the simulation parameters. Importantly, the type I error rate was not associated with the size of the genotyping errors for any of the methods.
Power
The following paragraphs consider the impact of genotyping errors on the power of rare variant tests. Each type of error (∊01, ∊10) is first considered separately (models 1 and 2), and then a more complex error model (model 3) is considered which allows both errors to occur simultaneously.
Impact of Common Homozygote to Heterozygote Errors
Table 1 shows the average power of each of the four methods across the 16 combinations of risk (1.25 and 2.00), sample size, number of SNVs and MAF for different error simulation settings. Considering situations where only common homozygote to heterozygote errors are present (∊01 > 0, ∊10 = 0; model 1), each of the four rare variant methods shows significant loss of power, even for low genotyping error rates. For example, the methods averaged about 2% loss in power from as little as a 0.1% homozygote to heterozygote error rate. As ∊01 increases, larger power losses are observed. For example, when ∊01 = 1%, power loss was approximately 10%, and when ∊01 = 5%, power loss was approximately 25–30%. This trend held true across the 16 genotype-phenotype simulation settings.
Table 1.
Average power by genotype error ratea
| Error rate | CMC |
WS |
PR |
CMAT |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ε01 only (model 1) | ε10 only (model 2) | both (model 3) | ε01 only (model 1) | ε10 only (model 2) | both (model 3) | ε10 only (model 1) | ε10 only (model 2) | both (model 3) | ε01 only (model 1) | ε10 only (model 2) | both (model 3) | |
| 0% | 60.2% | 60.4% | 60.3% | 76.8% | 76.6% | 76.8% | 72.4% | 72.4% | 72.5% | 71.4% | 71.4% | 71.6% |
| 0.1% | 58.7% | 60.5% | 58.4% | 74.8% | 76.8% | 75.0% | 70.9% | 72.4% | 71.1% | 70.0% | 71.4% | 70.1% |
| 0.5% | 53.5% | 60.3% | 53.4% | 70.2% | 76.6% | 69.9% | 67.2% | 72.4% | 66.9% | 66.3% | 71.4% | 66.0% |
| 1% | 47.7% | 60.1% | 47.1% | 65.4% | 76.4% | 65.2% | 62.9% | 72.1% | 62.8% | 62.2% | 71.1% | 62.1% |
| 1.5% | 42.4% | 60.1% | 41.9% | 61.9% | 76.3% | 61.7% | 59.8% | 72.1% | 59.4% | 59.1% | 71.1% | 58.7% |
| 2.0% | 38.2% | 59.9% | 37.6% | 59.2% | 76.4% | 58.5% | 57.2% | 72.0% | 56.4% | 56.6% | 71.1% | 55.7% |
| 2.5% | 36.2% | 60.0% | 35.5% | 56.9% | 76.3% | 56.3% | 55.0% | 72.1% | 54.3% | 54.3% | 71.1% | 53.6% |
| 3.0% | 34.4% | 59.9% | 33.6% | 54.6% | 76.1% | 54.0% | 52.7% | 71.7% | 52.0% | 52.1% | 70.8% | 51.4% |
| 4.0% | 32.4% | 60.0% | 31.4% | 51.4% | 76.1% | 50.3% | 49.6% | 71.7% | 48.4% | 49.2% | 70.7% | 47.8% |
| 5.0% | 30.3% | 59.6% | 29.2% | 48.6% | 76.0% | 47.2% | 46.7% | 71.6% | 45.1% | 46.2% | 70.6% | 44.7% |
Margin of error is ≤0.35% for all estimates.
Table 2 gives the results of the regression for five covariates explaining the change in power due to a 0.5% increase in genotyping error rate. Within the common homozygote to heterozygote only error model (model 1) there were some significant relationships between the covariates and power loss due to increased error. MAF showed a significant impact on power loss for WS, PR and CMAT. Because these coefficients are positive, this means that decreasing the MAF increases the impact of genotyping errors. While CMC did not have a significant change in power loss based on MAF, an increased number of SNVs and increased relative risk were both significantly associated with increased power loss due to error. Lastly, all methods showed significantly less effect of errors on power as error rates increased, meaning that, after controlling for overall power, the most substantial power loss for a given increase in error rates was observed moving from 0 to 0.5% genotyping errors.
Table 2.
Coefficients from regression models
| Parameter | ε01 only (model 1) |
ε10 only (model 2) |
Both (model 3) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CMC | WS | PR | CMAT | CMC | WS | PR | CMAT | CMC | WS | PR | CMAT | |
| Previous power | −0.080∗ | 0.020 | 0.009 | 0.011 | −0.002 | −0.001 | −0.001 | −0.000 | −0.085∗ | 0.015 | 0.007 | 0.010 |
| Error level | −0.027∗∗∗ | −0.016∗∗ | −0.012∗ | −0.011∗ | −0.000 | 0.001 | 0.000 | 0.000 | −0.029∗∗∗ | −0.017∗∗ | −0.013∗ | −0.012∗ |
| Sample sizea | −0.026 | 0.008 | 0.004 | 0.004 | 0.000 | 0.000 | 0.000 | 0.001 | −0.027 | 0.007 | 0.003 | 0.003 |
| Number of SNVsa | −0.026∗∗ | 0.004 | −0.000 | −0.001 | −0.001 | 0.000 | 0.000 | 0.000 | −0.027∗∗ | 0.002 | −0.002 | −0.002 |
| MAFa | −0.010 | 0.036∗∗ | 0.032∗ | 0.032∗ | −0.001 | 0.000 | 0.000 | 0.000 | −0.011 | 0.035∗ | 0.034∗ | 0.034∗ |
| Relative riska | −0.058∗∗ | 0.017 | 0.007 | 0.007 | −0.000 | −0.000 | 0.000 | 0.000 | −0.062∗∗ | 0.016 | 0.007 | 0.007 |
p < 0.05
p < 0.01
p < 0.001.
Coefficients for these terms are interpreted as the change in the reduction in power from moving from the high to the low simulation setting. For example, for sample size, values in the table indicate how power losses change as one moves from having 2,000 to 500 individuals in the study.
Impact of Heterozygote to Common Homozygote Errors
In general, the impact of heterozygote to common homozygote errors on power (model 2) is much lower than the impact of common homozygote to heterozygote errors (model 1). Table 1 illustrates only modest decreases in power as the value of ∊10 decreases. In summary, over the four methods and 16 sets of simulation parameters, the average decrease in power due to a 0.1% rate of misclassifying the heterozygote as the major homozygote was 0.1%. The average decrease in power due to a 5% rate of misclassifying the heterozygote as the major homozygote was 4.1%. While the loss in power due to heterozygote to major homozygote genotyping error is minimal, separate simple linear regressions (details not shown) of power versus the error rate indicate that power is significantly related to error rate for each of the four methods. So the impact of the error is small but real. Finally, our regression analysis (table 2) shows that under this error scheme, there was no covariate in any of the four models which had a significant impact on the effect on power of a 0.5% error increase.
Equal Errors Model
Overall, in the equal errors model (model 3), power is similar to that of model 1, since the unique impact of heterozygote to homozygote errors (model 2) is minimal (see table 1). Similarly, the coefficients of the regression model in table 2 for the equal errors model (model 3) are also very similar to their counterparts for the homozygote to heterozygote only error model (model 1).
Consideration of Other Genotyping Error Models
Table 3 gives results analogous to those in table 1, except it considers error models 4 and 5, which were designed to represent error models closer to those observed when using current genotype calling algorithms for NGS data. Namely, genotype error rates from the heterozygote to the common homozygote are much larger than the reverse, due to the fact that genotype callers for rare variant data make extremely rare variants difficult to detect. First, table 3 illustrates that when heterozygote to homozygote error rates increase to values 10 times greater than those considered in error model 2, there can be appreciable power loss. However, when these much larger error rates are combined with substantially lower (10 times smaller) common homozygote to heterozygote errors, the observed power loss is still mainly driven by the homozygote to heterozygote errors.
Table 3.
Average power by genotype error ratea
| Error rate (ε10) | ε10 only (model 4) |
Error rate (ε01,ε10) | Both (model 5; 10 (ε10) = ε10) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| CMC | WS | PR | CMAT | CMC | WS | PR | CMAT | ||
| 0.0% | 60.1% | 76.2% | 72.1% | 71.1% | 0.0%, 0.0% | 60.4% | 76.4% | 72.3% | 71.3% |
| 1.0% | 60.1% | 76.2% | 72.2% | 71.1% | 0.1%, 1.0% | 58.6% | 74.6% | 71.0% | 69.9% |
| 5.0% | 59.7% | 75.5% | 71.5% | 70.4% | 0.5%, 5.0% | 52.4% | 68.8% | 65.9% | 64.9% |
| 10% | 59.0% | 75.0% | 70.8% | 69.6% | 1.0%, 10% | 44.7% | 62.4% | 60.0% | 59.0% |
| 15% | 58.0% | 74.0% | 69.5% | 68.3% | 1.5%, 15% | 38.2% | 57.5% | 55.4% | 54.5% |
| 20% | 57.5% | 73.0% | 68.5% | 67.3% | 2.0%, 20% | 32.3% | 53.1% | 51.0% | 50.2% |
| 25% | 56.6% | 72.1% | 67.4% | 66.0% | 2.5%, 25% | 28.3% | 48.6% | 46.6% | 45.8% |
| 30% | 55.9% | 71.1% | 66.1% | 64.7% | 3.0%, 30% | 25.5% | 44.5% | 42.6% | 41.8% |
| 40% | 53.6% | 68.9% | 63.3% | 61.9% | 4.0%, 40% | 20.7% | 36.9% | 34.4% | 33.7% |
| 50% | 50.6% | 65.7% | 59.7% | 58.3% | 5.0%, 50% | 16.1% | 29.5% | 26.9% | 26.3% |
Margin of error is ≤0.35% in all cases.
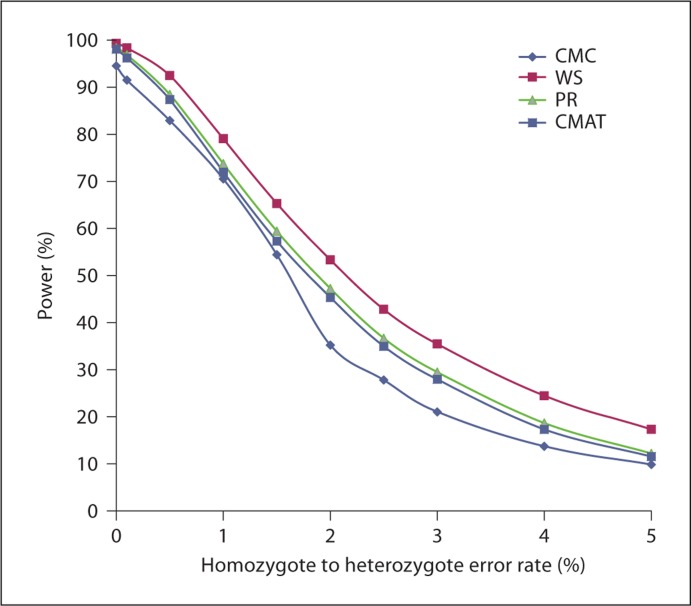
To further illustrate the significance of this power loss due to error, we consider one particular choice of simulation parameters. Figure 1 shows the power curves of the four methods as functions of the error level for the setting of 1,000 cases and 1,000 controls genotyped to test the association of a gene which includes 8 rare variants (6 SNVs with MAF 0.1% and 2 SNVs with MAF 1%) with the phenotype which follows an additive disease model with all SNVs causal with relative risk 2.00. Power decreases slowly as a function of error level when it is at the top and bottom of the range and decreases most quickly near the value 50%.
Fig. 1.
An example of how the power for each of the four methods diminishes as the error level increases in error model 5 for one choice of simulation settings (high sample size, low number of SNVs, low MAF and high relative risk). Heterozygote to homozygote error rates are 10 times larger than homozygote to heterozygote error rates. The margin of error is ≤1.41% in all cases.
Discussion
Our results confirm that all four rare variant methods maintain type I error in the presence of non-differential genotyping errors under all simulation settings considered here. However, power loss from non-differential genotyping errors can be substantial, even for low error rates, and is particularly problematic for errors misclassifying the common homozygote as a heterozygote, i.e. identifying an individual as possessing a risk allele when one is not actually present. Power loss can also be particularly substantial for variant sites with low MAF.
As has been documented for single marker tests of association, there can be substantial power loss from genotyping errors, especially errors calling an individual a heterozygote when they are actually the common homozygote. This substantial power loss is not surprising when put into context; for example, if the common homozygote to heterozygote error rate is 2% and the MAF of a SNV is ≤1%, then about two-thirds of heterozygote observations at that base pair in the data will be incorrect genotype calls. In fact, whenever the population MAF for a SNV is less than half the error rate, it is likely that the majority of minor alleles observed at that variant site are not actually minor alleles but genotype misclassifications. This point is readily seen when one realizes that the observed rare variants will be a mixture of errors from the common homozygote (observed at a rate of ∊01(1 – MAF)2) and true heterozygotes ((1 – ∊10)2MAF(1 – MAF)). For small MAF, the errors from the common homozygote will be approximately ∊01, while the true heterozygotes will be at most about 2MAF; thus, a reasonable rule of thumb is that whenever 2MAF <∊01, the majority of minor alleles observed are errors.
Power loss for heterozygote to homozygote errors is substantially less than that from homozygote to heterozygote errors of the same rate. Furthermore, power loss from a mix of error types (homozygote to heterozygote and heterozygote to homozygote) showed a semi-additive impact on power loss. These results are in line with the results by Kang et al. [24,25] who explored common variant tests of association.
However, observed rates for heterozygote to common homozygote errors can be very large because current genotype calling algorithms make it very difficult to call rare alleles. Genotype calling algorithms are tuned to minimize the type I error in the genotype calling process, making it difficult to discover rare novel variants. The choice of genotype caller can significantly affect the performance of rare variant testing methods. Population- and linkage disequilibrium-aware callers exacerbate the bias against discovery and genotyping of rare non-reference variants. This increases the heterozygote to common homozygote error rate. In case of imputed genotypes, the choice of reference panel for imputation can have a higher impact on imputation accuracy of rare variants. While the general strategy for genotype calling algorithms is reasonable in light of the finding that homozygote to heterozygote errors are so costly, it would be valuable to investigate whether current genotype calling algorithms are ‘tuned’ correctly so that genome-wide power loss is minimized.
One promising method for reducing the impact of genotyping errors on downstream analysis is the use of probabilistic genotypes, instead of genotype calls. Current genotype calling algorithms provide a vector of posterior probabilities for each individual at each of the three possible genotypes. These posterior probabilities capture the uncertainty in genotypes that exists after applying a genotype calling algorithm. In downstream tests of association on imputed genotypes, using posterior probabilities has been shown to provide increased power over the use of called genotypes [35], a result that has also been shown for measured common variants [36,37]. Similarly, another recently proposed approach [12] incorporates genotyping errors directly into the test statistic to handle the hazards outlined in this paper.
Table 1 suggests that, on average, power loss is similar across all four methods (WS, PR, CMAT and CMC), suggesting that no one method is more or less susceptible to genotyping error than the others. While the goal of our simulation was not to provide a comprehensive comparison of the four rare variant methods considered here, for our choices of simulation settings, WS almost always yielded the most power. In fact, the relative ordering of the power of the different methods (WS, PR, CMAT and CMC) stayed the same in almost all cases.
While our analysis has provided a broad overview of the impact of genotyping errors on rare variant tests of association, there are limitations to our analysis. First, while we have used a published simulation technique to generate genotypes and phenotypes, other more sophisticated models exist, including modeling other disease modes of inheritance, simulating haplotypes instead of distinct genotypes and assessing the impact of neutral (non-causal) variants on resulting tests of association. Furthermore, many more rare variant analysis methods have been proposed than were compared in this paper. While the impact of genotyping errors may be different in different tests and in different settings, at this point we have no reason to believe that the general pattern of results will not hold in these cases. Additionally, we considered a broad range of simulation parameters, but the novelty of the field of analysis of rare variants with real data limits our ability to say that parameters chosen in our models are necessarily realistic. This is a limitation of any current methodological paper related to rare variant analysis. Lastly, our analysis is limited to association studies which may be more prone to error and less powerful than some family-based association studies for rare variants, as well as to only four of the most common rare variant tests of association. Further work is necessary to explore these alternative tests.
Overall, we have found that genotyping errors can have substantial impact on the power of rare variant tests. Even small (0.1%) non-differential error rates can produce significant power loss when errors are made in identifying the more common homozygote as a heterozygote. Today's genotype calling algorithms generally have much higher error rates for heterozygotes to homozygotes, which produce less power loss in general but cause measurable power loss as rates increase. In all cases, power loss is magnified as the MAF decreases, a particularly concerning result for tests involving the rarest variants. Care should be taken in the design and analysis of rare variant studies to consider the potential impact of genotyping errors on rare variant tests of association.
Acknowledgements
This work was funded by the National Human Genome Research Institute (R15HG004543). We acknowledge the input of Alexander Luedtke and Airat Bekmetjev in early phases of this project, Paul Van Allsburg and the Hope College parallel computing cluster Curie for assistance in data simulation.
References
- 1.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin in Genet Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zawistowski M, Gopalakrishnan S, Ding J, Li Y, Grimm S, Zöllner S. Extending rare-variant testing strategies: analysis of noncoding sequence imputed genotypes. Am J Hum Genet. 2010;87:604–617. doi: 10.1016/j.ajhg.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Zhang H, Yu K. Approaches for evaluating rare polymorphisms in genetic association studies. Hum Hered. 2010;69:219–228. doi: 10.1159/000291927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Byrnes AE, Li M. To identify associations with rare variants, just WHaIT: weighted haplotype and imputation-based tests. Am J Hum Genet. 2010;87:728–735. doi: 10.1016/j.ajhg.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price AL, Kryukov GV, de Bakker PIW, Purcell SM, Staples J, Wei LJ, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell SM, Roeder K, Daly MJ. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan W, Shen X. Adaptive tests for association analysis of rare variants. Genet Epidemiol. 2011;35:381–388. doi: 10.1002/gepi.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon D, Finch SJ, De La Vega F. A new expectation-maximization statistical test for case-control association studies considering rare variants obtained by high-throughput sequencing. Hum Hered. 2011;71:113–125. doi: 10.1159/000325590. [DOI] [PubMed] [Google Scholar]
- 13.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Awadalla P, Gauthier J, Myers RA, Casals F, Hamdan FF, Griffing AR, Côté M, Henrion E, Spiegelman D, Tarabeux J, Piton A, Yang Y, Boyko A, Bustamante C, Xiong L, Rapoport JL, Addington AM, DeLisi JLE, Krebs MO, Joober R, Millet B, Fombonne É, Mottron L, Zilversmit M, Keebler J, Daoud H, Marineau C, Roy-Gagnon MH, Dubé MP, Eyre-Walker A, Drapeau P, Stone EA, Lafreniére RG, Rouleau GA. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am J Hum Genet. 2010;87:316–324. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilie L, Fazayeli F, Ilie S. HiTEC: accurate error correction in high-throughput sequencing data. Bioinformatics. 2011;27:295–302. doi: 10.1093/bioinformatics/btq653. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;12:443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ledergerber C, Dessimoz C. Base-calling for next-generation sequencing platforms. Brief Bioinform. 2011 doi: 10.1093/bib/bbq077. DOI: 10.1093/bib/bbq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tintle NL, Ahn K, Mendell NR, Gordon D, Finch SJ. Characteristics of replicated single-nucleotide polymorphism genotypes from COGA: affymetrix and center for inherited disease research. BMC Genet. 2005;6(suppl 1):S154. doi: 10.1186/1471-2156-6-S1-S154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bansal V, Libiger O, Torkamani A, Schork NJ. Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet. 2010;11:773–785. doi: 10.1038/nrg2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon D, Finch SJ. Factors affecting statistical power in the detection of genetic association. J Clin Invest. 2005;115:1408–1418. doi: 10.1172/JCI24756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moskvina V, Craddock N, Holmans P, Owen MJ, O'Donovan MC. Effects of differential genotyping error rate on the type I error probability of case-control studies. Hum Hered. 2006;61:55–64. doi: 10.1159/000092553. [DOI] [PubMed] [Google Scholar]
- 23.Ahn K, Gordon D, Finch SJ. Increase of rejection rate in case-control studies with differential genotyping error rates. Stat Appl Genet Mol Biol. 2009;8:25. doi: 10.2202/1544-6115.1429. [DOI] [PubMed] [Google Scholar]
- 24.Kang SJ, Gordon D, Finch SJ. What SNP genotyping errors are most costly for genetic association studies? Genet Epidemiol. 2004;26:132–141. doi: 10.1002/gepi.10301. [DOI] [PubMed] [Google Scholar]
- 25.Kang SJ, Finch SJ, Haynes C, Gordon D. Quantifying the percent increase in minimum sample size necessary for SNP genotyping errors in genetic model-based association studies. Hum Hered. 2004;58:139–144. doi: 10.1159/000083540. [DOI] [PubMed] [Google Scholar]
- 26.Ahn K, Haynes C, Kim W, St. Fleur R, Gordon D, Finch SJ. The effects of SNP genotyping errors on the power of the Cochran-Armitage linear trend test for case/control association studies. Ann Hum Genet. 2007;71:249–262. doi: 10.1111/j.1469-1809.2006.00318.x. [DOI] [PubMed] [Google Scholar]
- 27.Gordon D, Finch SJ, Nothnagel M, Ott J. Power and sample size calculations for case-control genetic association tests when errors are present: application to single nucleotide polymorphisms. Hum Hered. 2002;54:22–33. doi: 10.1159/000066696. [DOI] [PubMed] [Google Scholar]
- 28.Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, Scheet P. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L, Wang C, Rosenberg NA. The relationship between imputation error and statistical power in genetic association studies in diverse populations. Am J Hum Genet. 2009;85:692–698. doi: 10.1016/j.ajhg.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beecham GW, Martin ER, Gilbert JR, Haines JL, Pericak-Vance MA. APOE is not associated with Alzheimer disease: a cautionary tale of genotype imputation. Ann Hum Genet. 2010;74:189–194. doi: 10.1111/j.1469-1809.2010.00573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pluzhnikov A, Below JE, Konkashbaev A, Tikhomirov A, Kistner-Griffin E, Roe CA, Nicolae DL, Cox NJ. Spoiling the whole bunch: quality control aimed at preserving the integrity of high-throughput genotyping. Am J Hum Genet. 2010;87:123–128. doi: 10.1016/j.ajhg.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitry D, Campbell H, Charteris DG, Fleck BW, Tenesa A, Dunlop MG, Hayward C, Wright AF, Vitart V. SNP mistyping in genotyping arrays – an important cause of spurious association in case-control studies. Genet Epidemiol. 2011;35:423–426. doi: 10.1002/gepi.20559. [DOI] [PubMed] [Google Scholar]
- 33.Garner C. Confounded by sequencing depth in association studies of rare alleles. Genet Epidemiol. 2011;35:261–268. doi: 10.1002/gepi.20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Development Core Team: R: A Language and Environment for Statistical Computing. The R-Project, v2.11.1. www.r-project.org
- 35.Zheng J, Li Y, Abecasis GR, Scheet P. A comparison of approaches to account for uncertainty in analysis of imputed genotypes. Genet Epidemiol. 2011;35:102–110. doi: 10.1002/gepi.20552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tintle NL, Gordon D, McMahon FJ, Finch SJ. Using duplicate genotyped data in genetic analyses: testing association and estimating error rates. Stat App Genet Mol Biol. 2007;6:4. doi: 10.2202/1544-6115.1251. [DOI] [PubMed] [Google Scholar]
- 37.Borchers B, Brown M, McLellan B, Bekmetjev A, Tintle NL. Incorporating duplicate genotype data into linear trend tests of genetic association: methods and cost-effectiveness. Stat App Genet Mol Biol. 2009;8:24. doi: 10.2202/1544-6115.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]