Abstract
Objectives
Bile acids promoted the replication of hepatitis C virus (HCV) and compromised the anti-HCV effects of interferon-α (IFN-α) in replicon-harboring cells. To explore a potential mechanism for the observation, we studied the effects of bile acids on the epidermal growth factor receptor (EGFR) and the extracellular signal-regulated kinase (ERK) pathway in association with HCV replication in genotype 1a or 1b replicon-harboring cells.
Methods
Replicon-harboring cells were treated with various bile acids, IFN-α and small molecule inhibitors either individually or combined together. The effects of these treatments were measured using cell cycle analysis, qRT-PCR, and Western blot analysis.
Results
Bile acids induced the activation of EGFR/ERK pathway and extended S-phase of cells, which was correlated with the increased levels of viral replication. The inhibitors of EGFR (AG1478) or ERK (U0126) significantly mitigated the bile acid-mediated promotion of HCV replication. When AG1478 or U0126 were added to the treatment of bile acids and IFN-α, they were able to restore the anti-HCV effects of IFN-α.
Conclusion
Our data suggest that the addition of an EGFR or ERK inhibitor to the current IFN-α-based regimen may improve overall treatment efficacy by blocking the bile acid-mediated promotion of HCV replication.
Key Words: Bile acids, EGFR/ERK pathway, Hepatitis C virus, HCV replication, Replicon-harboring cells
Introduction
Hepatitis C virus (HCV) is a single-stranded, positive sense RNA virus with a genome length of approximately 10 kb [1,2,3,4]. HCV can persist as a chronic infection in hepatocytes, and the spontaneous clearance of an HCV infection is rare [4,5]. Furthermore, chronic HCV infections frequently lead to liver cirrhosis and cancer [6,7]. The current standard treatment for chronic HCV infection is a combination therapy consisting of pegylated interferon-α (IFN-α) and ribavirin [8,9]. Both IFN-α and ribavirin have been shown to be cost-effective and demonstrate significant antiviral effects against an HCV [10,11,12]. The combination therapy is very effective against HCV genotype 2 and 3 viruses resulting in sustained virological response rate of around 80%. However, the effectiveness of the combination treatment is low (40–50%) in patients infected with HCV genotype 1 [5]. The lack of an effective treatment for HCV genotype 1 serves as a motivation to gain further knowledge of the HCV replication process and its relationship with the host cells.
While several HCV strains have recently been isolated that have been successfully tissue culture adapted, the majority of HCV remains fastidious in cell culture systems [4,13,14,15,16]. As a solution to the lack of tissue culture adapted viruses in a cell-based virus replication system, replicon-harboring cells were established and have provided an excellent tool for the study of virus replication and the development of antivirals [17,18,19]. These replicon-harboring cells contain an RNA replicon composed of the HCV 5′ untranslated region, the neomycin phosphotransferase gene, the encephalomyocarditis virus (EMCV) internal ribosomal entry site, and the subgenomic portion of the HCV genome, thus constantly express both the subgenomic RNA and all of the associated non-structural HCV proteins [17,18,19]. In our previous study, we demonstrated that bile acids promoted HCV replication and compromised the anti-HCV action of IFN-α using replicon-haboring cells containing HCV genotype 1b (GS4.1 cells) [20]. Our study suggested that bile acids may be a reason for low efficacy of IFN-based treatment in a large percentage of patients with chronic HCV infections [20]. Various bile acids including chenodeoxycholic acid (CDCA), glycolchenodeoxycholic acid (GCDCA), deoxycholic acid (DCA), and ursodeoxycholic acid (UDCA) all promoted HCV replication and interfered with IFN-α action in the replicon-harboring cells [20]. Scholtes et al. [21] also demonstrated that bile acids enhanced the replication of genotype 1, but not genotype 2 HCV in a live viral system, but not in a replicon-harboring system.
Bile acids have been shown to promote the proliferation of various cells including hepatocytes via several pathways including the epidermal growth factor receptor (EGFR) [22,23,24,25,26,27]. Many reports suggest that the highest levels of HCV RNA and proteins are observed while the replicon-harboring cells were undergoing active division [28,29]. This suggests that HCV replication may be associated with the cell cycle progress which can be altered by the extracellular signal-regulated kinase (ERK) pathway. Therefore, we hypothesized that bile acids promote HCV replication in replicon-harboring cells by modulating the EGFR/ERK pathway, and that blocking this modulation will restore the anti-HCV effects of IFN. In this report, we found that bile acids led to the activation of the promoter activities of activator protein 1 (AP-1) and serum responding element (SRE) in replicon-harboring cells. We also demonstrated that bile acids modulated the cell cycle progression by extending S stage, which was associated with the phosphorylation of EGFR and enhanced levels of both HCV RNA and protein in the cells. In addition, the inhibitors of either EGFR (AG1478) or the mitogen-activated protein kinase kinase (MEK)1/2 (U0126) blocked bile acid-mediated promotion of HCV replication in the cells.
Finally, we found that the interference of IFN-α action by bile acids was mitigated in the presence of the EGFR or ERK inhibitor. Our data suggest that the addition of an EGFR or ERK inhibitor to the current IFN-α-based regimen may improve overall treatment efficacy by blocking the bile acid-mediated promotion of HCV replication.
Materials and Method
Cell Lines and Reagents
Huh-7, GS4.1 (replicon-harboring cells with the HCV 1b genotype, provided by Dr. C. Seeger at the Fox Chase Cancer Center, Philadelphia, Pa., USA), and 1A7 (replicon-harboring cells with the HCV 1a genotype, provided by Dr. S. Lemon at the University of Texas Medical Branch at Galveston, Galveston, Tex., USA) were maintained in Dulbecco's Minimum Essential Medium containing 10% fetal bovine serum. Both GS4.1 and 1A7 cells were maintained in the presence of G418 (Cellgro, Manassas, Va., USA) at 0.5 μg/ml. Antibodies against HCV NS5b, β-actin, phosphor-EFGR or EGFR were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif., USA). Bile acids, CDCA, GCDCA, DCA, and UDCA were obtained from Sigma-Aldrich (St. Louis, Mo., USA). GCDCA was resuspended in deionized-distilled water, while all unconjugated bile acids (CDCA, UDCA, and DCA) were resuspended in dimethyl sulfoxide. IFN-α (recombinant IFN-αA/D human), AG1478 and U0126 were also obtained from Sigma-Aldrich.
AP-1 and SRE Promoter Assay
Replicon-harboring cells (GS4.1 or 1A7) or the parental Huh-7 cells were transfected with 2 μg of either pAP1-TA-Luc or pSRE-TA-Luc (Clonetech, Palo Alto, Calif., USA) and 2 μg of pRL-CMV (Promega, Madison, Wisc., USA) using lipofectamine 2000 (Invitrogen, Carlsbad, Calif., USA). After 4 h of transfection, cells were incubated in the fresh media containing CDCA (100 μM), GCDCA (200 μM), DCA (100 μM), UDCA (200 μM), or a combination of CDCA (100 μM) and U0126 (20 μM). Cells were further incubated for 24 h before being lysed and subjected to luciferase assay using a Dual Glo luciferase assay kit (Promega). The level of firefly luciferase expression was normalized against the expression level of the renilla luciferase encoded for by pRL-CMV.
Cell Cycle Analysis
Semiconfluent 1A7 cells were synchronized using aphidicolin (Sigma-Aldrich) at a concentration of 5 μM for 24 h before being stimulated with CDCA (100 μM) or GCDCA (200 μM) for 0, 6, 12, and 24 h. For cell cycle analysis, cell monolayers were lysed with Vindelov's propidium iodide solution [30] and cell nuclei were strained through a cell strainer cap on 5-ml round-bottom tubes (BD Biosciences, San Jose, Calif., USA). Nuclei were analyzed on a Beckman Dickinson flow cytometer and the cell division stages were analyzed using ModFit LT (VSH Software, Topsham, Me., USA). The specific percentage of cells at a particular stage in the cell cycle was determined using ModFit LT.
Treatment of Replicon-Harboring Cells with Bile Acids, Inhibitors of EGFR (AG1478) or MEK1/2 (U0126), and IFN-α
First, we examined the cytotoxic effects of each bile acid and/or the inhibitor on GS4.1 or 1A7 cells using a CytoTox 96® non-radioactive cytotoxicity assay kit (Promega), to obtain the concentration of each agent with minimum cell toxicity. To examine the role of EGFR and the ERK pathway in bile acid-mediated promotion of HCV replication and/or interference with the anti-HCV action of IFN-α, AG1478 and U0126 were added in conjunction with various bile acid concentrations. Both GS4.1 and 1A7 cells were used in this study. First, the treatments were focused on the ability of EGFR or ERK inhibitors in blocking bile acid-mediated enhancement of the HCV replication. The treatments included AG1478 (15 or 30 μM), U0126 (10 or 20 μM), CDCA (100 μM), and CDCA in combination with AG1478 or U0126, using the concentrations listed above. Cells were incubated with the various treatments for 24 h before HCV RNA and protein levels were measured using real-time quantitative reverse transcription-PCR (qRT-PCR) and Western blot analysis. Second, the treatments were designed to examine the ability of the EGFR or ERK inhibitor in restoring the anti-HCV effects of IFN-α in the presence of bile acids. The treatments included AG1478 (15 or 30 μM), U0126 (10 or 20 μM), IFN-α (5 U/ml), CDCA (100 μM), CDCA + AG1478 or U0126, IFN-α + U0126 or AG1478, IFN-α + CDCA, and CDCA + IFN-α + AG1478 or U0126. The cells were incubated with the various treatments for 24 h before HCV RNA and protein levels were measured using qRT-PCR and Western blot analysis, respectively.
Detection of HCV RNA and NS5b Protein
The HCV RNA and NS5b protein levels were measured 24 h after semiconfluent GS4.1 or 1A7 cells were treated with various bile acids, IFN-α and/or AG1478, and/or U0126 by qRT-PCR or Western blot analysis, respectively. Total RNA was isolated from GS4.1 or 1A7 cells in 6-well plates following treatment using the RNeasy Mini Kit (Qiagen, Valencia, Calif., USA) and qRT-PCR was performed using a One-Step Superscript III qRT-PCR kit (Invitrogen) with primers and probes previously described [20]. Western blot analysis was performed on cells lysed with lithium dodecyl sulfate sample buffer containing 2% β-mercaptoethanol. NS5b protein was detected using a mouse monoclonal anti-NS5B antibody at a 1:1,000 dilution in 2% non-fat dry milk followed by a secondary HRP-conjugated goat-anti-mouse (Pierce Biosciences, Rockford, Ill., USA) at 1:1,000. Protein was visualized using a chemiluminescent substrate (Pierce Biosciences). β-Actin levels were measured by qRT-PCR and Western blot analysis for the normalization of RNA and protein levels of each sample, respectively. The expression of NS5B in cells with or without bile acid treatment was also examined by flow cytometry analysis as described above.
Detection of Phosphor-EGFR and EGFR
The activation of EGFR was detected using Western blot analysis with phosphor-EFGR and EGFR antibodies. Briefly, semiconfluent GS4.1 cells were incubated with CDCA (100 μM), GCDCA (200 μM), or UDCA (200 μM) for 30 min before lysis with lithium dodecyl sulfate sample buffer containing 2% β-mercaptoethanol. Western blot analysis was performed using an anti-phosphor-EGFR or EGFR antibody at 1:1,000 dilution in 2% non-fat dry milk and a secondary anti-mouse HRP-conjugated detection antibody at 1:500. Proteins were visualized using a chemiluminescent substrate as described above.
Statistical Analysis
Each experiment was repeated a minimum of three times. Statistical analysis was performed using GraphPad Prism 5.0 data analysis software.
Results
Bile Acids Increased HCV Replication in 1A7 Cells
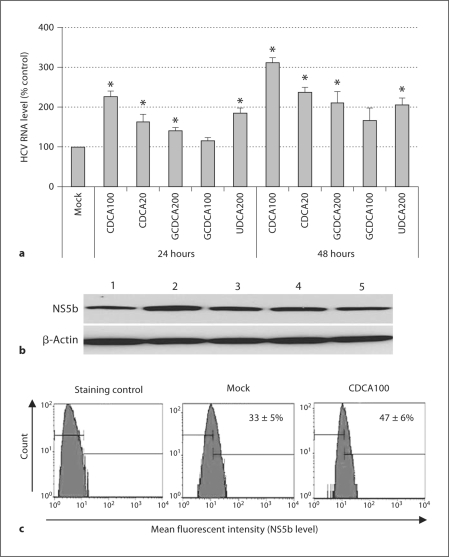
In a previous study, we demonstrated that bile acids promoted the replication of HCV RNA and proteins in GS4.1 cells containing a genotype 1b replicon [20]. In this report, we found that various bile acids also increased the levels of HCV RNA and protein in 1A7 cells containing a genotype 1a replicon. 1A7 cells were treated with mock medium, CDCA, GCDCA, or UDCA, and HCV RNA or NS5b was measured by real-time qRT-PCR, or Western blot analysis or flow cytometry analysis, respectively. After incubation with 100 or 20 μM of CDCA for 24 h, HCV RNA levels in 1A7 cells were significantly increased (p < 0.05) to 226 ± 14% or 163 ± 19% compared to mock treatment (100%). The levels of HCV RNA were further increased to 312 ± 12% and 237 ± 13% compared to mock treatment after a 48-hour incubation. The incubation of cells with 200 μM GCDCA or 100 μM UDCA also showed an increase in HCV RNA levels to 140 ± 9% or 185 ± 12% (24 h) and 166 ± 31% or 206 ± 17% (48 h) (p < 0.05), respectively (fig. 1a). The protein levels of HCV NS5b correlated with HCV RNA levels after the incubation with CDCA 100 μM (fig. 1b, lane 2), with a significant enhancement in protein level compared to mock-treated cells (fig. 1b, lane 1). Enhanced NS5b protein levels were observed in all other treatments including CDCA 20 μM (lane 3), GCDCA 200 μM (lane 4), and GCDCA 100 μM (lane 5). The flow cytometry analysis also confirmed the enhanced expression of NS5b by the treatment with bile acids: after treatment with 100 μM of CDCA for 24 h, cells expressing NS5b had increased compared to mock-treated cells (47 vs. 33%) (fig. 1c).
Fig. 1.
Enhancement of HCV replication after bile acid treatment in 1A7 cells. Semiconfluent cells were treated with mock medium, CDCA, GCDCA, or UDCA for 24 or 48 h. HCV RNA (a) or NS5b (b) was measured by real-time qRT-PCR or Western blot analysis, respectively. a qRT-PCR levels after treatment with mock medium, CDCA 100 μM (CDCA100), CDCA 20 μM (CDCA20), GCDCA 200 μM (GCDCA 200), GCDCA 100 μM (GCDCA100) and UDCA 200 μM (UDCA200) for 24 or 48 h. Asterisk (∗) indicates that the RNA levels by the treatment were significantly increased compared to those by control (mock medium) treatment (p < 0.05). b Western blot analysis detecting NS5b after the treatment for 24 h. Upper panel, lane 1: mock medium; lane 2: CDCA (100 μM); lane 3: CDCA (20 μM); lane 4: GCDCA (200 μM); lane 5: GCDCA (100 μM). Lower panel, as a loading control, Western blot analysis of β-actin was performed with the same samples. c Flow cytometry analysis of NS5b levels in 1A7 cells with the treatment of mock medium or CDCA (100 μM) for 24 h. Staining control was prepared using the same procedure without the incubation with the NS5b antibody.
Enhancement of Luciferase Activity under AP-1 or SRE Promoter after Bile Acid Treatment in GS4.1 Cells
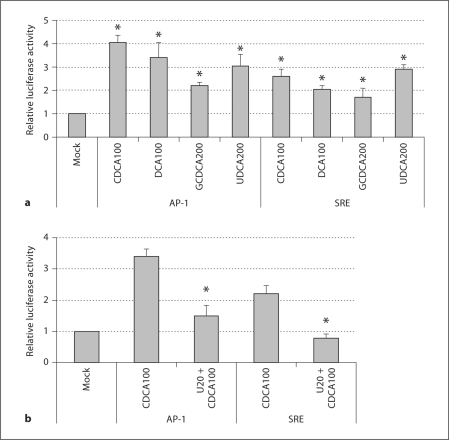
Luciferase activity under the control of the AP-1 or SRE promoter was significantly increased (>1.8-fold, p < 0.05) by the treatment with CDCA (100 μM), DCA (100 μM), GCDCA (200 μM), or UDCA (200 μM) for 24 h compared to mock-medium (negative) treatment (fig. 2a). CDCA was the most efficient bile acid tested in increasing luciferase activity under AP-1 or SRE, which was consistent with the promotion HCV replication in cells (fig. 1a) [20]. The treatment of the cells with U0126 blocked the bile acid-mediated enhancement of luciferase activity (fig. 2b), which further suggests that bile acids induce the activation of the signaling pathways of AP-1 or SRE in the cells. The expression of luciferase after the transfection of the plasmids with the AP-1 and SRE promoters and incubation with or without bile acids was similar in both the parental Huh-7 cells and 1A7 to GS4.1 cells.
Fig. 2.
Enhancement of luciferase activity under AP-1 or SRE promoter control after bile acid treatment in GS4.1 cells. The luciferase-based plasmid under AP-1 or SRE promoter control was transfected into semiconfluent GS4.1 cells. The cells were treated with CDCA 100 μM (CDCA-100), DCA 100 μM (DCA100), GCDCA 200 μM (GCDCA200), UDCA 200 μM (UDCA200), or CDCA100 + U0126 20 μM (U20) 4 h after transfection and incubated for an additional 24 h. Asterisk (∗) indicates that the luciferase activities by the treatment were significantly increased compared to those by the control treatment (p < 0.05). a Treatment of various bile acids in the luciferase activity under the control of signaling pathways, AP-1 or SRE. b The effects of U0126 on CDCA-mediated induction of luciferase expression under AP-1 or SRE. Asterisk (∗) indicates that the luciferase activities by co-treatment with CDCA and U0126 were significantly reduced compared to those by the treatment with CDCA alone (p < 0.05).
Bile Acids Induce a Prolonged S-Phase Period in Replicon-Harboring Cells
Since AP-1 and SRE are known to play a role in modulation of the cell cycle, we analyzed cell cycle progression of 1A7 cells after treatment with mock medium, CDCA, and GCDCA for up to 24 h (table 1). For 1A7 cells, cell cycle progression with G1, S, or G2 stages showed no significant difference between mock, CDCA, or GCDCA at 6 h post-treatment (table 1). However, 1A7 cells in S phase were significantly increased (p < 0.05) at 12 h post-treatment with CDCA or GCDCA when compared to mock-treated cells: 86% (CDCA) and 81% (GCDCA) versus 67% (Mock) (table 1). At 24 h, 1A7 cells in S phase were also significantly increased (p < 0.05) by treatment with CDCA compared to mock treatment (40 versus 14%) (table 1).
Table 1.
Cell cycle analysis of 1A7 cells incubated with mock medium, CDCA (100 μM) or GCDCA (200 μM) for 6,12, or 24 h
| Treatment | Percentage of cells1 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 hour |
6 hours |
12 hours |
24 hours |
|||||||||
| G1 | S | G2 | G1 | S | G2 | G1 | S | G2 | G1 | S | G2 | |
| Mock | 75 ± 3 | 25 ± 3 | 0 | 25 ± 16 | 75 ± 15 | 1 ± 1 | 10 ± 1 | 67 ± 10 | 23 ± 10 | 67 ± 2 | 13 ± 3 | 20 ± 2 |
| CDCA | – | – | – | 41 ± 28 | 59 ± 27 | 1 ± 1 | 9 ± 2 | 86 ± 62 | 6 ± 4 | 26 ± 4 | 40 ± 42 | 38 ± 3 |
| GCDCA | – | – | – | 24 ± 16 | 75 ± 16 | 1 ± 1 | 8 ± 1 | 81 ± 12 | 11 ± 0 | 61 ± 4 | 14 ± 4 | 25 ± 1 |
The percentages of cells in each stage were calculated based on the analysis of 50,000 cell nuclei using ModFit 5.0. These results are the average of three independent experiments.
Numbers of cells in S stage were significantly higher compared to those by the mock treatment (p < 0.05).
Bile Acid-Mediated Promotion of HCV Replication Is Correlated with EGFR Pathway
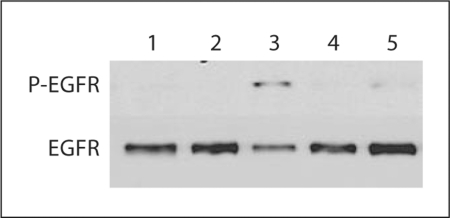
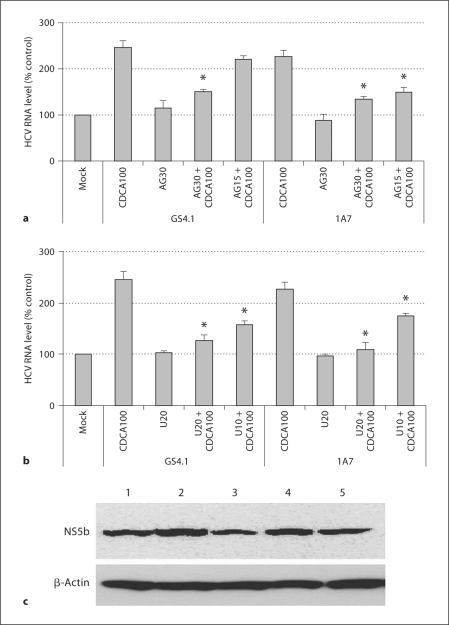
We determined bile acids induced the activation of EGFR in replicon-harboring cells. When GS4.1 cells were incubated with medium containing mock medium, CDCA, UDCA, or GCDCA for 30 min, the phosphorylation of EGFR was evident in cells with CDCA or UDCA (fig. 3). To examine the role of an EGFR inhibitor in the bile acid-mediated enhancement of HCV replication, semiconfluent GS4.1 or 1A7 cells were incubated with mock medium, CDCA, AG1478 or CDCA + AG1478 for 24 h, and HCV replication was measured. The treatment with AG1478 alone did not change HCV RNA levels in the cells, but when AG1478 was co-treated with CDCA (100 μM), it significantly reduced (p < 0.05) CDCA-mediated enhancement of HCV RNA levels at the concentrations above 15 μM. The treatment with CDCA increased HCV RNA levels to over 200% compared with the mock treatment in both GS4.1 and 1A7 cells. However, the HCV RNA levels were significantly (p < 0.05) decreased to 151 ± 4% or 135 ± 5% in GS4.1 and 1A7 cells, respectively, in the presence of 30 μM of AG1478 (+CDCA) (fig. 4a). Like AG1478, U0126 mitigated bile acid-mediated promotion of HCV replication in GS4.1 or 1A7 cells, while U1026 (10 or 20 μM) alone did not significantly alter the HCV RNA level (fig. 4b). While the treatment with CDCA increased HCV RNA levels to over 200%, in the presence of U0126 (+ CDCA), the HCV RNA levels were significantly (p < 0.05) decreased to 126 ± 11% or 158 ± 7% (GS4.1) and 109 ± 14% or 175 ± 5% (1A7) at 20 or 10 μM, respectively (fig. 4b). The levels of HCV NS5b were well correlated with HCV RNA levels after treatment with AG1478 and/or bile acids in replicon-harboring cells (fig. 4c).
Fig. 3.
The activation of EFGR by the treatment with bile acids in GS4.1 cells. Confluent GS4.1 cells were treated with mock medium, CDCA, GCDCA, or UDCA for 30 min, and then cell lysates were prepared and Western blot analysis detecting phosphor-EGFR or EGFR. Lane 1: parental Huh-7 cells with mock treatment; lane 2: mock treatment; lane 3: CDCA 100 μM; lane 4: GCDCA 200 μM; lane 5: UDCA 200 μM.
Fig. 4.
Inhibitors of EGFR or ERK in bile acid-mediated promotion of HCV replication. Semiconfluent GS4.1 or 1A7 cells were incubated with mock medium, CDCA, AG1478 or CDCA + AG1478 for 24 h, and HCV replication was measured by detecting HCV RNA (a, b) or protein (c). a Real-time qRT-PCR analysis of HCV RNA levels after the treatment with CDCA 100 μM (CDCA100), AG1478 30 μM (AG30), CDCA100 + AG30, or CDCA100 + AG1478 15 μM (AG15). b Real-time qRTPCR analysis of HCV RNA levels after the treatment with CDCA100, U0126 20 μM (U20), CDCA100 + U20, or CDCA100 + U0126 10 μM (U10). Asterisk (∗) indicates that the RNA levels by double treatment with CDCA and AG1478 or U0126 were significantly reduced compared to those by the treatment with CDCA alone (p < 0.05). c Western blot analysis of HCV protein NS5B in GS4.1 cells after treatment with CDCA, AG1478 or CDCA + AG1478. Lane 1: Mock; lane 2: CDCA 100 μM; lane 3: AG1478 30 μM + CDCA 100 μM; lane 4: AG1478 5 μM + CDCA 100 μM, and lane 5: AG1478 30 μM. As a loading control, Western blot analysis of β-actin was performed with the same samples.
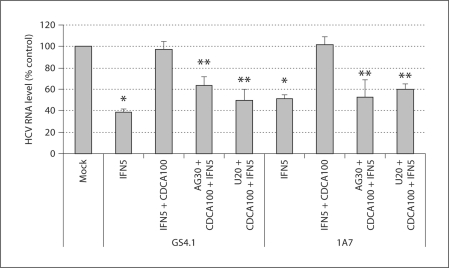
Inhibitor of EGFR or ERK Restores the Anti-HCV Action of IFN-α in the Presence of Bile Acids
Because bile acids compromise the anti-HCV action of IFN-α, we examined if AG1478 or U0126 could restore IFN-α action in the presence of bile acids. GS4.1 or 1A7 cells were treated with IFN-α alone (5 U/ml), double treatment of IFN-α and CDCA (100 μM) or triple treatment of IFN-α, CDCA and AG1478 (30 μM) or U0126 (20 μM). The levels of HCV RNA were measured after 24 h of treatments. In GS4.1 cells, the treatment with IFN-α reduced the HCV RNA levels to 39 ± 3% of those with mock treatment, but in the presence of CDCA it was reduced to only 97 ± 8% (p < 0.05 compared to IFN-α treatment). With the triple treatment with A1478 or U0126, the HCV RNA levels were 64 ± 8% or 50 ± 10% (p < 0.05 compared to double treatment of IFN-α and CDCA) of those treated with mock treatment (fig. 5). We found similar results with 1A7 cells. While the treatment with IFN-α (5 U/ml) reduced the HCV RNA levels to 51 ± 4% of those with mock treatment, with the double treatment of IFN-α and CDCA, HCV RNA levels were 102 ± 7%. The triple treatment of IFN-α, CDCA and A1478 or U0126 resulted in the HCV RNA levels to 53 ± 16% or 60 ± 5% of those receiving mock treatment (fig. 5). These results suggest the inhibitor of EGFR or ERK restores the anti-HCV action of IFN-α in the presence of bile acids.
Fig. 5.
Inhibitors of EGFR or ERK in anti-HCV action of IFN in the presence of CDCA. HCV RNA levels were measured after 24 h treatments with IFN 5 U/ml (IFN5), CDCA 100 μM (CDCA100) + IFN5, or CDCA100 + IFN5 + (AG1478 30 μM [AG30] or U0126 20 μM [U20]). Asterisk (∗) indicates that the RNA levels by IFN 5 U/ml treatment were significantly reduced compared to those by mock treatment (p < 0.05). Double asterisk (∗∗) indicates that the RNA levels by triple treatment were significantly reduced compared to those by double treatments of IFN5 + CDCA100 (p < 0.05).
Discussion
While several putative receptors of HCV, including CD81 [31,32,33,34] and human scavenger receptor class B1 [35,36,37,38], are distributed throughout the body, HCV mainly replicates within hepatocytes in the liver. This suggests there are hepatocyte-specific co-factors that determine the liver tropism of HCV. Previously we demonstrated that bile acids, both conjugated and unconjugated, can play an important role in the upregulation of HCV replication in replicon-harboring cells [20]. There are the primary bile acids (cholic acid and CDCA which are two major bile acids) and secondary bile acids (lithocholic acid and DCA) in the body. While the primary bile acids are synthesized from cholesterol, secondary bile acids are derived from the primary bile acids within the small intestine [39]. In the body, most bile acids present as conjugated forms with taurine or glycine. Although the exact composition of bile acids varies among individuals dependent on the genetic and cultural (food) background, taurocholic acid and glycocholic acid represent approximately 80% of all bile salts in adults [39]. Bile acids are collected and stored in the gallbladder at concentrations reaching as high as 320 mM before being released into the upper small intestine (duodenum) [39]. Most bile acids are returned to the liver through the enterohepatic circulation via the portal vein where the concentration of bile acids can reach up to 80 μM as it passes into the liver [40]. Within the systemic circulation the concentration of bile acids is typically below 10 μM [39,40]. The enterohepatic circulation requires for the expression of bile acid transporters small intestines and liver cells for efficient uptakes of conjugated bile acids. Because the Huh-7 cell line and Huh-7-based replicon-harboring cells do not express the bile acid transporter, we used primarily non-conjugated bile acids such as CDCA for this study. However, we found that most of conjugated or non-conjugated bile acids showed similar results in HCV replication as CDCA with varying efficiency [20]. The conjugated bile acids required higher concentrations for the same effects as the non-conjugated bile acids [20]. Furthermore, we found that the minimum concentration of CDCA for the bile acid-mediated promotion was approximately 50 μM, which may be relevant for the physiologic concentrations for the returning bile acids to liver cells via the portal vein. We used 100 μM of CDCA throughout for the current study because there was greater consistency with this concentration in the study.
In this report, we demonstrated that the activation of the EGFR/ERK pathway may play an important role in the bile acid-mediated enhancement of HCV replication. In 2004, Carloni et al. [41] demonstrated that binding to CD81, a putative receptor for HCV, has the ability to activate the ERK pathway. In addition, Brazzoli et al. [42] demonstrated that activation of the ERK pathway by CD81 was necessary for specific cellular events required for the replication HCV. When the authors blocked the ERK signaling cascade using the MEK1/2 inhibitor (U0126) at a post-entry step, viral replication was significantly reduced. Previous studies have shown that bile acids are important in the normal regeneration of the liver [22,23], and they activate the ERK pathway in primary rat hepatocytes with the EGFR receptor [27]. The ERK pathway stimulates both AP-1 and the SRE through a series of intermediate proteins, including the ternary complex factor subfamily of proteins [43]. In accordance with the literature, we demonstrated that bile acids activate the ERK pathway and found that bile acids led to the enhanced expression of luciferase under both the AP-1 and SRE promoters in replicon-harboring cells with genotype 1b (GS4.1 cells) (fig. 2a). In addition, the total luciferase activity was significantly reduced when the replicon-harboring cells (GS4.1 cells) were treated with 100 μM CDCA in combination with 20 μM U0126 (fig. 2b). This suggests that signaling through the ERK pathway is induced by bile acids and leads to the activation of both AP-1 and SRE. The AP-1 complex is comprised of components from the jun oncogene (JUN), v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS), activating transcription factor (ATF), and v-maf musculoaponeurotic fibrosarcoma oncogene homolog (MAF) protein families [44]. FOS and JUN are the main proteins in mammalian systems which form AP-1 complexes and regulate gene expression. AP-1 has the ability to be both oncogenic and anti-oncogenic through the regulation of different sets of genes, including those involved in cell proliferation [44,45].
Because both AP-1 and SRE lead to cell proliferation, we examined the cell cycle progression of replicon-harboring cells in the presence of bile acids using flow cytometry analysis. It has been previously reported that cell cycle progression was important in HCV replication, with the highest levels of HCV RNA being observed while cells are actively undergoing division in replicon-harboring cells [13]. We observed that upon stimulation with various bile acids, 1A7 and GS4.1 cells entered into the extended S phase (table 1), which correlated with the enhanced expression levels of HCV RNA and proteins (fig. 1a–c). The cell cycle progression of GCDCA-treated cells had a shorter S phase hold than its CDCA counterpart. This shorter S phase hold correlates with the lower overall levels of HCV RNA and protein seen after stimulation with bile acids. The extended S phase that is induced by bile acids may be caused by the continuous stimulation of the AP-1 and SRE promoters mediated by the constitutive activation of the ERK pathway in the presence of bile acids. The activation of the ERK pathway upon binding of HCV to CD81 and the continuous stimulation by bile acids suggest that the ERK pathway may play an important role in the overall life cycle of HCV. Furthermore, both the HCV core and NS5b proteins have been demonstrated to induce a high percentage of cells to hold in S phase when overexpressed in a non-neoplastic human hepatocyte cell line [46,47], suggesting that active replication of HCV may enhance cells in S phase, and consequently a further increase in virus replication. However, it is not clear what exact mechanism(s) is (are) involved in enhanced virus replication in S-staged cells. It is possible that proteins expressed in cells in S stage may act as co-factors for virus replicase, and we will examine this possibility in the future. The studies by Nelson and Tang [48] demonstrated that HCV expression was significantly reduced by the cell confluence, but cell growth arrest by serum starvation or the treatment with aphidicolin was not the mechanism of inhibition in the replicon-harboring cells. They also suggested that the reduced HCV expression at the cell confluence may be due to the lack of nucleoside availability with limited de novo pathways [48]. Thus, it is possible that the extended S stage with active de novo pathways of nucleosides may be a reason for the enhanced HCV replication.
When GS4.1 cells were treated with either CDCA, or UDCA, the phosphorylation of EGFR was seen in the presence of each bile acid after 30 min (fig. 3), suggesting an important role of EGFR in AP-1 and SRE activation and the cell cycle progress in the presence of bile acids. A 30-min time point was chosen because previously published data shows high levels of EGFR phosphorylation after bile acid stimulation in rat hepatocytes [27]. To study the effect of the activation of the EGFR/ERK pathway on bile acid-mediated enhancement of HCV replication and the interference with the anti-HCV effects of IFN-α, we used AG1478 and U0126. While AG1478 alone had little effect on HCV replication, it significantly reduced CDCA-mediated promotion of HCV replication at concentrations above 15 μM in both GS4.1 and 1A7 cells (fig. 4a). Similar to AG1478, U0126 had little effect on the HCV replication alone, but when combined with CDCA it significantly mitigated the bile acid-mediated enhancement of HCV (fig. 4b). This is contrary to the data published by Brazzoli et al. [42] in which the virus replication was significantly reduced by the treatment with U0126, and this could be due to the differences between the complete replication system and replicon systems. As Brazzoli et al. [42] demonstrated, the viral binding to CD81 initiated the ERK pathway, which may promote virus replication. In the replicon-harboring cells, without the initial virus–CD81 interaction, the addition of bile acids may lead to the same effects on ERK pathway and virus replication. Currently, we are studying the effects of bile acids on HCV replication using the complete virus replication system.
These data suggest that the ERK pathway plays an important role in the bile acid-mediated enhancement of HCV RNA in replicon-harboring cells. Since we previously demonstrated that bile acids compromise the anti-HCV action of IFN-α, we hypothesized that AG1478 and/or U0126 can restore the IFN-α action in replicon-harboring cells in the presence of bile acids by reducing the overall bile acid-mediated enhancement of the HCV replicon. The anti-HCV effects of IFN-α were significantly improved when GS4.1 and 1A7 cells were treated with the combination of CDCA and IFN-α with either U0126 or AG1478 (fig. 5). These data suggest that the addition of an EGFR or ERK inhibitor, such as AG1478 or U0126, to the current IFN-α-based treatment may improve overall efficacy by blocking the bile acid-mediated increase in HCV replication. In summary, we demonstrated that bile acids induce an EGFR/ERK pathway in genotype 1a and 1b replicon-harboring cells, which was associated with increased levels of HCV replication. Disruption of this signaling by either AG1478 or U0126 can significantly reduce the bile acid-mediated promotion of HCV and restore the anti-HCV effects of IFN-α in the cells in both genotypes.
Acknowledgements
This work was partly supported by NIH COBRE, P20RR016443. We thank Tammy Koopman for the assistance in flow cytometry analysis.
References
- 1.Dubuisson J. Hepatitis C virus proteins. World J Gastroenterol. 2007;13:2406–2415. doi: 10.3748/wjg.v13.i17.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jubin R. Hepatitis C internal ribosomal entry site: translating translation into a therapeutic target. Curr Opin Mol Ther. 2001;3:278–287. [PubMed] [Google Scholar]
- 3.Kato N. Genome of human hepatitis C virus: gene organization, sequence diversity, and variation. Microb Comp Genomics. 2000;5:129–151. doi: 10.1089/omi.1.2000.5.129. [DOI] [PubMed] [Google Scholar]
- 4.Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 5.Major E, Rehermann B, Feinstone SM. Hepatitis C Viruses. ed 4. Philadelphia: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 6.Alter HJ. HCV natural history: the retrospective and prospective in perspective. J Hepatol. 2005;43:550–552. doi: 10.1016/j.jhep.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Pawlotsky J. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 2004;12:96–102. doi: 10.1016/j.tim.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 9.Muir AJ, Bornstein JD, Killenberg PG. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med. 2004;350:2265–2271. doi: 10.1056/NEJMoa032502. [DOI] [PubMed] [Google Scholar]
- 10.Buti M, Casado MA, Fosbrook L, Wong JB, Esteban R. Cost-effectiveness of combination therapy for naive patients with chronic hepatitis C. J Hepatol. 2000;33:651–658. doi: 10.1034/j.1600-0641.2000.033004651.x. [DOI] [PubMed] [Google Scholar]
- 11.Wong JB, Poynard T, Ling MH, Albrecht JK, Pauker SG. Cost-effectiveness of 24 or 48 weeks of interferon-α2b alone or with ribavirin as initial treatment of chronic hepatitis C. International Hepatitis Interventional Therapy Group. Am J Gastroenterol. 2000;95:1524–1530. doi: 10.1111/j.1572-0241.2000.2089_1.x. [DOI] [PubMed] [Google Scholar]
- 12.Younossi ZM, Singer ME, McHutchison JG, Shermock KM. Cost effectiveness of interferon-α2b combined with ribavirin for the treatment of chronic hepatitis C. Hepatology. 1999;30:1318–1324. doi: 10.1002/hep.510300518. [DOI] [PubMed] [Google Scholar]
- 13.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi M, Lemon SM. Genotype 1a HCV (H77S) infection system. Methods Mol Biol. 2009;510:337–346. doi: 10.1007/978-1-59745-394-3_25. [DOI] [PubMed] [Google Scholar]
- 15.Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartenschlager R, Lohmann V. Novel cell culture systems for the hepatitis C virus. Antiviral Res. 2001;52:1–17. doi: 10.1016/s0166-3542(01)00164-4. [DOI] [PubMed] [Google Scholar]
- 18.Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 19.Trujillo-Murillo KDC, Garza-Rodríguez MDL, Martínez-Rodríguez HG, Barrera-Saldaña HA, Bosques-Padilla F, Ramos-Jiménez J, et al. Experimental models for hepatitis C virus: new opportunities for combating hepatitis C. Ann Hepatol. 2004;3:54–62. [PubMed] [Google Scholar]
- 20.Chang K, George DW. Bile acids promote the expression of hepatitis C virus in replicon-harboring cells. J Virol. 2007;81:9633–9640. doi: 10.1128/JVI.00795-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholtes C, Diaz O, Icard V, Kaul A, Bartenschlager R, Lotteau V, et al. Enhancement of genotype 1 hepatitis C virus replication by bile acids through FXR. J Hepatol. 2008;48:192–199. doi: 10.1016/j.jhep.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Geier A, Trautwein C. Bile acids are ‘homeotrophic’ sensors of the functional hepatic capacity and regulate adaptive growth during liver regeneration. Hepatology. 2007;45:251–253. doi: 10.1002/hep.21521. [DOI] [PubMed] [Google Scholar]
- 23.Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 24.Jaiswal K, Lopez-Guzman C, Souza RF, Spechler SJ, Sarosi GA. Bile salt exposure increases proliferation through p38 and ERK MAPK pathways in a non-neoplastic Barrett's cell line. Am J Physiol. 2006;290:G335–G342. doi: 10.1152/ajpgi.00167.2005. [DOI] [PubMed] [Google Scholar]
- 25.Jaiswal K, Tello V, Lopez-Guzman C, Nwariaku F, Anthony T, Sarosi GA. Bile salt exposure causes phosphatidyl-inositol-3-kinase-mediated proliferation in a Barrett's adenocarcinoma cell line. Surgery. 2004;136:160–168. doi: 10.1016/j.surg.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Z, Gong J, Zhang Z, Qiao Z. Influence of acid and bile acid on ERK activity, PPAR-γ expression and cell proliferation in normal human esophageal epithelial cells. World J Gastroenterol. 2006;12:2445–2449. doi: 10.3748/wjg.v12.i15.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao Y, Studer EJ, Stravitz RT, Gupta S, Qiao L, Dent P, et al. Activation of the Raf-1/MEK/ERK cascade by bile acids occurs via the epidermal growth factor receptor in primary rat hepatocytes. Hepatology. 2002;35:307–314. doi: 10.1053/jhep.2002.31104. [DOI] [PubMed] [Google Scholar]
- 28.Aardema KL, Nakhleh RE, Terry LK, Burd EM, Ma CK, Moonka DK, et al. Tissue quantification of hepatitis C virus RNA with morphologic correlation in the diagnosis of recurrent hepatitis C virus in human liver transplants. Mod Pathol. 1999;12:1043–1049. [PubMed] [Google Scholar]
- 29.Pietschmann T, Lohmann V, Rutter G, Kurpanek K, Bartenschlager R. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J Virol. 2001;75:1252–1264. doi: 10.1128/JVI.75.3.1252-1264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson J. Handbook of Flow Cytometry Methods. ed 1. West Lafayette: Purdue University; 1990. [Google Scholar]
- 31.Cormier EG, Tsamis F, Kajumo F, Durso RJ, Gardner JP, Dragic T. CD81 is an entry coreceptor for hepatitis C virus. Proc Natl Acad Sci USA. 2004;101:7270–7274. doi: 10.1073/pnas.0402253101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flint M, von Hahn T, Zhang J, Farquhar M, Jones CT, Balfe P, et al. Diverse CD81 proteins support hepatitis C virus infection. J Virol. 2006;80:11331–11342. doi: 10.1128/JVI.00104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meola A, Sbardellati A, Bruni Ercole B, Cerretani M, Pezzanera M, Ceccacci A, et al. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J Virol. 2000;74:5933–5938. doi: 10.1128/jvi.74.13.5933-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 35.Grove J, Huby T, Stamataki Z, Vanwolleghem T, Meuleman P, Farquhar M, et al. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J Virol. 2007;81:3162–3169. doi: 10.1128/JVI.02356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maillard P, Huby T, Andréo U, Moreau M, Chapman J, Budkowska A. The interaction of natural hepatitis C virus with human scavenger receptor SR-BI/Cla1 is mediated by ApoB-containing lipoproteins. FASEB J. 2006;20:735–737. doi: 10.1096/fj.05-4728fje. [DOI] [PubMed] [Google Scholar]
- 37.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voisset C, Callens N, Blanchard E, Op De Beeck A, Dubuisson J, Vu-Dac N. High-density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J Biol Chem. 2005;280:7793–7799. doi: 10.1074/jbc.M411600200. [DOI] [PubMed] [Google Scholar]
- 39.Johnson L. Secretion. In: Johnson L, editor. Essential Medical Physiology. New York: Lippincott-Raven; 1998. pp. 445–472. [Google Scholar]
- 40.Legrand-Defretin V, Juste C, Corring T, Rerat A. Enterohepatic circulation of bile acids in pigs: diurnal pattern and effect of a re-entrant biliary fistula. Am J Physiol. 1986;250:G295–G301. doi: 10.1152/ajpgi.1986.250.3.G295. [DOI] [PubMed] [Google Scholar]
- 41.Carloni V, Mazzocca A, Ravichandran KS. Tetraspanin CD81 is linked to ERK/MAPKinase signaling by Shc in liver tumor cells. Oncogene. 2004;23:1566–1574. doi: 10.1038/sj.onc.1207287. [DOI] [PubMed] [Google Scholar]
- 42.Brazzoli M, Bianchi A, Filippini S, Weiner A, Zhu Q, Pizza M, et al. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol. 2008;82:8316–8329. doi: 10.1128/JVI.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 44.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 45.Angel P, Karin The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 46.Naka K, Dansako H, Kobayashi N, Ikeda M, Kato N. Hepatitis C virus NS5B delays cell cycle progression by inducing interferon-β via Toll-like receptor-3 signaling pathway without replicating viral genomes. Virology. 2006;346:348–362. doi: 10.1016/j.virol.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 47.Ruggieri A, Murdolo M, Harada T, Miyamura T, Rapicetta M. Cell cycle perturbation in a human hepatoblastoma cell line constitutively expressing hepatitis C virus core protein. Arch Virol. 2004;149:61–74. doi: 10.1007/s00705-003-0202-x. [DOI] [PubMed] [Google Scholar]
- 48.Nelson HB, Tang H. Effect of cell growth on hepatitis C virus (HCV) replication and a mechanism of cell confluence-based inhibition of HCV RNA and protein expression. J Virol. 2006;80:1181–1190. doi: 10.1128/JVI.80.3.1181-1190.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]