Abstract
The Shigella outer membrane protease IcsP removes the actin assembly protein IcsA from the bacterial surface, and consequently modulates Shigella actin-based motility and cell-to-cell spread. Here, we demonstrate that IcsP expression is undetectable in mutants lacking either of two transcriptional activators, VirF and VirB. In wild-type Shigella spp., virB expression is entirely dependent on VirF; therefore, to circumvent this regulatory cascade, we independently expressed VirF or VirB in Shigella strains lacking both activators and measured both IcsP levels and transcription from the icsP promoter. Our results show that VirB significantly enhanced icsP transcription, even in the absence of VirF. In contrast, when VirF was induced in the absence of VirB, VirF had variable effects. The regulation of icsP is distinctly different from the regulation of the gene encoding its major substrate, icsA, which is activated by VirF and not VirB. We propose that the different pathways regulating icsA and icsP may be critical to the modulation of IcsA-mediated actin-based motility by IcsP.
Shigella spp., gram-negative bacterial pathogens cause severe and bloody diarrhea in their human hosts by invading and spreading through the colonic epithelium. Shigella movement within the host cell cytoplasm is dependent on the ability of the bacterium to recruit host cell actin to its surface to form an “actin tail,” which propels the bacterium from one cell to another (5, 16, 29). Actin tail assembly is mediated by a single bacterial protein, IcsA, which is found on the outer surface at one pole of the bacterium (17). This asymmetric localization of IcsA ensures that actin assembly occurs in a directional manner. In its mature form, IcsA is comprised of two domains: the α domain (residues 53 to 758) contains the determinant for actin assembly (14) and extends from the bacterial surface into the extracellular environment, whereas the β domain (residues 759 to 1102) is embedded in the outer membrane (33). The amount of IcsA α domain exposed on the bacterial surface correlates with the efficiency of actin tail formation in the cytoplasm of infected cells (21).
IcsP, an outer membrane protease of Shigella, cleaves IcsA between Arg758 and Arg759, removing the entire IcsA α domain from the bacterial surface (8, 13, 15a, 31). Overexpression of IcsP leads to complete removal of the IcsA α domain from the bacterial cell surface (32), whereas genetic disruption of icsP increases the total amount of cell associated IcsA α domain, leading to an increase in the rate of actin-based movement of Shigella (31). Although IcsP is not required for polar localization of IcsA (6, 28), it contributes to the maintenance of a tight polar cap of IcsA on the bacterial surface (31). Furthermore, as Shigella enter stationary phase, the amount of cell-associated IcsA α domain decreases dramatically, an effect due at least in part to IcsP (18, 32).
These data demonstrate that IcsP plays an important role in modulating the amount of the IcsA α domain present on the bacterial surface and indicate that the amount of IcsA expressed on the bacterial surface correlates with the efficiency of Shigella actin-based motility. Given the importance of actin-based motility in Shigella pathogenesis, we postulate that it would be advantageous for IcsP to be tightly regulated. Here, we investigate the regulation of IcsP by two regulators of Shigella virulence protein expression, VirF and VirB.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in the present study are listed in Table 1. Bacteria were grown routinely at 37°C in Luria-Bertani (LB) broth (23) with aeration or on LB agar (LB broth containing 1.5% [wt/vol] agar). Antibiotics were added at the following final concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 25 μg ml−1; kanamycin, 50 μg ml−1; and tetracycline, 12.5 μg ml−1. Where appropriate, to ensure that Shigella strains had maintained the large virulence plasmid during manipulation, Congo red binding was tested on Trypticase soy broth agar plates containing 0.01% (wt/vol) Congo red (Sigma Chemical Co., St. Louis, Mo.).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| MC4100 | Wild type | 25 |
| MC4100 hns | MC4100 hns::Knr | 38 |
| S. flexneri | ||
| 2457T | Wild type serotype 2a | 15 |
| BS103 | 2457T cured of the virulence plasmid | 22 |
| AWY3 | 2457T virB::Tn5 | This work |
| AWY7 | 2457T virF::pMBG326 virB::Tn5 | This work |
| AWY8 | 2457T virF::pMBG326 virB::Tn5 PicsP-lacZ | This work |
| MBG338 | 2457T virF::pMBG326 | This work |
| MBG341 | 2457T icsP::Ampr | 31 |
| SY327 λpir | SY327 with integrated pir | 24 |
| YSH6000 | Wild type serotype 2a | 1 |
| YSH6000 virB | YSH6000 virB::Tn5 | 1 |
| Plasmids | ||
| pACYC184 | Cloning vector; Tetr/Cmr | 26 |
| pAM4 | pBC-KS-icsP | 31 |
| pATM324 | pBAD-virB; Ampr | 30 |
| pBAD18 | Arabinose-inducible pBAD expression vector, pBRori; Ampr | 19 |
| pAWY7 | pFSV-1-PicsP-lacZ Tetr | This work |
| pHJW4 | pBAD18-virF; Ampr | This work |
| pCVD442 | suicide vector; Ampr | 9 |
| pFSV-1 | suicide vector; Tetr | 4 |
| pHJW6 | icsP promoter and gene cloned into pACYC184 | This work |
| pHJW7 | icsP promoter transcriptionally fused to lacZ in pACYC184 | This work |
| pHJW14 | pBAD18-cat; Cmr | This work |
| pHJW16 | pHJW14-virB; Cmr | This work |
| pHJW17 | pBAD18-tet; Tetr | This work |
| pMBG326 | Suicide vector pCVD442 containing internal fragment of virF; Ampr | This work |
Ampr, ampicillin resistance; Tetr, tetracycline resistance; Cmr, chloramphenicol resistance; Knr, kanamycin resistance.
Construction of reporter plasmids.
The icsP reporter plasmid pHJW6 was constructed as follows. The icsP promoter (the 1,256-bp sequence located upstream of the icsP transcription start site) and icsP gene were isolated from the high-copy-number plasmid pAM4 (31) and cloned into the lower-copy-number plasmid pACYC184, so that the icsP gene is in the opposite orientation to the disrupted tetracycline resistance cassette. pHJW7, which is derived from pHJW6, carries the icsP promoter and the first 48 bp of the icsP coding region, cloned upstream of a translation stop site and a promoterless lacZ gene, so that expression of lacZ is directly regulated by the icsP promoter.
Construction of S. flexneri strains.
The S. flexneri 2457T virF mutant MBG338 was created as follows. A 570-bp fragment internal to the coding sequence of virF (extending from 131 bp to 701 bp of the open reading frame) was amplified by PCR from 2457T template and cloned into the ampicillin-resistant suicide vector pCVD442. The resultant plasmid (pMBG326) was introduced into the tetracycline-resistant S. flexneri strain BS109 by conjugation, and transconjugants were selected on ampicillin and tetracycline plates. Integration of the vector into the virF locus was verified by Southern blotting. The targeted virF disruption was then transduced into the S. flexneri wild-type strain by using P1L4 phage transduction and, again, integration at the virF locus was verified by Southern blotting.
The S. flexneri 2457T virB mutant AWY3 was created by moving the kanamycin-resistant locus from YSH6000 virB::Tn5 (gift of C. Sasakawa [1]) into the S. flexneri wild-type strain 2457T by transduction (as described above). To create the virF virB Shigella mutant AWY7, the ampicillin-resistant locus from MBG338 was transduced into the virB mutant AWY3. The icsP transcriptional reporter strain AWY8 was derived from AWY7 as follows. A fragment carrying the icsP-lacZ fusion from pHJW7 was cloned into the suicide plasmid pFSV-1 (a gift of J. Bliska [4]). The resultant plasmid, pAWY7, was introduced into the Escherichia coli strain SY327 λpir and then mobilized into AWY7 by conjugation. Transconjugants were selected by plating on tetracycline.
For strains generated in the present study, integration into the appropriate locus and, where appropriate, the presence of virF, virB, icsA, and icsP were routinely verified by PCR.
Construction of pBAD derivatives and inducible virF and virB expression constructs.
Where necessary, virF and virB were supplied in trans on the arabinose-inducible expression vector pBAD18. The virF or virB gene was amplified by PCR from 2457T template and cloned into the multiple cloning site so that its expression was under the control of the l-arabinose-inducible araBAD promoter, thereby generating pBAD-virF and pBAD-virB. To introduce pBAD-virF or pBAD-virB into ampicillin-resistant strain backgrounds, tetracycline or chloramphenicol resistance derivatives were generated by cloning the respective resistance genes into the ScaI site in the bla gene of pBAD18. All pBAD plasmids carrying virF or virB were checked for their ability to complement the Congo red minus phenotype of a virF mutant or a virB mutant, respectively, when grown in the presence of 0.2% (wt/vol) l-arabinose.
Quantification of IcsP levels in Shigella.
Throughout this study, IcsP expression was measured in mid-exponential-phase cultures because preliminary experiments had shown that IcsP is first detected in wild-type cells under these conditions (data not shown). Cells were routinely back-diluted 1:100 from an overnight culture and grown for 4 to 5 h in LB (at which point the A600 was ∼2.5). To examine the effect of expressing virF or virB from the pBAD vectors, cells were instead back-diluted 1:100 in 5 ml of LB medium containing 0.2% (wt/vol) glucose and, after 4 to 5 h were harvested, washed with an equivalent volume of LB medium and diluted 5-fold into LB medium containing either 0.08% (wt/vol) l-arabinose or 0.08% (wt/vol) d-arabinose. Cultures were then grown for an additional 2 h before being harvested.
Whole-cell protein extracts were prepared as described previously (32). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Equivalent amounts of protein were loaded by normalizing the harvest volume to cell density. Western blot analysis was performed with an affinity-purified IcsP rabbit antiserum (32) and by enhanced chemiluminescence (Pierce). Integrated density measurements were performed by using QuantityOne software on a Bio-Rad Gel Doc 2000 system.
Quantification of transcription from the icsP promoter.
Transcription from the icsP promoter was determined by measuring β-galactosidase activity (as described previously [20] by using the Miller protocol) in AWY8 or strains carrying pHJW7. Routinely, transcription was analyzed in mid-exponential-phase cultures. Cells were back-diluted 1:100 and grown for 4 to 5 h in LB medium. To examine the effect of expressing virF or virB from the pBAD vectors, cells were instead grown as described above before being harvested for β-galactosidase quantification.
RESULTS AND DISCUSSION
IcsP expression is dependent on virulence plasmid encoded loci.
IcsP is encoded by a monocistronic operon on the large (230-kb) virulence plasmid of Shigella (13, 31). Many virulence plasmid loci are regulated by factors encoded by the virulence plasmid. To examine IcsP expression in strains with or without the virulence plasmid, a low-copy-number plasmid carrying the icsP operon (pHJW6) was introduced into both S. flexneri 2a strain 2457T cured of the virulence plasmid (BS103) and a 2457T icsP mutant. Since both strains lacked the native icsP gene but carried the icsP reporter plasmid (pHJW6), direct comparison of IcsP expression could be made. Approximately fivefold more IcsP was detected in the strain carrying the virulence plasmid (Fig. 1, lane 1 compared to lane 2). The plasmid copy number was comparable in the two strains. These results indicate that virulence plasmid encoded factors positively regulate IcsP expression.
FIG. 1.
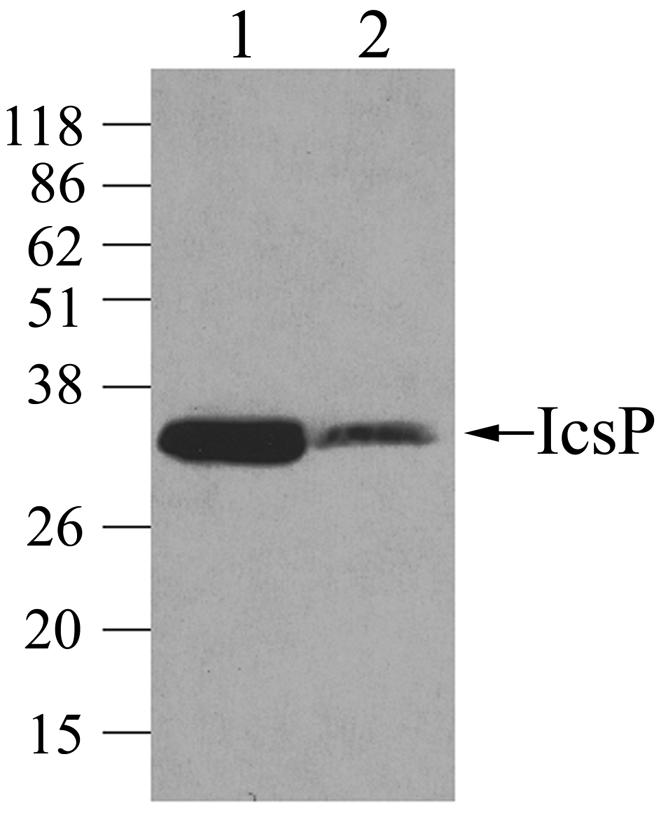
Comparison of IcsP levels in Shigella strains with or without the virulence plasmid. Western blot analysis of the icsP mutant (MBG341) (lane 1) and virulence plasmid-cured Shigella (BS103) (lane 2), each carrying the icsP reporter plasmid (pHJW6), was performed with IcsP antiserum. Equivalent amounts of total cellular protein were loaded onto the gel for the two strains. The experiment was repeated three times, and representative data are shown. Apparent molecular masses are indicated in kilodaltons.
IcsP is undetectable in virF and virB mutants of Shigella.
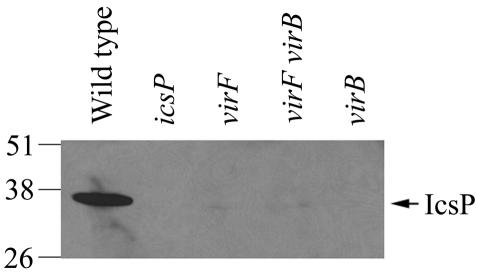
VirF and VirB (InvE) are virulence plasmid encoded transcription factors that regulate many genes encoded by the Shigella virulence plasmid (12). Virulence gene expression is thermoregulated and maximal at 37°C. At this temperature, VirF activates the transcription of icsA and virB (invE). VirB, whose expression is completely dependent on VirF, in turn activates additional genes, most of which are found within the 31-kb invasion locus of the virulence plasmid (12), which encodes the Shigella type III secretion apparatus. Since we had demonstrated that the absence of the virulence plasmid results in a decrease in IcsP expression, we investigated whether this phenotype was due to the loss of virF and/or virB. In mutants lacking virF (MBG338), virB (AWY3), or both (AWY7), IcsP was markedly reduced (Fig. 2). The absence of IcsP in these strains was due to the lack of either VirF or VirB, because IcsP expression was restored in the virF and virB mutants by complementation with virF or virB, respectively (data not shown). Of note, the band corresponding to IcsP in the virulence plasmid cured strain (Fig. 1, lane 2) was more prominent than those in the virF (MBG338), virB (AWY3), and virF virB mutants (AWY7) (Fig. 2). This was likely due to differences in exposure of the blots and the presence of icsP on the multicopy reporter plasmid (pHJW6) in the virulence plasmid-cured strain (Fig. 1). Since virF mutants do not express VirB (1, 35), these results suggest that VirF has no significant effect on IcsP expression under these experimental conditions (Fig. 2, virB mutant) and that VirB increases IcsP expression.
FIG. 2.
IcsP levels in the absence of the transcriptional activators VirF and VirB. Western blot analysis of total cellular protein harvested from wild-type Shigella (2457T), the icsP mutant (MBG341), the virF mutant (MBG338), the virF virB mutant (AWY7), and the virB mutant (AWY3) was performed with IcsP antiserum. Apparent molecular masses are indicated in kilodaltons.
Expression of VirF or VirB independently increases IcsP levels.
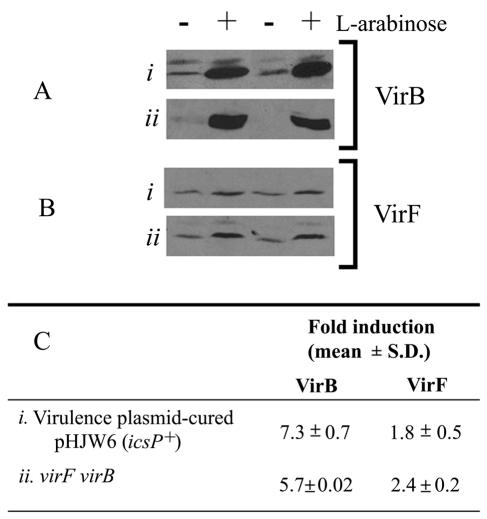
To investigate whether each activator contributes independently to the expression of IcsP, IcsP levels were measured in strains lacking both virF and virB but supplied with either virF or virB in trans. Virulence plasmid-cured Shigella (BS103) containing the icsP reporter plasmid (pHJW6) and the virF virB Shigella mutant (AWY7) were each supplied with either virF or virB under the control of the arabinose promoter on a multicopy vector (PBAD-virF and PBAD-virB). When virB was induced, IcsP levels were increased 7.3-fold in virulence plasmid-cured Shigella carrying the icsP reporter plasmid, BS103(pHJW6), and by 5.7-fold in the virF virB mutant (AWY7; Fig. 3, showing the results of duplicate experiments). These data confirm that VirB positively regulates IcsP in the absence of VirF.
FIG. 3.
Effect of independent expression of VirF or VirB on IcsP levels. Western blot analysis of Shigella strains deficient for both virF and virB but supplied in trans with either virF or virB expressed from an arabinose-inducible promoter was carried out. IcsP expression was measured in (i) virulence plasmid-cured Shigella (BS103) carrying the icsP reporter plasmid (pHJW6) or (ii) the virF virB mutant (AWY7). Cells were grown either with (+) or without (−) induction of virB (A) or virF (B), and data from duplicate experiments are shown. (C) Quantification of bands by integrated densitometry. Approximately fourfold less protein was loaded in the top blot of each panel, due to the presence of icsP on a multicopy plasmid in the virulence plasmid-cured strain.
Surprisingly, when virF was expressed at high levels in either strain background, IcsP levels increased significantly, albeit by small amounts [1.8-fold in BS103(pHJW6) and 2.4-fold in AWY7; Fig. 3C]. These results contrast with those shown in Fig. 2, where native levels of VirF did not significantly increase IcsP expression in the absence of VirB, and indicate that VirF may regulate IcsP expression independently of VirB. Although in this experiment, the increase in IcsP expression may be caused by supraphysiological levels of VirF, it is possible that regulation of IcsP by VirF may be physiologically relevant under experimental conditions not examined in the present study or may have been relevant at some prior point in Shigella evolution. Regardless of whether IcsP was expressed from the icsP reporter plasmid (pHJW6 in BS103) or from the native gene (in AWY7), regulation by VirF and VirB followed a similar pattern, indicating the low-copy plasmid encoding icsP (pHJW6) was a reasonable reporter of IcsP expression.
VirB significantly enhances transcription of icsP.
To examine the effect of VirB on icsP transcription, we measured β-galactosidase production from a low-copy-number icsP-lacZ transcriptional reporter (pHJW7) in virulence plasmid-cured Shigella and in the virF virB mutant (AWY7), each carrying PBAD-virB. When virB was induced in each strain, transcription from the icsP promoter was significantly increased, indicating that VirB activates icsP transcription (Table 2).
TABLE 2.
Activation of icsP-lacZ transcriptional fusions by VirB
| Strain | Genotypea | β-Galactosidase expressionb
|
Fold activation (mean ± SD) | |
|---|---|---|---|---|
| Uninduced | Induced | |||
| BS103(pHJW7) | 2457T vir. plasmid-cured PicsP-lacZ | 260 | 3,996 | 15.4 ± 0.7 |
| AWY7(pHJW7) | 2457T virF virB PicsP-lacZ | 1,284 | 10,119 | 7.9 ± 0.9 |
| AWY8 | AWY7 PicsP-lacZ | 5,384 | 13,749 | 2.6 ± 0.1 |
In all backgrounds, virB was carried on a pBAD18 derivative.
In Miller units. Data are the means of three experiments.
DNA topology has been shown to influence transcription (11). Therefore, we proceeded to measure transcription from the icsP promoter in its natural context. An icsP-lacZ transcriptional reporter was integrated onto the Shigella virulence plasmid at the native icsP locus in a virF virB mutant. The resultant strain, AWY8, carries both an icsP-lacZ transcriptional fusion at the locus normally occupied by the icsP gene and a second copy of the icsP promoter controlling the native gene downstream of the integrant. Transcription from the icsP promoter in AWY8 was increased by 2.6-fold in the presence of VirB (Table 2). Taken together, these data demonstrate that VirB positively regulates the icsP at the level of transcription.
Interestingly, activation of the icsP promoter by VirB was significantly greater in virulence plasmid-cured Shigella (BS103) than in the virF virB mutant (AWY7), suggesting that the presence of the virulence plasmid inhibits transcription from the icsP promoter. One possibility is that VirB activation of icsP is modulated by a virulence plasmid factor. Alternatively, since the intracellular concentration of VirB has been shown to correlate with levels of virulence gene expression (3), reduced levels of activation by VirB in the presence of the virulence plasmid could be caused by titration of VirB by other VirB binding sites. It was also notable that icsP promoter activation from the integrated reporter (in AWY8) was lower than icsP promoter activation from the plasmid-borne reporters (in BS103 and AWY7), suggesting the context of the icsP promoter modulates its activation by VirB.
Although our data do not distinguish whether the effect of VirB on the icsP promoter is direct or indirect, VirB increases transcription from the icsP promoter in both virulence plasmid-cured Shigella (BS103; Table 2) and E. coli (data not shown), a finding consistent with a direct effect. Recently, a consensus binding site has been described for VirB in S. sonnei (34). Our analysis of the icsP promoter region has revealed five sites that are similar to those identified in S. sonnei, with one (GAGAAAT), located 172 bp upstream of the proposed transcription start site, having a complete match to the consensus (A/G)(A/T)G(G)AAAT sequence (13). It is not yet known whether any of these putative VirB binding sites is required for the regulation of icsP by VirB.
The icsP promoter is repressed by H-NS and derepressed by VirB.
The virB gene lies immediately adjacent to the ipa-mxi-spa region of the virulence plasmid. Previously, VirB has been shown to increase transcription from three promoters in the ipa-mxi-spa region of the virulence plasmid (PicsB, PipgD, and Pspa [1, 10, 37]), as well as from the virA promoter, which is found in a distinct region on the virulence plasmid (36). The icsP gene is located close to the origin of replication of the virulence plasmid, which is distant from and directly opposite the ipa-mxi-spa locus on a circular map. Therefore, whereas virB and the ipa-mxi-spa locus were likely incorporated into the virulence plasmid as a single evolutionary event, it seems likely that icsP and virA were acquired in distinct events, implying that VirB regulation of icsP and virA may have evolved recently.
Each of the previously described VirB-regulated promoters is repressed by the nucleoid structuring protein H-NS, leading to the proposal that the role of VirB at these promoters is one of derepression rather than activation (2). To examine whether the icsP promoter was also repressed by H-NS and derepressed by VirB, we examined both icsP transcription in the presence or absence of H-NS and the effect of VirB on icsP transcription under these conditions. β-Galactosidase production from the icsP::lacZ fusion plasmid (pHJW7) was compared for an E. coli MC4100 hns mutant (MC4100 hns::Kn) and the wild-type strain MC4100 after growth at either 30 or 37°C with or without induction of VirB expression from the PBAD-virB plasmid. At both temperatures, regardless of whether VirB was present, icsP transcription was increased in the hns mutant compared to the wild type (Table 3), a finding consistent with H-NS repression of the promoter. Furthermore, in the wild-type background when VirB was induced transcription was significantly increased at both 30 and 37°C (5.4- and 1.8-fold, respectively). Interestingly, in the hns strain no additional increase in promoter activity was observed when VirB was induced, a finding consistent with VirB derepressing the icsP promoter rather than activating it per se. It has been reported that the copy number of at least some plasmids is lower in an hns mutant background (7); since icsP expression was increased in the hns mutant, such a difference in copy number could not be responsible for these results. We conclude, therefore, that the icsP promoter is repressed by H-NS at both 30 and 37°C and that VirB can overcome this repression to some extent at both temperatures, although more effectively at 30°C (Table 3; 5.4-fold compared to 1.8-fold at 37°C). Thus, the regulation of the icsP promoter by H-NS is similar to that previously described for other VirB-regulated promoters.
TABLE 3.
Repression of icsP-lacZ by H-NS and derepression by VirB
| Strain | Genotypea | Fold activationb (mean ± SD) at:
|
|||
|---|---|---|---|---|---|
| 30°C
|
37°C
|
||||
| Uninduced | Induced | Uninduced | Induced | ||
| MC4100 (pHJW7) | Wild type PicsP-lacZ | 1.0 ± 0.1 | 5.4 ± 0.3 | 0.8 ± 0.1 | 1.8 ± 0.1 |
| MC4100 hns (pHJW7) | hns::Kanr PicsP-lacZ | 8.8 ± 0.3 | 7.5 ± 0.4 | 7.2 ± 0.01 | 6.0 ± 0.3 |
In both backgrounds, virB was carried on a pBAD18 derivative.
Data have been normalized to the activity obtained in the wild-type strain in the absence of VirB at 30°C and are displayed as the fold activation. The mean and standard deviation of three experiments are presented.
Role of VirF in transcription of icsP.
We also examined whether expression of VirF enhanced transcription of icsP in the absence of VirB. We observed variable and nonreproducible effects of VirF on expression of the icsP-lacZ reporters in each of the strain backgrounds described above. In the presence of VirF, icsP transcription was unchanged or increased up to 1.5-fold. Nevertheless, in control experiments, induction of the same virF construct increased expression of an icsA-lacZ fusion 2.9-fold, similar to the 2.5- to 5-fold activation of icsA by VirF that has been reported previously (27), indicating that the virF construct was functional. We were unable to define experimental conditions in which activation of icsP-lacZ was reproducible. Since small increases in transcription can lead to significant increases in steady-state levels of protein, a minor effect on icsP transcription could possibly account for the observed increases in IcsP protein (Fig. 3). Moreover, overall protein expression was not significantly altered in the presence of VirF (data not shown), suggesting increases in IcsP expression by VirF are specific.
Since the possibility remained that VirF might contribute to the regulation of icsP in the presence of VirB, we examined whether VirF might have a more significant effect on icsP transcription in the presence of VirB. We compared icsP-lacZ transcription after induction of virF alone, virB alone, or the two together in AWY7. When virF and virB were induced simultaneously, transcription from the icsP promoter was increased slightly and reproducibly, but not significantly, compared to when virB was induced alone (data not shown), indicating VirF and VirB do not cooperatively regulate the icsP promoter.
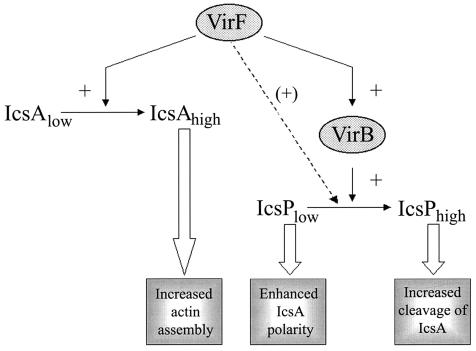
The regulation of IcsP by VirB, and possibly VirF, is distinctly different from the regulation of the major substrate of IcsP, IcsA. IcsA is transcriptionally activated by VirF but is unaffected by VirB (1, 27). Because VirB depends on VirF for its own activation, the different pathways that lead to the expression of IcsA and IcsP may reflect subtle differences in the timing and levels of expression of the two proteins during infection. Since the balance between levels of IcsA and levels of IcsP is a critical determinant of the ability of the organism to undergo actin-based motility, differential regulation of the two genes enables the organism to fine-tune this balance, thereby modulating actin-based motility. We propose a model in which VirB activation of IcsP expression leads to increased cleavage of the IcsA actin assembly domain (α domain) from the bacterial surface (Fig. 4). We postulate that this, in conjunction with the distinct pathway of IcsA activation, leads to precise modulation of Shigella actin-based motility during infection.
FIG. 4.
Model of the distinct regulatory pathways that modulate IcsP and IcsA expression in Shigella. VirF positively regulates transcription of icsA and virB. This increases the amount of IcsA on the bacterial surface and leads to increased levels of VirB. VirB positively regulates transcription of icsP, leading to an increase in IcsP from its basal level of expression. VirF may, under certain circumstances, also increase IcsP expression (see the text).
Acknowledgments
This study was supported by NIH grants AI35817 (M.B.G.) and AI43562 (M.B.G.), the Massachusetts General Hospital Fund for Medical Discovery (H.J.W.), and the Charles H. Hood Foundation, Inc., Boston, Mass. (H.J.W.).
We thank C. Sasakawa, A. Maurelli, S. Busby, and J. Bliska for providing plasmids and strains and J. Butterton and C. Lesser for critical reading of the manuscript.
REFERENCES
- 1.Adler, B., C. Sasakawa, T. Tobe, S. Makino, K. Komatsu, and M. Yoshikawa. 1989. A dual transcriptional activation system for the 230-kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3:627-635. [DOI] [PubMed] [Google Scholar]
- 2.Beloin, C., and C. J. Dorman. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47:825-838. [DOI] [PubMed] [Google Scholar]
- 3.Beloin, C., S. McKenna, and C. J. Dorman. 2002. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J. Biol. Chem. 277:15333-15344. [DOI] [PubMed] [Google Scholar]
- 4.Bliska, J. B., K. L. Guan, J. E. Dixon, and S. Falkow. 1991. Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc. Natl. Acad. Sci. USA 88:1187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandon, L. D., and M. B. Goldberg. 2000. Exploitation of mammalian host cell function by Shigella spp., p. 175-187. In K. A. Brogden et al. (ed.), Virulence mechanisms of bacterial pathogens, 3rd ed. ASM Press, Washington, D.C.
- 6.Charles, M., M. Perez, J. H. Kobil, and M. B. Goldberg. 2001. Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae and Vibrio. Proc. Natl. Acad. Sci. USA 98:9871-9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deighan, P., A. Free, and C. J. Dorman. 2000. A role for the Escherichia coli H-NS-like protein StpA in OmpF porin expression through modulation of micF RNA stability. Mol. Microbiol. 38:126-139. [DOI] [PubMed] [Google Scholar]
- 8.d'Hauteville, H., R. Dufourc Qlagelouse, F. Nato, and P. J. Sansonetti. 1996. Lack of cleavage of IcsA in Shigella flexneri causes aberrant movement and allows demonstration of a cross-reactive eukaryotic protein. Infect. Immun. 64:511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorman, C. J., S. McKenna, and C. Beloin. 2001. Regulation of virulence gene expression in Shigella flexneri, a facultative intracellular pathogen. Int. J. Med. Microbiol. 291:89-96. [DOI] [PubMed] [Google Scholar]
- 11.Dorman, C. J., and N. Ni Bhriain. 1993. DNA topology and bacterial virulence gene regulation. Trends Microbiol. 1:92-99. [DOI] [PubMed] [Google Scholar]
- 12.Dorman, C. J., and M. E. Porter. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29:677-684. [DOI] [PubMed] [Google Scholar]
- 13.Egile, C., H. d'Hauteville, C. Parsot, and P. J. Sansonetti. 1997. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol. Microbiol. 23:1063-1073. [DOI] [PubMed] [Google Scholar]
- 14.Egile, C., T. P. Loisel, V. Laurent, R. Li, D. Pantaloni, P. J. Sansonetti, and M. F. Carlier. 1999. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 146:1319-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Formal, S. B., G. J. Dammin, E. H. LaBrec, and H. Schneider. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J. Bacteriol. 75:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Fukuda, I., T. Suzuki, H. Munakata, N. Hayashi, E. Katayama, M. Yoshikawa, and C. Sasakawa. 1995. Cleavage of Shigella surface protein VirG occurs at a specific site, but the secretion is not essential for intracellular spreading. J. Bacteriol. 177:1719-1726. [DOI] [PMC free article] [PubMed]
- 16.Goldberg, M. B. 2001. Actin-based motility of intracellular microbial pathogens. Microbiol. Mol. Biol. Rev. 65:595-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg, M. B., O. Barzu, C. Parsot, and P. J. Sansonetti. 1993. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. Infect. Agents Dis. 2:210-211. [PubMed] [Google Scholar]
- 18.Goldberg, M. B., J. A. Theriot, and P. J. Sansonetti. 1994. Regulation of surface presentation of IcsA, a Shigella protein essential to intracellular movement and spread, is growth phase dependent. Infect. Immun. 62:5664-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayaraman, P. S., T. C. Peakman, S. J. Busby, R. V. Quincey, and J. A. Cole. 1987. Location and sequence of the promoter of the gene for the NADH-dependent nitrite reductase of Escherichia coli and its regulation by oxygen, the Fnr protein and nitrite. J. Mol. Biol. 196:781-788. [DOI] [PubMed] [Google Scholar]
- 21.Magdalena, J., and M. B. Goldberg. 2002. Quantification of Shigella IcsA required for bacterial actin polymerization. Cell Motil. Cytoskeleton 51:187-196. [DOI] [PubMed] [Google Scholar]
- 22.Maurelli, A. T., B. Blackmon, and R. Curtiss III. 1984. Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect. Immun. 43:397-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae require. toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pogliano, J. A., and J. Beckwith. 1994. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 13:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rose, R. E. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakai, T., C. Sasakawa, and M. Yoshikawa. 1988. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kiloDalton virF protein. Mol. Microbiol. 2:589-597. [DOI] [PubMed] [Google Scholar]
- 28.Sandlin, R. C., and A. T. Maurelli. 1999. Establishment of unipolar localization of IcsA in Shigella flexneri 2a is not dependent on virulence plasmid determinants. Infect. Immun. 67:350-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sansonetti, P. J., and C. Egile. 1998. Molecular bases of epithelial cell invasion by Shigella flexneri. Antonie Leeuwenhoek 74:191-197. [DOI] [PubMed] [Google Scholar]
- 30.Schuch, R., R. C. Sandlin, and A. T. Maurelli. 1999. A system for identifying post-invasion functions of invasion genes: requirements for the Mxi-Spa type III secretion pathway of Shigella flexneri in intercellular dissemination. Mol. Microbiol. 34:675-689. [DOI] [PubMed] [Google Scholar]
- 31.Shere, K. D., S. Sallustio, A. Manessis, T. G. D'Aversa, and M. B. Goldberg. 1997. Disruption of IcsP, the major Shigella protease that cleaves IcsA, accelerates actin-based motility. Mol. Microbiol. 25:451-462. [DOI] [PubMed] [Google Scholar]
- 32.Steinhauer, J., R. Agha, T. Pham, A. W. Varga, and M. B. Goldberg. 1999. The unipolar Shigella surface protein IcsA is targeted directly to the bacterial old pole: IcsP cleavage of IcsA occurs over the entire bacterial surface. Mol. Microbiol. 32:367-377. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, T., M. C. Lett, and C. Sasakawa. 1995. Extracellular transport of VirG protein in Shigella. J. Biol. Chem. 270:30874-30880. [DOI] [PubMed] [Google Scholar]
- 34.Taniya, T., J. Mitobe, S. Nakayama, Q. Mingshan, K. Okuda, and H. Watanabe. 2003. Determination of the InvE binding site required for expression of IpaB of the Shigella sonnei virulence plasmid: involvement of a ParB boxA-like sequence. J. Bacteriol. 185:5158-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobe, T., S. Nagai, N. Okada, B. Adler, M. Yoshikawa, and C. Sasakawa. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5:887-893. [DOI] [PubMed] [Google Scholar]
- 36.Uchiya, K., T. Tobe, K. Komatsu, T. Suzuki, M. Watarai, I. Fukuda, M. Yoshikawa, and C. Sasakawa. 1995. Identification of a novel virulence gene, virA, on the large plasmid of Shigella, involved in invasion and intercellular spreading. Mol. Microbiol. 17:241-250. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe, H., E. Arakawa, K. Ito, J. Kato, and A. Nakamura. 1990. Genetic analysis of an invasion region by use of a Tn3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of invE with ParB of plasmid P1. J. Bacteriol. 172:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada, H., T. Yoshida, K. Tanaka, C. Sasakawa, and T. Mizuno. 1991. Molecular analysis of the Escherichia coli gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol. Gen. Genet. 230:332-336. [DOI] [PubMed] [Google Scholar]