Abstract
Early studies of a ureB mutant derivative of Helicobacter pylori had suggested that urease is needed for motility and that urease action helps energize flagellar rotation. Here we report experiments showing that motility is unaffected by deletion of ureA and ureB (urease genes) or by inactivation of ureB alone, especially if H. pylori strains used as recipients for transformation with mutant alleles are preselected for motility. This result was obtained with the strain used in the early studies (CPY3401) and also with 15 other strains, 3 of which can colonize mice. We conclude that urease is not needed for H. pylori motility.
Helicobacter pylori is a gram-negative pathogen that is implicated in peptic ulcer disease and gastric cancer (for reviews, see references 6, 9, and 23). Mutational analyses have shown that urease, an enzyme that H. pylori makes in abundance, is needed for mammalian infection but is not essential in laboratory culture (7, 22, 33, 35). Urease might have several in vivo roles, including (i) protection from gastric acidity, via the ammonia that it generates from the urea in host tissues (22, 26); (ii) nutrition, by ammonia-elicited tissue damage and a consequent release of host metabolites that promote H. pylori growth (4, 11, 28); and/or (iii) generation of proton motive force through urea hydrolysis (26), which in turn helps drive flagellar rotation and bacterial motility (20, 25, 36). The third role was postulated, based in large part, on the finding that a ureB insertion mutant derivative of H. pylori strain CPY3401 was nonmotile in standard soft agar medium (25, 36). Because motility itself is needed for H. pylori colonization (e.g., see references 3, 8, 15, and 35), a dependence of motility on urease action would complicate efforts to examine urease's other possible roles.
H. pylori's motility can be quite unstable, in part because of a repetitive, frameshift mutation-prone sequence in fliP, a flagellar biosynthetic gene (14, 32). In addition, flagellar synthesis and motility probably impose a physiologic cost, estimated at some 2% of total energy expenditure for enteric species (19). As a result, nonmotile strains would tend to outgrow isogenic motile strains when motility is not needed, as in standard laboratory culture. H. pylori is also an extremely diverse species genetically (1, 2, 10), and recent experiments have shown that background genotype can influence whether inactivation of certain metabolic genes will affect particular phenotypes. For example, depending on the H. pylori strain used, inactivation of the fdxA ferredoxin gene can be lethal or be tolerated; fdxA can help downregulate the frxA nitroreductase gene, but only in certain strains; and in a few cases, the lethality of fdxA inactivation can be reversed by prior frxA inactivation (24). We therefore wondered if urease-dependent motility might also be a strain-specific property.
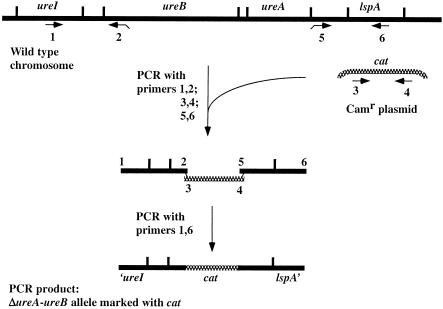
We were intrigued by Nakazawa's proposed connection between urease, motility, and intracellular metabolic flux (25, 36). The present mutational studies were begun in the hope of confirming her interpretation, testing its generality, and characterizing its basis. Transformable, motile H. pylori strains from our collection (Table 1) were cultured at 37°C in a microaerobic incubator with 5% O2 and 10% CO2, using brain heart infusion (BHI) agar with 7% horse blood (standard conditions) (13). Motility was assayed by inoculating cultures into 0.35% agar medium containing brucella broth with serum (motility agar) and incubating the inoculated plates as described previously (25). In testing single (transformant) colonies for motility, reproducibility was improved by first streaking colonies onto ∼1- to 2-cm2 patches on BHI agar, growing these patches for 24 h, and then using cells from them to inoculate motility test agar. To generate urease-deficient derivatives of motile and transformable strains, we generated an allele in which ureA and most of ureB was replaced by a cat (chloramphenicol resistance [Camr]) cassette by a PCR method that does not involve recombinant DNA plasmid cloning (illustrated in Fig. 1 and described in reference 5). The PCR product containing this ΔureAB allele was used to transform motile H. pylori strains, with selection for Camr (15 μg/ml) (13). PCR tests of representative Camr transformants demonstrated the expected replacement of ureA and ureB by cat in each case. Motility tests were carried out on at least four single colonies and also on pools containing 20 or more colonies from each transformation. In control experiments with parental strains, motile halos of growth were evident around points of inoculation on the 3rd or 4th day of incubation with 13 of 16 strains and on the 5th day with the other 3 strains; these halos continued increasing in size for at least 3 more days. Accordingly, the motility of ΔureAB transformants was also scored on each of several days, beginning on the 4th day after inoculation into motility agar.
TABLE 1.
List of H. pylori strains used in this study
| Strain name | Country of origin | Source (reference) |
|---|---|---|
| A28-1 | Lithuania | Limas Kupcinskas, Kaunas |
| A66-1 | Lithuania | Limas Kupcinskas |
| PeCan28 | Peru | Robert H. Gilman (16) |
| Chen13 | India | Usha Anand Rao, Chennai |
| F28 | Japan | Takeshi Azuma, Fukui |
| GS5 | Japan | Teruko Nakazawa, Ube |
| HK192 | Hong Kong | Benjamin Wong |
| PCM4 | Hong Kong | Benjamin Wong |
| R64 | South Africa | Issy Segal, Johannesburg |
| R66 | South Africa | Issy Segal |
| R76 | South Africa | Issy Segal |
| R82 | South Africa | Issy Segal |
| CPY3401 | Japan | Junko Akada and Teruko Nakazawa (33) |
| SS1 | Australia | Adrian Lee (18) |
| X47 | United States | Harry Kleanthous (17) |
| 88-3887 | United Kingdom | Kate Eaton (12, 14) |
FIG. 1.
Construction of ΔureA-ureB (deletion) allele marked with a resistance determinant. This construction involved PCR without cloning, essentially as described previously (5). The following six primers were used (see the figure for positions): 1 (ureABR2), 5′-TCCCTAAAGGGATTTTCAAGATGT; 2 (ureCAMF2), 5′-CCCAGTTTGTCGCACTGATAACCATGTGTTCGTGGATGGCAA; 3 (C2), 5′-TTATCAGTGCGACAAACTGGG; 4 (C1), 5′-GATATAGATTGAAAAGTGGAT; 5 (ureCAMR1), 5′-ATCCACTTTTCAATCTATATCATTCTCCTATTCTTAAAGTGTTTT; and 6 (ureABF1), 5′-CATGGGGGCGTGGTGGATTA. Overlaps between primers 2 and 3 and between primers 4 and 5 are underlined. PCR product sizes were 797 bp (primers 1 and 2), 742 bp (primers 3 and 4), 513 bp (primers 5 and 6), and 2,010 bp (primers 1 and 6).
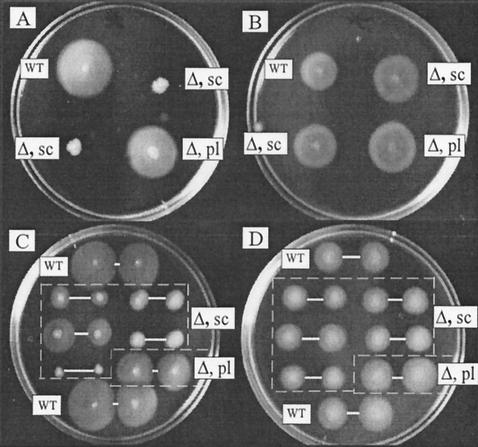
In an initial survey of 12 strains (Table 1, first 12 strains listed), nonmotile single transformant colonies were obtained from strains Chen13, R64, and GS5, as illustrated in Fig. 2A for strain Chen13. The lack of motility of the two single transformant colonies (labeled “sc”) matches that reported earlier with a ureB derivative of strain CPY3401 (25). However, pools of Camr (ΔureAB) transformants (labeled “pl”) seemed fully motile in each of these three cases. In addition, all tested transformants of the nine other strains remained motile. With four of these strains, however, single transformant colonies produced subtly smaller halos than did their wild-type parents, whereas with a fifth strain (R76), ΔureAB transformants produced slightly larger halos (Fig. 2B). Such subtle differences were sometimes difficult to reproduce or quantify reliably: they might be explained by changes in properties distinct from the strength or rate of flagellar rotation (e.g., effects on cell shape, clumping, or surface texture), and they have not been studied further to date.
FIG. 2.
Motility tests of representative H. pylori wild-type (WT) strains and isogenic ΔureAB transformants (Δ, sc, single colony; Δ, pl, pool) stabbed into motility agar and incubated for 6 days. The strains shown are Chen13 (A), R76 (B), R64 (C), and SS1 (D). In panels C and D, each culture was stabbed twice to score reproducibility of the motility assay. Duplicate stabs are designated by white bands.
To test whether differences in motility of single colony transformants versus pools from 3 of 12 strains (as in Fig. 2A) could be ascribed to heterogeneity in the recipient population, we prepared subpopulations of strains Chen13, R64, and GS5 from pools of bacterial cells recovered from edges of halos after 4 days of growth in motility agar. These “motility-selected” cell populations were then used as recipients in transformation. Single Camr (ΔureAB) transformant colony derivatives of Chen13 prepared in this way were fully as motile as their parents (data not shown). In further tests, two of four colonies from the original nontransformed Chen13 stock were nonmotile. Thus, our initial finding of urease-deficient derivatives of the original Chen13 stock that were nonmotile can be ascribed to heterogeneity in this stock, to variants that had accumulated before transformation. In contrast, marked diversity in halo size was again seen after transformation of motility-selected populations of strains R64 and GS5. This is illustrated in Fig. 2C for strain R64, in which each transformant colony tested was picked twice into motility agar to monitor reproducibility (duplicate stabs identified by white connecting bars). Of five transformants scored here, the halo size of one was about two-thirds that of its R64 wild-type parent, the halos of three others were more sharply reduced, and a fifth transformant seemed completely nonmotile. Similar heterogeneity was also seen in a motility-selected nontransformed R64 wild-type population. In further tests, two of six ΔureAB transformants (single colonies) of motility-selected GS5 also seemed completely nonmotile, whereas the other four formed halos that were about 80% as large as those of their wild-type parent. We infer that metastability of motility can be characteristic of certain H. pylori clinical isolates, as exemplified by strains R64 and GS5, independent of urease gene status.
With these results in hand, we investigated effects of deletion of ureA and ureB on an additional four strains, using subpopulations selected for high motility, as just described. These strains were CPY3401, early study of which (25) had suggested that motility was urease dependent; and SS1, X47, and 88-3887, each of which can colonize mice (12, 17, 18). In our hands, urease gene inactivation had no effect on halo size of the motility-selected CPY3401 recipient population, nor did it cause loss of motility in any of the other three motility-selected strains. With SS1, however, single colony transformants exhibited subtly reduced halo size (Fig. 2D). We do not yet know if the smaller halos of ΔureAB transformants of SS1 and 5 other strains (of the 16 tested) stem from urease gene inactivation per se, versus rapid accumulation of changes in other genes that affect motility or other determinants of halo size. In this context, we note that derivatives of strain X47 selected directly for high motility form halos approximately 20% larger in diameter than do derivatives recovered from infected mice (D. Dailidiene and D. E. Berg, unpublished observations), in which motility is also required. Subtle differences in halo size could be affected by many factors, such as cell shape and texture, as well as speed and pattern of flagellar rotation.
Two further control experiments were carried out. First, the effect of specific urea supplementation on H. pylori motility was tested, even though H. pylori can also derive urea from arginine in culture media (21). No difference in motility of ΔureAB transformants versus isogenic parents was seen in any of four lineages (CPY3401, SS1, X47, or 88-3887) on medium that was either free of additional urea or contained 1 or 5 mM urea (concentrations that have been used in other studies and that are in the range of those in the human gastric mucosa) (26, 27, 31, 34).
Second, we also studied the effect on motility of a ureB-null insertion allele, very similar to the one Nakazawa and collaborators had used (25). These ureB alleles each contained an aphA-3 (kanamycin resistance) gene inserted without deletion at the BamHI site in ureB (nucleotide position 1013, some 60% from this gene's 5′ end). (Our allele was generated by PCR in vitro, essentially as shown in Fig. 1 [primer sequences available on request].) ureB-deficient derivatives of strains CPY3401, SS1, X47 and 88-3887 were then generated by DNA transformation. In each case, the motility halos of ureB-deficient transformants matched those of isogenic ΔureAB strains and also those of their wild-type parents in three cases (CPY3401, X47, and 88-3887); halo sizes of ureB insertion derivatives of SS1 were about 80% of those of the parental wild type, as was also the case with SS1's ΔureAB derivatives (noted above). This outcome argued against a possible physiologic imbalance model, involving the UreA urease subunit without UreB, as an explanation for the reported nonmotility of a ureB H. pylori strain (25).
We suggest that the reported (25, 33, 36) nonmotility of one ureB derivative of CPY3401 stems from an unanticipated heterogeneity in the bacterial population used as transformation recipients and the unfortunate choice of a ureB transformant of a preexisting nonmotile subclone. Nonmotility would, in this case, be due to genetic change at another locus distinct from ureB: perhaps a locus such as fliP, which is metastable due to a frameshift-prone, repetitious DNA motif (14, 32). We had also seen nonmotility in some transformants of our original stock of strain Chen13. This result was interpreted to be spurious, because such transformants were not found with a motility-selected Chen13 recipient population. We suggest that the availability of numerous motile, urease-deficient, mouse-adapted strains of H. pylori and also of special achlorhydric mice (29, 30) should contribute to the definition of the H. pylori urease's true role or roles in vivo: e.g., critical tests of whether urease is needed solely to neutralize gastric acidity or whether this abundant enzyme also has additional vital roles.
Acknowledgments
We thank Junko Akada, Daiva Dailidiene, Keiji Ogura, Asish Mukhopadhyay, and Teruko Nakazawa for stimulating discussions and many collaborators for the H. pylori strains used here.
This work was supported by NIH grants AI38166, DK53727, DK63041, and P30 DK52574. S.T. was supported in part by a WU/HHMI Summer Undergraduate Research Fellowship funded by an Undergraduate Biological Sciences Education Program grant from the Howard Hughes Medical Institute to Washington University, by an undergraduate research fellowship from the American Society for Microbiology, and by a Florence Moog scholarship.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrutis, K. A., J. G. Fox, D. B. Schauer, R. P. Marini, X. Li, L. Yan, C. Josenhans, and S. Suerbaum. 1997. Infection of the ferret stomach by isogenic flagellar mutant strains of Helicobacter mustelae. Infect. Immun. 65:1962-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calam, J. 1995. Pathogenic mechanisms. Bailliere's Clin. Gastroenterol. 9:487-506. [DOI] [PubMed] [Google Scholar]
- 5.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cover, T. L., D. E. Berg, M. J. Blaser, and H. L. T. Mobley. 2001. H. pylori pathogenesis, p. 509-558. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, New York, N.Y.
- 7.Eaton, K. A., and S. Krakowka. 1994. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect. Immun. 62:3604-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst, P. B., and B. D. Gold. 2000. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu. Rev. Microbiol. 54:615-640. [DOI] [PubMed] [Google Scholar]
- 10.Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens, M. Kidd, M. J. Blaser, D. Y. Graham, S. Vacher, G. I. Perez-Perez, Y. Yamaoka, F. Megraud, K. Otto, U. Reichard, E. Katzowitsch, X. Wang, M. Achtman, and S. Suerbaum. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582-1585. [DOI] [PubMed] [Google Scholar]
- 11.Gobert, A. P., B. D. Mersey, Y. Cheng, D. R. Blumberg, J. C. Newton, and K. T. Wilson. 2002. Cutting edge: urease release by Helicobacter pylori stimulates macrophage inducible nitric oxide synthase. J. Immunol. 168:6002-6006. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman, P. S., N. Vats, D. Hutchison, J. Butler, K. Chisholm, G. Sisson, A. Raudonikiene, J. S. Marshall, and S. J. O. Veldhuyzen van Zanten. 2003. Development of an interleukin-12-deficient mouse model that is permissive for colonization by a motile KE26695 strain of Helicobacter pylori. Infect. Immun. 71:2534-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong, J.-Y., A. K. Mukhopadhyay, D. Dailidiene, Y. Wang, B. Velapatiño, R. H. Gilman, A. J. Parkinson, G. B. Nair, B. C. Y. Wong, S. K. Lam, R. Mistry, I. Segal, Y. Yuan, H. Gao, T. Alarcon, M. L. Brea, Y. Ito, D. Kersulyte, H.-K. Lee, Y. Gong, A. Goodwin, P. S. Hoffman, and D. E. Berg. 2000. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182:5082-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Josenhans, C., K. A. Eaton, T. Thevenot, and S. Suerbaum. 2000. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect. Immun. 68:4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Josenhans, C., and S. Suerbaum. 2001. Helicobacter motility and chemotaxis, p. 171-184. In M. Achtman and S. Suerbaum (ed.), Helicobacter pylori: molecular and cellular biology. Horizon Scientific Press, Wymondham, United Kingdom.
- 16.Kersulyte, D., B. Velapatiño, G. Dailide, A. K. Mukhopadhyay, Y. Ito, L. Cahuayme, A. J. Parkinson, R. H. Gilman, and D. E. Berg. 2002. Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity. J. Bacteriol. 184:992-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleanthous, H., T. J. Tibbitts, H. L. Gray, G. A. Myers, C. K. Lee, T. H. Ermak, and T. P. Monath. 2001. Sterilizing immunity against experimental Helicobacter pylori infection is challenge-strain dependent. Vaccine 19:4883-4895. [DOI] [PubMed] [Google Scholar]
- 18.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 19.MacNab, R. M. 1987. Motility and chemotaxis, p. 732-759. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C.
- 20.MacNab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C
- 21.McGee, D. J., F. J. Radcliff, G. L. Mendz, R. L. Ferrero, and H. L. T. Mobley. 1999. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J. Bacteriol. 181:7314-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mobley, H. L. T. 2001. Urease, p. 179-191. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 23.Mobley, H. L. T., G. L. Mendz, and S. L. Hazell (ed.). 2001. Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 24.Mukhopadhyay, A. K., J.-Y. Jeong, D. Dailidiene, P. S. Hoffman, and D. E. Berg. 2003. The fdxA ferredoxin gene can down-regulate frxA nitroreductase gene expression and is essential in many strains of Helicobacter pylori. J. Bacteriol. 185:2927-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura, H., H. Yoshiyama, H. Takeuchi, T. Mizote, K. Okita, and T. Nakazawa. 1998. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect. Immun. 66:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sachs, G., D. R. Scott, D. L. Weeks, M. Rektorscheck, and K. Melchers. 2001. Regulation of urease for acid habitation, p. 277-283. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 27.Sidebotham, R. L., M. L. Worku, Q. N. Karim, N. K. Dhir, and J. H. Baron. 2003. How Helicobacter pylori urease may affect external pH and influence growth and motility in the mucus environment: evidence from in-vitro studies. Eur. J. Gastroenterol. Hepatol. 15:395-401. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki, M., S. Miura, M. Suematsu, D. Fukumura, I. Kurose, H. Suzuki, A. Kai, Y. Kudoh, M. Ohashi, and M. Tsuchiya. 1992. Helicobacter pylori-associated ammonia production enhances neutrophil-dependent gastric mucosal cell injury. Am. J. Physiol. 263:G719-G725. [DOI] [PubMed] [Google Scholar]
- 29.Syder, A. J., J. L. Guruge, Q. Li, Y. Hu, C. M. Oleksiewicz, R. G. Lorenz, S. M. Karam, P. G. Falk, and J. I. Gordon. 1999. Helicobacter pylori attaches to NeuAc alpha 2, 3Gal beta 1, 4 glycoconjugates produced in the stomach of transgenic mice lacking parietal cells. Mol. Cell 3:263-274. [DOI] [PubMed] [Google Scholar]
- 30.Syder, A. J., J. D. Oh, J. L. Guruge, D. O'Donnell, M. Karlsson, J. C. Mills, B. M. Bjorkholm, and J. I. Gordon. 2003. The impact of parietal cells on Helicobacter pylori tropism and host pathology: an analysis using gnotobiotic normal and transgenic mice. Proc. Natl. Acad. Sci. USA 100:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Testerman, T. L., D. J. McGee, and H. L. T. Mobley. 2001. Helicobacter pylori growth and urease detection in the chemically defined medium Ham's F-12 nutrient mixture. J. Clin. Microbiol. 39:3842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 33.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen, Y., E. A. Marcus, U. Matrubutham, M. A. Gleeson, D. R. Scott, and G. Sachs. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 71:5921-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirth, H.-P., M. H. Beins, M. Yang, K. T. Tham, and M. J. Blaser. 1998. Experimental infection of Mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect. Immun. 66:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshiyama, H., and T. Nakazawa. 2000. Unique mechanism of Helicobacter pylori for colonizing the gastric mucus. Microbes Infect. 2:55-60. [DOI] [PubMed] [Google Scholar]