Abstract
Listeria monocytogenes is well known for its durable physiological characteristics, which allow the organism to grow at low temperature and pH and high osmolarity. Growth under high osmolarity depends on the accumulation of compatible solutes, among which glycine betaine and carnitine are the preferred solutes for this organism. Three different transport systems, Gbu, BetL, and OpuC, have been identified in L. monocytogenes which serve to scavenge the preferred compatible solutes. The general stress response regulator σB has been shown to play an important role in osmotic adaptation in L. monocytogenes, presumably by directing transcription from one or more of the solute transport genes. In the studies presented here, we have used primer extension analyses to identify the promoter elements responsible for transcription of the opuC, gbuA, and betL genes. All three genes are osmotically inducible to some degree. betL is transcribed from a σB-independent promoter, while gbuA is transcribed from dual promoters, one of which is σB dependent. opuC is transcribed exclusively from a σB-dependent promoter. The betL promoter is similar in sequence to the σB-independent gbuAP1 promoter. Kinetic analysis of transcript accumulation after osmotic upshift demonstrated that σB-dependent transcripts from gbuAP2 and sigB accumulate for an extended period after upshift, suggesting that σB activity may provide a mechanism for sustained high-level expression during osmotic challenge. In contrast to osmotic upshift, expression from the σB-dependent opuC and gbuAP2 promoters after temperature upshift and ethanol stress was minimal, suggesting that additional mechanisms may also participate in regulating transcription from these σB-dependent promoters.
Listeria monocytogenes is a gram-positive food-borne pathogen that is ubiquitous in the environment and problematic to the food production industry due to its ability to grow at refrigeration temperatures, at low pH, and elevated osmolarity (10, 20, 22, 35). Studies on growth and survival of L. monocytogenes under elevated osmolarity have shown a crucial role for transport of organic compounds such as glycine betaine (N,N,N-trimethylglycine) (7, 18) and carnitine (β-hydroxy-γ-N-trimethylaminobutyrate) (7, 32). These compounds, known as compatible solutes or osmolytes, can accumulate to high intracellular concentrations and relieve turgor pressure without significantly affecting the activity or function of cellular components (36). Although several structural analogs of glycine betaine, including dimethylhetin, β-butyrobetaine, acetylcarnitine proline, proline betaine, glutamate, choline, small peptides, and trehalose can be used as osmoprotectants by different bacterial species (17), betaine and carnitine are the preferred compatible solutes in L. monocytogenes (7, 18, 32). After osmotic upshift, these osmolytes, which are found primarily in plant and animal environments, accumulate to high intracellular concentrations via active transport in L. monocytogenes rather than by de novo synthesis (18, 19, 31, 32).
Transport of betaine and carnitine in L. monocytogenes has been studied at both the biochemical and genetic levels. Physiological studies have demonstrated the existence of an ATP-dependent carnitine transport system (32) and an ATP-dependent, salt-inducible betaine transporter in strain Scott A (19). In addition, a sodium ion-driven betaine transport system was reported in membrane vesicles (14). These biochemical observations have been confirmed recently by cloning of the three distinct transporters, BetL, Gbu, and OpuC (3, 11, 19, 26), and demonstration that these three genes likely encode the entire complement of betaine and carnitine transporters in L. monocytogenes (2, 34). The BetL protein of strain LO28 is a betaine transporter, belonging to a Na+-dependent symport family of secondary transporters (26). Deletion of betL in L. monocytogenes results in slow growth in media with elevated osmolarity but no significant effect on virulence (27). Gbu, encoded by the gbuABC operon, is an ATP-dependent transporter belonging to the ATP binding cassette (ABC) superfamily of transporters, and it is homologous to OpuA in Bacillus subtilis and ProU in Escherichia coli (19). Analysis of the gbu operon revealed the presence of three protein subunits. GbuA is an ATPase, GbuB is a transmembrane protein, and GbuC is a substrate-binding protein. Disruption of gbuA results in reduced growth and a low rate of accumulation of betaine when cells are grown in minimal media with 8% NaCl or at low temperatures (19). Finally, two laboratories independently identified a gene encoding a third osmolyte transporter, opuC, which is an ABC transporter specific for carnitine (3, 11). The opuCABCD operon is homologous to opuC and opuB of B. subtilis and is composed of genes encoding two permeases, one extracellular solute-binding protein, and one ATPase. OpuC is activated at low temperatures, and accumulation of carnitine is severely reduced in opuC null mutants (3, 12, 28). Studies with opuC mutants and strains carrying multiple mutations in betL, gbuA, and opuC have recently shown that OpuC is the primary carnitine transporter in L. monocytogenes (2).
In addition to the direct function of compatible solute transporters in the osmotic response of L. monocytogenes, the general stress response regulator, σB, also participates in the response to osmotic upshift. Osmotic upshift is one of the most potent stimulators of σB activity, and absence of σB impairs the ability of cells to accumulate both betaine and carnitine as osmoprotectants and cryoprotectants (5, 6, 13). DNA sequences upstream of the betL and opuC genes show some similarity to putative σB-dependent promoters (11, 26, 29), and recent studies using promoter fusions and reverse transcription-PCR (RT-PCR) have confirmed the dependence of opuC and gbuA expression on σB, providing a simple explanation for the participation of σB (13). To more precisely examine the role of σB in the expression of these osmolyte transporters, we have used primer extension and RT-PCR analyses to map transcription start sites from the three transporter genes and to examine the kinetics of transcript accumulation during osmotic upshift. We show in strain 10403S that gbuA and opuC are transcribed from osmotically inducible, σB-dependent promoters. The gbuA operon is also transcribed from a σB-independent promoter that is highly similar in sequence to the σB-independent promoter upstream of the betL gene. Kinetic analysis of transcript accumulation after osmotic upshift suggests that σB activity is maintained for long periods of time after osmotic upshift and may therefore be the mechanism used for expression of gbuA during prolonged periods of osmotic stress.
MATERIALS AND METHODS
Cloning of betL, opuC, and gbuA from L. monocytogenes.
Prior to identification of betL (26) and publication of the L. monocytogenes strain EGDe genome sequence (15), we cloned the betL gene from strain 10403S using the B. subtilis opuD gene as a probe. A 1,084-base fragment of the opuD gene of B. subtilis was amplified by Taq DNA polymerase using primers BSOPUDF (5′-AAACAGAAACCCCCCAAG-3′) and BSOPUDR (5′-GCAATCATCAGCAGGATAAC-3′). The product was blunt ended and cloned to the EcoRV site of the pBluescript KS(+) cloning vector. Preliminary Southern blot hybridization experiments with the cloned B. subtilis gene as a probe indicated that a single 4.5-kb HindIII fragment of the L. monocytogenes 10403S chromosome hybridized under conditions of moderate stringency. A plasmid library of 4- to 5-kb HindIII fragments was then constructed from gel-purified digested chromosomal DNA using the pUC19 vector. Approximately 400 colonies were screened using the B. subtilis opuD PCR product as a probe. One clone harboring a 5-kb HindIII fragment was obtained and designated p4B3 and used for further studies.
The opuC and gbuA promoter regions were amplified by PCR using OpuCF (5′-ATCGATGTCACACGTGCCACAAGTAC-3′), OpuCR (5′-TCTCGATAATGTCCATAACAGAAG-3′), Gbu5 (5′-TTTGTCCACATCCCTTTGA-3′), and Gbu3 (5′-ATATTGATCGCCGTAACCAG-3′). Primer design was based on the sequences from strain EGDe (15). The primers corresponded to positions −367 to −348 upstream and +427 to +447 downstream of the initiation codon of gbuA and −181 to −154 and +948 to + 926 of opuC, respectively. The opuC and gbuA amplicons were subsequently cloned into the pTOPO cloning vector using the topoisomerase-mediated ligation reaction (Invitrogen, Carlsbad, Calif.), and the plasmids were designated as pOPUC and pGBUA, respectively.
DNA sequence analysis.
Plasmid p4B3 was digested by ClaI, and the resulting two fragments were ligated separately to the ClaI site of pBluescript KS(+), designated as pCla2 and pCla3, and sequenced using universal primers. The remaining vector was self ligated, designated as pCla1, and sequenced using M13 forward and reverse primers. The PCR products from gbuA and opuC were also confirmed by DNA sequencing using the T3 and T7 primers. All DNA sequence analyses were performed using end-labeled sequencing in conjunction with a LiCor 4200 automated sequencer (LiCor, Inc., Lincoln, Nebr.), and the contigs were assembled using Sequencher (Gene Codes, Inc., Ann Arbor, Mich.).
Growth conditions and preparation of RNA.
Cells of L. monocytogenes 10403S were inoculated into 200 ml of brain heart infusion (BHI) medium with 1% of an overnight BHI-grown culture and incubated at 37°C with shaking at 200 rpm. At mid-logarithmic phase (optical density at 600 nm [OD600] of ∼0.5), the cells were harvested at room temperature and washed twice in 50 mM potassium phosphate buffer, pH 6.8. Cells were resuspended in the same buffer with 5 mM glucose for 20 min. Separate aliquots of the cells were then treated for 20 min by the addition of NaCl (3% final concentration). After 20 min, the cells were harvested by centrifugation at 6,000 × g for 5 min, and the cell pellets were frozen at −70°C overnight. The cells were subsequently resuspended in 1 ml of Tri reagent (Molecular Research Center, Inc., Cincinnati, Ohio) solution and disrupted by homogenization with beads four times at 1-min intervals on ice. Total RNA was extracted according to the manufacturer's directions.
Primer extension analysis of betL, opuC, and gbuA transcripts.
Primers (100 pmol each) complementary to the 5′ ends of the betL (PEX3, 5′-ATTCTCTGTATCTATTTCCCCATG-3′), opuCA (CEX3, 5′-AATTCTCCTTTATCGATGTTAAGTG-3′), and gbuA (AEX3, 5′-CACCAATTGTCGCGCCTGTTTTCTTTC-3′) coding regions were end labeled with 50 μCi of [γ-32P]ATP. For each extension reaction, 10 pmol of labeled primer was then mixed with 50 μg of RNA, heated to 80°C for 5 min, chilled on ice for 5 min, and incubated at 42°C with 3 U of SuperScript II RNase H-reverse transcriptase (Invitrogen). After 1 h of extension, the products were precipitated with ethanol, washed in 70% ethanol, and dissolved in loading buffer, as described previously (5). Sequencing ladders were generated by double-strand sequencing reactions using Sequenase 2.0 (U.S. Biochemicals, Cincinnati, Ohio). Ladders were primed with unlabeled PEX3, CEX3, and AEX3 primers using the pCla1, pOPUC, and pGBUA plasmids, respectively, as templates. Each of the primer extension reactions was performed on RNA samples extracted from at least two independent cultures or treatments. For band quantification, bands were excised using an autoradiogram as a guide. Portions of the lanes with no apparent band were also excised and used to estimate background (three slices averaged for each background measurement). The quantification was performed at least twice on independent RNA samples. The graphs shown are derived from a single experiment and are representative of the results from independent experiments.
RT-PCR analyses.
For each sample, 10 μg of total RNA was treated twice to remove the trace contaminating DNA using the DNA-free kit (Ambion, The RNA Company, Austin, Tex.) according to the manufacturer's instructions. cDNA was synthesized from 5 μg of total RNA using SuperScript II RNase H-reverse transcriptase (Gibco, BRL, Cleveland, Ohio) and 250 ng of random hexamer primers according to the manufacturer's instructions. One microliter of the cDNA reaction mixture was used as a template for amplification in PCR. The specific primers used for PCR amplification were the following: SigB1, 5′-GCGCCGAATCAAAGAGTTAGG-3′; SigB2, 5′-CCACTTCCTCATTCTGCAACG-3′; BetL1, 5′-AAGTCCGATTGGCTCGATTC-3′, BetL2, 5′-ATCAAGTCCGGACATAGCCG-3′; OpuCA1, 5′-AATGGAGGTGTGTAGGCGTG-3′ OpuCA2, 5′-GTAATTGGATCTAGCGCGCC-3′; GbuAP1-1, 5′-TGGGCCGAATTTTTGACCTAG-3′ GbuAP1-2, 5′-CGCTCTTCTTTGTCCATTCC-3′; GbuAP2-1, 5′-CTAATTGAGCCTACGAGCGG-3′; and GbuAP2-2, 5′-TGAACGACAGAACCATCACG-3′.
The PCRs were performed in a total volume of 20 μl containing 20 pmol of each primer, 100 μM deoxynucleoside triphosphate, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 1 U of Taq DNA polymerase (Sigma, St. Louis, Mo.). The PCR cycling protocol was 95°C for 2 min; 21 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 45 s; and 72°C for 5 min for final extension. PCR products were electrophoresed on 0.7% agarose gels, poststained with ethidium bromide (0.5 μg/ml), and visualized under UV. Control reactions to confirm absence of DNA were performed using each of the RNA samples with no cDNA synthesis step and yielded no distinguishable amplification products (data not shown). Each experiment was repeated at least twice using independent RNA samples.
RESULTS
BetL expression is constitutive and independent of σB.
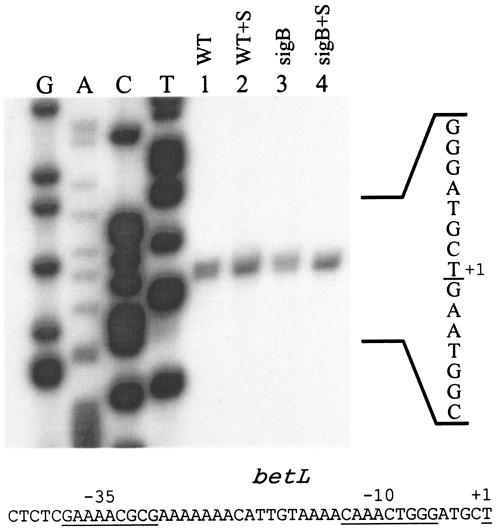
In a previous study, our investigators showed that the absence of σB in L. monocytogenes impaired the ability of these cells to use betaine as an osmoprotectant and specifically that the sigB mutation blocked osmotically inducible betaine accumulation (5). Slot blot and RT-PCR analysis of betL expression in strains LO28 and 10403S, respectively, showed that expression was slightly osmotically inducible and independent of σB (13, 26). To identify the promoter element, we used primer extension with a primer (PEX3) that is complementary to positions −39 to −17 relative to the start codon. As shown in Fig. 1, extension of this primer on mRNA extracted from logarithmically growing 10403S and LMA2B (sigB::Kan) cells yielded a 134-base product before and after the strains were subjected to an osmotic upshift. Although a prominent product was present prior to osmotic upshift, addition of 3% salt in both strains increased the abundance of the primer extension product, indicating that accumulation of the transcript is osmotically inducible. Since the primer extension product was present in the sigB::Kan mutant, we conclude that it is not dependent on σB. Using the PEX3 and four additional primers, we were never able to observe a primer extension product in the 10403S strain mapping to the putative σB-dependent promoter proposed by Sleator et al. (26). Inspection of the sequences at −10 and −35 relative to the transcription start site (Fig. 1) revealed sequence elements that do not display substantial similarity to known promoters in other gram-positive bacteria.
FIG. 1.
Primer extension analyses of transcripts originating from the betL promoter. Cells were grown in BHI (OD600, ∼0.5), harvested, washed, and resuspended in 50 mM phosphate potassium buffer with 50 mM glucose. The washed cell suspension from each strain was divided into two equal volumes, and NaCl was added to a 3% final concentration in one portion. After a 20-min incubation, RNA was harvested from the cells, and 50 μg of each RNA sample was used as template for extension of the labeled PEX3 primer. Strain designations are as follows: 10403S (WT); 10403S plus 3% NaCl (WT+S); LMA2B (sigB::Kan) (sigB); LMA2B plus 3% NaCl (sigB + S). The sequence ladder was derived from reactions using the PEX3 primer, and the top strand sequence is shown to the right of the ladder (reverse complement of the sequence ladder). The nucleotide corresponding to the 3′ end of the extension product is underlined. The corresponding top strand sequence is shown below, with the +1, −10, and −35 regions underlined.
gbuA is transcribed from dual promoters.
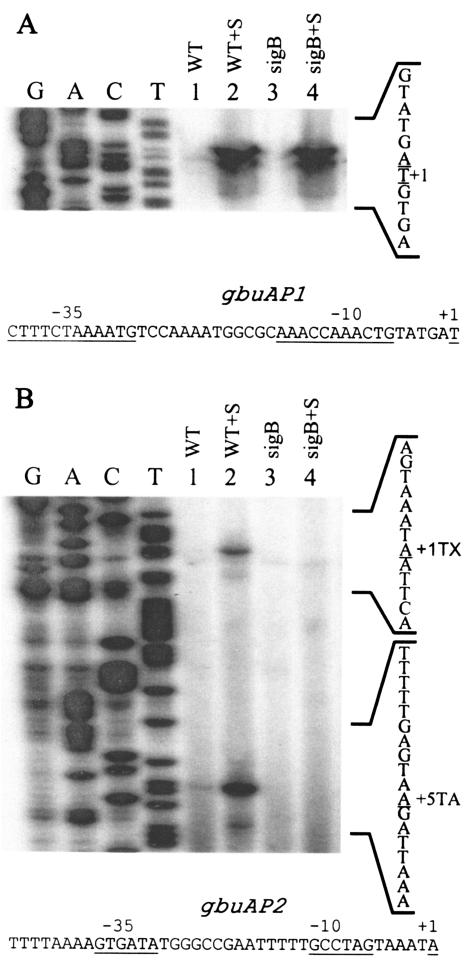
Since betL transcription is not dependent on σB, we postulated that the sigB phenotype of impaired betaine transport must be due to its effect on gbuA. We therefore investigated transcription of the gbuA operon in wild-type and sigB mutant cells using primer AEX3, which is complementary to positions +100 to +126 relative to the start codon. As shown in Fig. 2, three different primer extension products were detected. The largest product was induced after osmotic upshift (compare lanes 1 and 2 in Fig. 2A) and was present in the sigB mutant background, indicating that it is osmotically inducible but does not depend on σB. Inspection of the sequences upstream of this start site showed that this promoter region, termed gbuAP1, is unique in sequence and does not conform to a consensus σA-like promoter from the closely related species B. subtilis (Fig. 2A).
FIG. 2.
Primer extension analyses of transcripts originating from the gbuA promoter. Cells were grown in BHI (OD600, ∼0.5), harvested, washed, and resuspended in 50 mM potassium phosphate buffer with 50 mM glucose (final concentration). NaCl was added to a 3% final concentration to one portion of the cells. After a 20-min incubation, RNA was extracted, and 50 μg of each RNA sample was used as a template for extension of the labeled AEX3 primer. (A) Transcripts originating from the gbuAP1 promoter; (B) transcripts originating from the σB-dependent gbuAP2 promoter. Both panels were derived from the same autoradiogram. Lane 1,10403S (WT); lane 2, 10403S plus 3% NaCl (WT+S); lane 3, LMA2B (sigB::Kan) (sigB); lane 4, LMA2B(sigB::Kan) plus 3% NaCl (sigB+S). The sequence ladder was derived from sequencing reactions using the AEX3 primer, and the reverse complement sequence is shown to the right. The nucleotides corresponding to the 3′ end of the primer extension products (transcription initiation site) are underlined. The extension products in panel B are labeled +1TX (σB-dependent transcription initiation site) and +5TA (extension product mapping to +5 relative to translation). The top strand sequences of the regions 5′ to the gbuAP1 and gbuAP2 transcription start sites are shown below each panel with the relevant +1, −10, and −35 regions indicated.
In contrast to gbuAP1, the remaining extension products (Fig. 2B) were osmotically inducible but absent in the sigB null mutant background (compare lanes 3 and 4). Mapping of the σB-dependent primer extension products to the gbuA sequence showed that the longer product maps to a position −20 relative to the initiation codon, while the shorter product maps to +5 relative to the start codon. Alignment of sequences 5′ to these extension products showed that sequences at −10 and −35 relative to the −20 extension product aligned with σB-dependent promoters, while the sequences relative to the +5 extension product did not. Therefore, we conclude that the +5 extension product is likely derived from premature termination of the reverse transcriptase or due to a breakdown product derived from the longer transcript. Identification of the σB-dependent promoter, which we have termed gbuAP2, therefore provides an explanation for the phenotype of impaired osmotically inducible accumulation of betaine in sigB mutant strains.
Mapping of the σB-dependent opuC promoter.
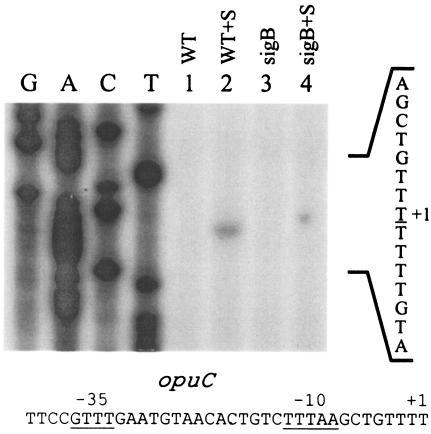
Genetic and expression analyses of the opuC transport system, which is the primary carnitine transport system in L. monocytogenes, have previously shown that carnitine transport depends on σB and that opuC expression is also σB dependent (6, 13). Sequences upstream of the coding region also display similarity to known σB promoters (13), supporting the idea that σB may be solely responsible for expression of the primary carnitine transporter in L. monocytogenes. Indeed, RT-PCR analyses in sigB mutant strains showed that the product from the opuC gene is nearly undetectable (13). To confirm the location of the start site, and to assess whether other promoters may drive opuC transcription, we mapped transcripts originating upstream of the opuCA gene using primer extension. A primer (CEX3) that is complementary to positions +63 to +88 relative to the start codon was end labeled and used for primer extension analysis. As shown in Fig. 3, a primer extension product was observed mapping to position −100 relative to the opuCA-coding region. This was the only product that was observed in our experiments, and it was not observed in the sigB mutant background. Alignments of the sequences upstream of this transcription start site revealed −10 and −35 elements that resembled known σB-dependent promoters but did not correspond to the start site predicted from the sequence analysis by Fraser et al. (11). Based on the results of our primer extension analyses, we conclude that the opuCABCD operon is transcribed exclusively from a σB-dependent promoter in the 10403S strain, which would explain the primary dependence of carnitine transport on sigB (6, 13).
FIG. 3.
Primer extension analyses of transcripts originating from the opuC promoter. Cells were grown in BHI (OD 600∼0.5), harvested, washed, and resuspended in 50 mM potassium phosphate buffer with 50 mM glucose (final concentration). NaCl was added to a final concentration of 3% to a portion of the cells, where appropriate. After a 20-min incubation, RNA was extracted, and 50 μg of each RNA sample was used as a template for extension of labeled CEX3 primer. Lanes: 1,10403S (WT); 2, 10403S plus 3% NaCl (WT+S); 3, LMA2B (sigB::Kan) (sigB); 4, LMA2B plus 3% NaCl (sigB+S). The sequence ladder was derived from reactions using the CEX3 primer, and the sequence shown to the right corresponds to the reverse complement. The nucleotide corresponding to the 3′ end of the extension product is underlined. The top strand sequence of the region immediately upstream of the transcription start site is shown at the bottom with the relevant + 1, −10, and −35 regions indicated.
Response of opuC and gbuA to heat and ethanol shock.
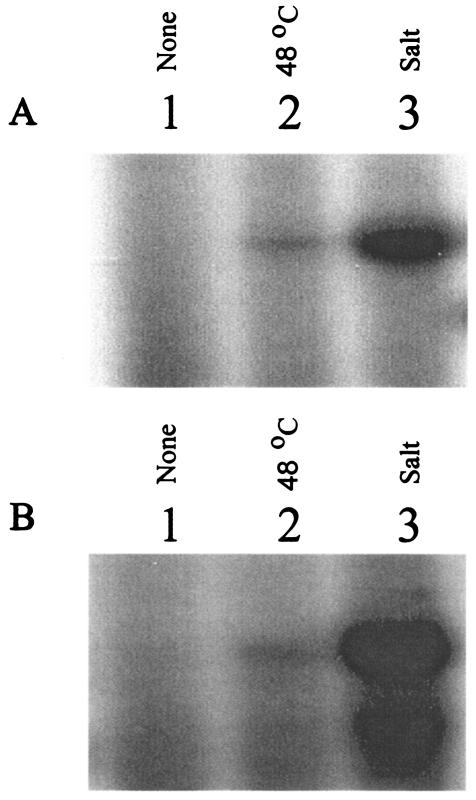
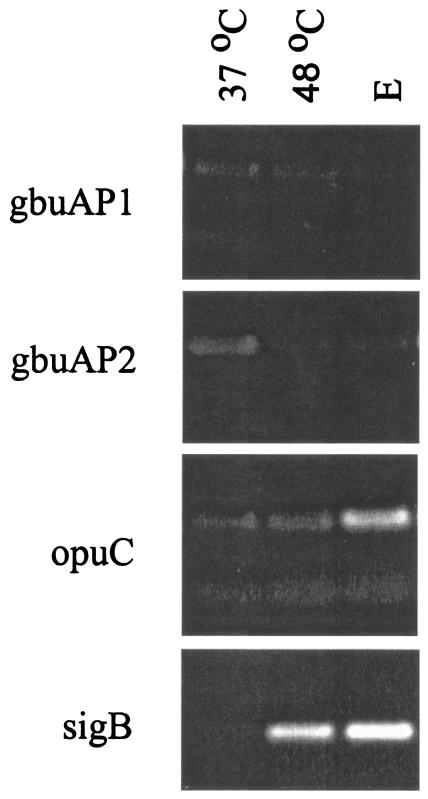
Because σB activity is induced by a number of different stress conditions (5), we hypothesized that induction of σB activity by nonosmotic stress would also lead to σB-dependent transcription of the gbuA and opuC osmolyte transporters, unless additional mechanisms were present to prevent induction. To test this hypothesis, we used primer extension and RT-PCR to examine transcripts originating from the σB-dependent gbuAP2 and opuC promoters after cells had been subjected to osmotic or temperature upshift. Cells for these experiments were grown in BHI at 37°C to mid-log phase, washed, and resuspended in transport buffer with glucose. After equilibration, the cells were subjected to osmotic upshift or temperature upshift to 47°C for 20 min. As shown in Fig. 4, transcripts originating from the σB-dependent opuC and gbuAP2 accumulated substantially after osmotic upshift but were barely detectable after temperature upshift. Since σB activity is induced significantly by heat shock (5), it is surprising that its activities at opuC and gbuAP2 were not detectable after the temperature upshift. To confirm the primer extension results, we also used RT-PCR to examine transcripts from gbuA, opuC, and sigB. To differentiate transcription originating at gbuAP1 from gbuAP2, we compared RT-PCR products using two different sets of primers. Primers gbuAP1-1 and gbuAP1-2, which are immediately 3′ to gbuAP1, were used to detect transcripts originating at gbuAP1. Primers gbuAP2-1 and gbuAP2-2, positioned 3′ to gbuAP2, were used to detect transcripts originating at both gbuAP1 and gbuAP2 promoters. As shown in Fig. 5, none of the products from gbuAP1 or gbuAP2 increased significantly in abundance when log-phase cells were compared to cells subjected to temperature upshift or addition of ethanol. RT-PCR products from opuC showed a slight increase in abundance after addition of ethanol but not after temperature upshift. In contrast, RT-PCR products from sigB increased substantially after either temperature upshift or addition of ethanol. Therefore, both the primer extension and the RT-PCR data suggest that σB is necessary for directing transcription from opuC and gbuAP2, but additional mechanisms likely control the ability of the σB RNA polymerase holoenzyme to form transcription-competent complexes at these promoters. These mechanisms presumably serve to restrict upregulation of transcription to osmotic stress conditions.
FIG. 4.
Transcripts originating from opuC and gbuA promoters after temperature upshift. Mid-logarithmic-phase cells of strain 10403S (wild type) grown in BHI were washed and resuspended in 50 mM potassium phosphate buffer with 50 mM glucose and divided into three equal portions. One portion was untreated (lane 1, 37°C), one was subjected to temperature upshift to 48°C for 15 min (lane 2), and the third was subjected to addition of 3% NaCl for 20 min (lane 3). (A) Extension products from opuC; (B) products from gbuAP2.
FIG. 5.
RT-PCR analysis of transcripts from opuC, gbuA, and sigB after temperature upshift and ethanol stress. Mid-logarithmic-phase cells of strain 10403S (WT) grown in BHI were washed and resuspended in 50 mM potassium phosphate with 50 mM glucose and divided into equal portions. The portions were subjected to the following treatments: no treatment (lane 1, 37°C), temperature upshift to 48°C for 30 min (lane 2, 48°C) and addition of 4% ethanol for 30 min (lane 3, E). cDNA was prepared from 5 μg of RNA extracted from each of the samples and amplified with primers adjacent to the σB-dependent promoter of the sigB operon (sigB), the gbuAP1 promoter (gbuAP1), the gbuAP2 promoter (gbuAP2), and the opuC promoter (opuC). After 21 cycles of PCR, the products were electrophoresesd and stained with ethidium bromide. Negative control reactions, consisting of RNA samples without cDNA synthesis, were subjected to PCR and showed no detectable amplification products (data not shown).
Kinetics of gbuA and opuC transcription.
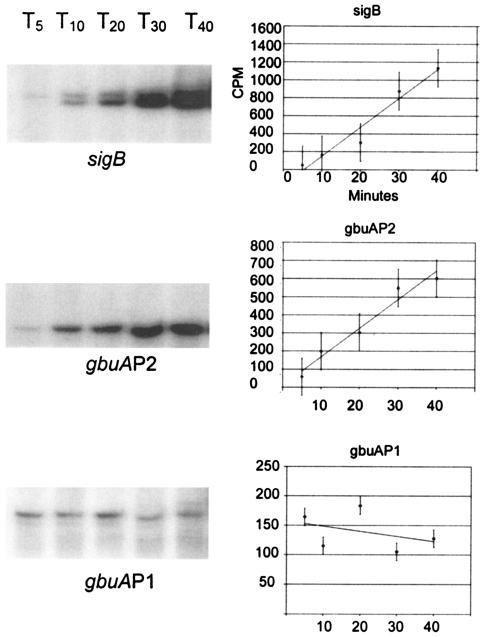
In B. subtilis, measurement of σB activity from promoter fusions and accumulation by Western blot analyses suggests that its activity at most promoters peaks between 10 and 20 min after exposure to activating conditions and then falls to a new level that is higher than before the shock (9). The σB-dependent promoter of the proline transporter, opuE, in B. subtilis follows this temporal pattern of activity after osmotic upshift, since transcripts accumulate for up to 20 min but are undetectable after 40 min (30). In contrast, transcripts originating from the osmotically inducible σA-dependent promoter upstream of opuE continue to accumulate for an extended period after the upshift (30). Our discovery of the dual gbuAP1 and gbuAP2 promoters provided an opportunity to similarly compare kinetics of osmotic activation of σB and another holoenzyme form in L. monocytogenes. We therefore measured transcripts originating from these promoters and the σB-dependent transcript from the sigB operon over time after an osmotic upshift. Surprisingly, the σB-dependent transcript from both sigB and the gbuAP2 promoters in L. monocytogenes did not accumulate transiently but continued to accumulate even up to 40 min postinduction (Fig. 6). Measurement of the band intensity by excision and scintillation counting demonstrated that the intensity of the labeled bands indeed increased significantly throughout the measurement period, suggesting that the transcript accumulated due to prolonged σB activity. In contrast, transcript levels from the osmotically inducible gbuAP1 promoter remained relatively constant after induction. Thus, kinetic analysis of transcript accumulation was consistent with the conclusion that the σB-dependent gbuAP2 is largely responsible for expression of gbuA during long-term osmotic stress. Similar experiments at the opuC promoter also demonstrated continued accumulation of σB-dependent transcripts after osmotic upshift, indicating that the response is not peculiar to the gbuAP2 or sigB promoters (data not shown).
FIG. 6.
Kinetic analyses of transcripts originating from sigB, opuC, and gbuA promoters. Cells of strain 10403S (wild type) were grown in BHI to an OD600 of ∼0.5, washed, and resuspended in 50 mM potassium phosphate with 50 mM glucose. NaCl was added to a final concentration of 3% at time zero, and aliquots were removed at 5 (T5), 10 (T10), 20 (T20), 30 (T30), and 40 (T40) minutes after addition. RNA was extracted from each aliquot, and primer extension reactions were performed on 50 μg of RNA using the primer VRPOM2 (5) to measure transcripts originating from the σB-dependent promoter of the sigB operon and the primer AEX to measure the σB-dependent gbuAP2 and the σB-independent gbuAP1 promoters from the gbuA operon. Graphs to the right of each image show the background-subtracted counts per minute from the respective labeled transcripts. Lines were plotted using linear regression.
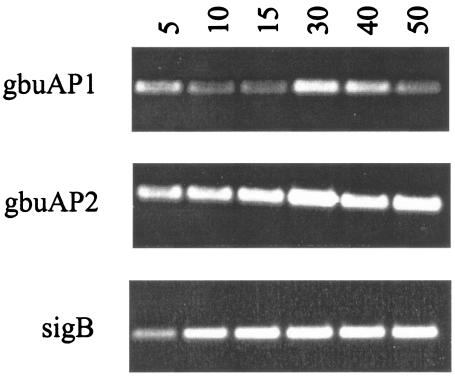
To confirm the kinetics of transcript accumulation observed by primer extension, we also performed RT-PCR on RNA samples from cells subjected to osmotic upshift. As shown in Fig. 7, RT-PCR analysis using primers 3′ to the σB-dependent promoter of the sigB operon showed product accumulation for the first 20 min, with saturated signal for the latter time points. Likewise, RT-PCR products amplified using primers 3′ to the gbuAP2 promoter (which would include transcripts initiating from gbuAP1 and gbuAP2) showed increasing signal intensity during the 5- to 30-min time period and saturation thereafter. In contrast, RT-PCR products amplified from segments of transcripts immediately 3′ to the gbuAP1 promoter were relatively constant during the upshift, with a minor peak occurring 30 min after upshift. These results suggest that the accumulating product derived from the gbuAP2 primers must be due to transcripts originating at gbuAP2. This supports the findings from the primer extension assays and further supports our hypothesis that σB activity remains high for extended periods after osmotic upshift.
FIG. 7.
Kinetic analysis of transcription from sigB, gbuA, and opuC by RT-PCR analysis. Cells of strain 10403S were grown in BHI to an OD600 of ∼0.5, washed, and resuspended in 50 mM potassium phosphate with 50 mM glucose. Aliquots were removed before and after addition of 3% NaCl. RNA was extracted, and 5-μg aliquots from the time points at 5, 10, 15, 30, 40, and 50 min were subjected to cDNA synthesis with reverse transcriptase. PCRs were then performed on the cDNA using primers 3′ to the σB-dependent promoter of the sigB operon, the gbuAP1 promoter, and the gbuAP2 promoter. The amplification products were electrophoresed on agarose gels and stained with ethidium bromide. Negative control reactions were performed by subjecting RNA samples without cDNA synthesis to PCR amplification and did not yield detectable products from any of the samples (data not shown).
DISCUSSION
The ability of L. monocytogenes to grow at high osmolarity and to accumulate compatible osmoprotectant molecules favors its growth in contaminated food products and promotes survival during transit through the gastrointestinal tract of animal and human hosts. In this report, we used primer extension analyses and physiological studies to examine how the general stress sigma factor, σB, controls accumulation of osmolytes through its activity in directing transcription from the genes encoding the primary osmolyte transporters. In strain 10403S, we showed that transcription of betL originates from a single promoter and that expression is modestly osmotically inducible, but not dependent on σB. Sequences upstream of the transcription start site showed some similarity to those recognized by the housekeeping sigma factor, σA, in B. subtilis but also deviated significantly from the consensus (Fig. 8B). It is therefore unclear if this promoter is recognized by the σA ortholog in L. monocytogenes. Although the betL extension product accumulated after osmotic upshift (Fig. 1), the fact that the transcript was quite detectable prior to osmotic upshift suggests that BetL is the primary transporter synthesized in unshocked cells and is likely to be the primary mechanism for transport of betaine during the early stages of osmotic upshift.
FIG. 8.
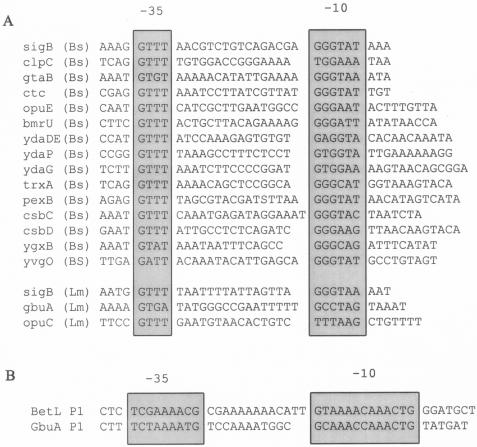
Promoter alignments. (A) Alignment of known σB-dependent promoters. The sequences immediately upstream of the σB-dependent transcription start site of the L. monocytogenes rsbV gene, opuC, and gbuA are aligned with several σB-dependent promoters from B. subtilis. The most highly conserved residues centering on the −10 and −35 regions are boxed. The alignment of B. subtilis promoters was derived from Petershon et al. (24), Helmann et al. (16), Scharf et al. (25), Antelmann et al. (4), Akbar et al. (1) and von Blohn et al. (33). Bs, B. subtilis; Lm, L. monocytogenes. (B) Alignments of the betL and gbuAP1 promoters. The sequences upstream of the start sites of betL and gbuA are aligned, and the most highly conserved residues centering on the −10 and −35 regions are boxed.
Unlike betL, expression of the gbuA operon was almost exclusively osmotically inducible. After osmotic upshift, the gbuA transporter was expressed from both σB-independent (gbuAP1) and σB-dependent (gbuAP2) promoters (Fig. 2). This finding is congruent with the fact that sigB mutants are defective only in osmotically inducible betaine transport.
In contrast to the betaine transport systems, expression of the carnitine transporter gene, opuC, was primarily dependent on σB in the 10403S strain background. This finding is particularly interesting, since a majority of stress response genes are typically under control of dual promoters to sustain flexible transcription under all conditions, particularly for those genes that are essential (25). Only a few genes, in fact, are known to rely solely upon σB for their transcription (1, 21, 24). The dependence of opuC on σB provides a mechanism for preferential use of betaine to maintain osmotic balance under normal growth conditions, since σB activity is minimal under those conditions. However, once the cells encounter osmotic stress, gbuA and opuC expression increases, providing the cells with an enhanced array of transporters and potential osmolytes.
Structure of σB-dependent gbuAP2 and opuC promoters.
Alignment of the gbuAP2 and opuC promoters with known σB-dependent promoters showed that both are quite deviant from consensus σB recognition sequences (Fig. 8A). The opuC −35 region is well aligned, whereas the −10 region deviates from consensus σB promoter sequences. In contrast, the −35 and −10 regions of gbuA both deviate somewhat from the consensus σB promoter sequences. This could imply that formation of closed complexes or isomerization to open complexes at these promoters is relatively inefficient. Since osmotic strength and counter ion concentration are known to affect the properties of RNA polymerase (8), it seems possible that changes in osmotic strength of the cytoplasm alter the affinity of σB holoenzyme for these promoters or the kinetics of open complex formation. While speculative, this could provide one explanation for why temperature upshift, which is a potent inducer of σB activity in L. monocytogenes, does not lead to significant transcript accumulation from gbuAP2 and opuC.
Perhaps of equal interest is the similarity between the σB-independent betL and gbuAP1 promoters. Alignment showed that the −10 and −35 regions are nearly identical (Fig. 8B). It is not yet clear if these promoters are recognized by the housekeeping form of RNA polymerase, since its specificity in L. monocytogenes has not been characterized. However, based on the canonical −10 and −35 sequences of B. subtilis σA promoters, the gbuAP1 and betL promoters show only limited similarity and thus may be utilized by a secondary form of RNA polymerase. Since the genome sequence from L. monocytogenes strain EGDe identified only five sigma factor paralogs, including the previously identified rpoD and sigB genes, the list of possible candidates is not overly large.
Distinct roles of σB in L. monocytogenes.
Relative to the well-studied σB regulon in B. subtilis, much less is known about the role of σB in the biology of L. monocytogenes. Perhaps the most significant differences identified to date are the relative magnitude and roles of σB in response to different stress in these two species. Our studies in this report now extend these observations to show that σB plays a role in long-term osmotic stress in L. monocytogenes (Fig. 6 and 7). Primer extension studies on the promoters of the opuE gene in B. subtilis have shown that the σB-dependent transcript accumulates only transiently after induction and is almost undetectable 40 min later, while the σA-dependent transcript remains at high levels 40 min postinduction (30, 33). In contrast, our studies showed that the σB-dependent transcript of the gbuAP2 promoter continued to accumulate at 40 min after induction, while the σB-independent gbuAP1 transcript remained constant. The similarity in transcript accumulation profiles at both sigB and gbuAP2 implies that σB activity may remain at high levels for long periods after upshift. Thus, the difference in σB kinetics at the opuE and gbuAP2 promoters in B. subtilis and L. monocytogenes, respectively, could imply that σB plays a primary role in directing gene expression in response to osmotic shock in L. monocytogenes but only an ancillary role in osmotic responses in B. subtilis. In support of an extended role for σB in directing stress response gene transcription in L. monocytogenes, we note that its activity profile during temperature downshift also displays continued transcript accumulation for hours after the downshift (6), suggesting that the long-term activity postinduction may be a general feature of σB induction in this organism. If this is indeed the case, then the role of σB in L. monocytogenes may be as a primary response to stress, whereas its role in B. subtilis is relegated to an ancillary and transient role to augment the many additional adaptive response pathways available in that organism.
Understanding the role and function of stress adaptation systems from a phylogenetic perspective may also provide significant insight as to the adaptive characteristics of different microorganisms. Indeed, this may provide an understanding of important characteristics of different subpopulations of the same species. Moorhead and Dykes (23) recently suggested that variations in the responses of different L. monocytogenes serotypes to osmotic stress may be due to differences in the magnitude of σB activation. Collectively, these studies underscore the importance of comparative analyses of the function and activation of σB in different species of gram-positive bacteria as well as within phylogenetic lineages of a single species, such as L. monocytogenes.
Acknowledgments
This work was supported by HATCH funds (project number NEB-16-077) from the United States Department of Agriculture Cooperative State Research, Education, and Extension Service to A.K.B.
We thank John Wise for assistance in preparation of the figures.
Footnotes
A contribution of the University of Nebraska Agricultural Research Division, Lincoln (journal series number 14326).
REFERENCES
- 1.Akbar, S., S. Y. Lee, S. A. Boylan, and C. W. Price. 1999. Two genes from Bacillus subtilis under the sole control of the general stress transcription factor σB. Microbiology 145:1069-1078. [DOI] [PubMed] [Google Scholar]
- 2.Angelidis, S. A., and G. M. Smith. 2003. Three transporters mediate uptake of glycine betaine and carnitine by Listeria monocytogenes in response to hyperosmotic stress. Appl. Environ. Microbiol. 69:1013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angelidis, S. A., L. T. Smith, L. M. Hoffman, and G. M. Smith. 2002. Identification of opuC as a chill-activated and osmotically activated carnitine transporter in Listeria monocytogenes. Appl. Environ. Microbiol. 68:2644-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antelmann, H., S. Engelmann, R. Schmid, A. Sorokin, A. Lapidus, and M. Hecker. 1997. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor σB in Bacillus subtilis. J. Bacteriol. 179:7251-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, L. A., M. S. Çetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, L. A., S. N. Evans, R. W. Hutkins, and A. K. Benson. 2000. Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J. Bacteriol. 182:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beumer, R. R., M. C. Te Giffel, L. J. Cox, F. M. Rombouts, and T. Abee. 1994. Effect of exogenous proline, betaine, and carnitine on growth of Listeria monocytogenes in a minimal medium. Appl. Environ. Microbiol. 60:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding, Q., S. Kusano, M. Villarejo, and A. Ishihama. 1995. Promoter selectivity control of Escherichia coli RNA polymerase by ionic strength: differential recognition of osmoregulated promoters by E sigma D and E sigma S holoenzymes. Mol. Microbiol. 16:649-656. [DOI] [PubMed] [Google Scholar]
- 9.Dufour, A., U. Voelker, A. Voelker, and W. G. Haldenwang. 1996. Relative levels and fractionation properties of Bacillus subtilis σB and its regulators during balanced growth and stress. J. Bacteriol. 178:3701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farber, J. M., and B. E. Brown. 1990. Effect of prior heat shock on heat resistance of Listeria monocytogenes in meat. Appl. Environ. Microbiol. 56:1584-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser, K. R., D. Harvie, P. J. Coote, and C. P. O'Byrne. 2000. Identification and characterization of an ATP binding cassette l-carnitine transporter in Listeria monocytogenes. Appl. Environ. Microbiol. 66:4696-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser, K. R., and C. P. O'Byrne. 2002. Osmoprotection by carnitine in a Listeria monocytogenes mutant lacking the OpuC transporter: evidence for a low affinity carnitine uptake system. FEMS Microbiol. Lett. 211:189-194. [DOI] [PubMed] [Google Scholar]
- 13.Fraser, K. R., D. Sue, M. Wiedmann, K. Boor, and C. P. O'Byrne. 2003. Role of σB in regulating the compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC is σB dependent. Appl. Environ. Microbiol. 69:2015-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerhardt, P. N., L. T. Smith, and G. M. Smith. 1996. Sodium-driven, osmotically activated glycine betaine transport in Listeria monocytogenes membrane vesicles. J. Bacteriol. 178:6105-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser, P., L. Frangeul, C. Buchrieser, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durand, O. Dussurget, K.-D. Entian, H. Fsihi, F. Garcia-Del Portillo, P. Garrido, L. Bautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L.-M. Jones, U. Karst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. Mata Vincente, E. Ng, G. Nordsiek, S. Novella, B. de Pablos, J.-C. Perez-Diaz, B. Remmel, M. Rose, C. Rusniok, T. Schlueter, N. Simoes, A. Tierrez, J.-A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 16.Helmann, J. D., M. F. W. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Narve, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress response to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 18.Ko, R., L. T. Smith, and G. M. Smith. 1994. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J. Bacteriol. 176:426-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko, R., and L. T. Smith. 1999. Identification of an ATP-driven, osmoregulated glycine betaine transport system in Listeria monocytogenes. Appl. Environ. Microbiol. 65:4040-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroll, R. G., and P. A. Patchett. 1992. Induced acid tolerance in Listeria monocytogenes. Lett. Appl. Microbiol. 14:224-227. [Google Scholar]
- 21.Maul, B., U. Völker, S. Riethdorf, S. Engelmann, and M. Hecker. 1995. σB-dependent regulation of gsiB in response to multiple stimuli in Bacillus subtilis. Mol. Gen. Genet. 248:114-120. [DOI] [PubMed] [Google Scholar]
- 22.Miller, A. J. 1992. Combined water activity and solute effects on growth and survival of Listeria monocytogenes Scott A. J. Food Prot. 55:414-418. [DOI] [PubMed] [Google Scholar]
- 23.Moorhead, S. M., and G. A. Dykes. 2003. The role of the sigB gene in the general stress response of Listeria monocytogenes varies between a strain of serotype 1/2a and a strain of serotype 4c. Curr. Microbiol. 46:461-466. [DOI] [PubMed] [Google Scholar]
- 24.Petershon, A., H. Antelmann, U. Gerth, and M. Hecker. 1999. Identification and transcriptional analysis of new members of the σB regulon in Bacillus subtilis. Microbiology 145:869-880. [DOI] [PubMed] [Google Scholar]
- 25.Scharf, C., S. Riethdorf, H. Ernst, S. Engelmann, U. Volker, and M. Hecker. 1998. Thioredoxin is an essential protein induced by multiple stress in Bacillus subtilis. J. Bacteriol. 180:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sleator, R. D., C. G. M. Gahan, T. Abee, and C. Hill. 1999. Identification and disruption of BetL, a secondary glycine betaine transport system linked to the salt tolerance of Listeria monocytogenes LO28. Appl. Environ. Microbiol. 65:2078-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sleator, R. D., C. G. M. Gahan, B. O'Driscoll, and C. Hill. 2000. Analysis of the role of betL in contributing to the growth and survival of Listeria monocytogenes LO28. Int. J. Food Microbiol. 60:261-268. [DOI] [PubMed] [Google Scholar]
- 28.Sleator, R. D., J. Wouters, C. G. M. Gahan, T. Abee, and C. Hill. 2001. Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl. Environ. Microbiol. 67:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sleator, R. D., C. G. M. Gahan, and C. Hill. 2003. A postgenomic appraisal of osmotolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 69:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiegelhalter, F., and E. Bremer. 1998. Osmoregulation of the opuE proline transport gene from Bacillus subtilis: contributions of the sigma A- and sigma B-dependent stress responsive promoters. Mol. Microbiol. 29:285-296. [DOI] [PubMed] [Google Scholar]
- 31.Verheul, A., E. Glaasker, B. Poolman, and T. Abee. 1997. Betaine and l-carnitine transport by Listeria monocytogenes Scott A in response to osmotic signals. J. Bacteriol. 179:6979-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verheul, A., F. M. Rombouts, R. R. Beumer, and T. Abee. 1995. An ATP-dependent l-carnitine transporter in Listeria monocytogenes Scott A is involved in osmoprotection. J. Bacteriol. 177:3205-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Blohn, C., B. Kempf, R. M. Kappes, and E. Bremer. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175-187. [DOI] [PubMed] [Google Scholar]
- 34.Wemekamp-Kamphuis, H. H., J. A. Wouters, R. D. Sleator, C. G. M. Gahan, C. Hill, and T. Abee. 2002. Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl. Environ. Microbiol. 68:4710-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkins, P. O., R. Bourgeois, and R. G. E. Murray. 1972. Psychrotrophic properties of Listeria monocytogenes. Can. J. Microbiol. 18:543-551. [DOI] [PubMed] [Google Scholar]
- 36.Yancey, P. H., M. E. Clark, S. C. Hand, R. D. Bowlus, and G. N. Somero. 1982. Living with water stress: evolution of osmolyte systems. Science 217:1214-1222. [DOI] [PubMed] [Google Scholar]