Abstract
We designed a selection strategy for the isolation of Escherichia coli mutants exhibiting enhanced protein disulfide isomerase activity. The folding of a variant of tissue plasminogen activator (v-tPA), a protein containing nine disulfide bonds, in the bacterial periplasm is completely dependent on the level of disulfide isomerase activity of the cell. Mutations that increase this activity mediate the formation of catalytically active v-tPA, which in turn cleaves a p-aminobenzoic acid (PABA)-peptide adduct to release free PABA and thus allows the growth of an auxotrophic strain. Following chemical mutagenesis, a total of eight E. coli mutants exhibiting significantly higher disulfide isomerization activity, not only with v-tPA but also with two other unrelated protein substrates, were isolated. This phenotype resulted from significantly increased expression of the bacterial disulfide isomerase DsbC. In seven of the eight mutants, the upregulation of DsbC was found to be related to defects in RNA processing by RNase E, the rne gene product. Specifically, the genetic lesions in five mutants were shown to be allelic to rne, while an additional two mutants exhibited impaired RNase E activity due to lesions in other loci. The importance of mRNA stability on the expression of DsbC is underscored by the short half-life of the dsbC transcript, which was found to be only 0.8 min at 37°C in wild-type cells but was two- to threefold longer in some of the stronger mutants. These results (i) confirm the central role of DsbC in disulfide bond isomerization in the bacterial periplasm and (ii) suggest a critical role for RNase E in regulating DsbC expression.
The formation of disulfide bonds is a critical step in the biogenesis of many secreted proteins. Disulfide bonds are introduced into proteins when newly synthesized polypeptides are translocated from the more reducing environment of the cytoplasm into the oxidizing environment of secretory compartments, such as the periplasmic space of gram-negative bacteria or the endoplasmic reticulum of eukaryotic cells. In Escherichia coli, oxidation of protein thiols in newly secreted proteins is catalyzed by the periplasmic enzyme DsbA (5, 21). Disulfide bonds between cysteines that are not linked in the native structure must be rearranged, a function that is catalyzed by two homologous, homodimeric enzymes, DsbC and DsbG (2, 19, 20, 23). Even though the periplasm is a highly oxidized environment due to the action of DsbA, which is a very potent catalyst of thiol oxidation, both DsbC and DsbG are maintained in a reduced state in the periplasm through the action of the integral membrane protein DsbD, which transfers electrons from thioredoxin in the cytoplasm to the active site thiols of DsbC and DsbG in the periplasmic space through a complex and novel electron transport pathway (8).
dsbC was originally isolated as a suppressor of dsbA mutants deficient in protein thiol oxidation and as a gene that confers dithiothreitol sensitivity (13, 22). It took several years of subsequent biochemical studies to establish the true physiological role of DsbC as the major disulfide isomerase in the E. coli periplasm (19, 23, 27). In fact, although disulfide isomerases were discovered almost 40 years ago by C. Anfinsen and have been the subject of intensive research ever since (6), there have been no reports of any genetic analyses specific for disulfide bond isomerization.
Here we report the design of a selection strategy in which E. coli growth is directly linked to disulfide isomerization activity in the periplasm. Using this selection strategy, we isolated eight mutant strains that exhibited significantly higher disulfide isomerization activity than the wild-type strain. All the mutations conferred significantly elevated DsbC protein expression. Unexpectedly, in seven of the eight mutants the upregulation of DsbC was mediated by defects in RNA processing by RNase E, which is encoded by the rne gene; mutations in five mutants were allelic to rne, whereas two other mutants were shown to exhibit impaired RNase E activity due to lesions in other loci.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and growth condition.
Bacterial strains, plasmids, and primers used in this work are listed in Table 1. E. coli strain MCZ4 was constructed by introducing a ΔpabB::Kn allele from strain BN117 into DH5α by P1 transduction. To construct plasmid pWKS30-rne, the wild-type rne gene, including its original promoter, 5′ untranslated region, and coding sequence, was amplified by PCR from strain MCZ4 by using primers RNE-EcoRI and RNE-XbaI. The PCR product was digested with XbaI and EcoRI and ligated into plasmid pWKS30 (26).
TABLE 1.
Strains, plasmids, and DNA oligonucleotides used in this work
| Strain, plasmid, or primer | Relevant genotype or feature | Source or reference |
|---|---|---|
| DH5α | F− (φ80dlacZΔM15) Δ(lacIZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK−mK+) supE44 thi-1 gyrA96 relA1 | Laboratory collection |
| BN117 | his-4 proA2 argE3 pheA1 tyrA4 Δ(trpEa)2 trpr (Tn10)pabA1 Δ(pabB::kan) rpsL 704 | Laboratory collection |
| MCZ4 | DH5α Δ(pabB::kan) | This study |
| M8 | MCZ4 rne8 | This study |
| M14 | MCZ4 m14 | This study |
| M18 | MCZ4 m18 | This study |
| M31 | MCZ4 rne31 | This study |
| M32 | MCZ4 rne32 | This study |
| M39 | MCZ4 rne39 | This study |
| M63 | MCZ4 rne63 | This study |
| M65 | MCZ4 m65 | This study |
| CH1827 | MC1061 zce-726::Tn10 | 27 |
| CH1828 | CH1827 ams-1 | 27 |
| pEZ201 | pSC101 derivative containing an rne::lacZ reporter gene comprising the rne promoter, 5′ untranslated region, and the first 181 codons of the rne coding region fused to the 10th codon of lacZ | 18 |
| pACYCBPTI | OmpA leader-BPTI gene fusion cloned into pACYC184 | 17 |
| pBAD-vtPA33 | h-tPA (Δ6-175) with stII leader sequence cloned in pBAD33 | 6 |
| pWKS30 | pSC101 origin low-copy-number plasmid for cloning | 12 |
| pWKS30-rne | Wild-type rne gene including its original promoter, 5′ untranslated region, and coding sequence cloned in pWKS30 | This study |
| DsbC-Up | ATGAAGAAAGGTTTTATGTTGTTTACTTTG | This study |
| DsbC-Dn | TTGGTGACATTGACCGGAGCCGTGCCA | This study |
| RNE-XbaI | GCTCTAGAGCCCTGGCAGTTACCAGGGCTTGATTACTTTGAGCTA | This study |
| RNE-EcoRI | GCGAATTCGAAACCGCCTCTTCTTCCCGCTCAGCAACGCGAACC | This study |
Unless otherwise specified, cells were grown at 37°C in Luria-Bertani (LB) medium with ampicillin (100 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (30 μg/ml), as required.
Chemical mutagenesis and selection.
E. coli MCZ4(pBAD33-vtPA) cells were grown in LB medium containing 30 μg of chloramphenicol per ml to an A600 of ∼0.3. Five milliliters of cells was collected by centrifugation at 2,800 × g for 10 min, washed twice, resuspended in 1.9 ml of cold citrate buffer (0.1 M sodium citrate, pH 5.5), and treated with 50 μg of 1-methyl-3-nitro-1-nitrosoguanidine (MNNG) (Sigma, St. Louis, Mo.) per ml at 37°C for 30 min. After MNNG treatment, the cells were washed twice and resuspended in 2 ml of phosphate buffer (0.1 M KH2PO4, pH 7.0). Then 200-μl portions of the 1:100 diluted cell samples were plated on M9 minimal medium supplemented with 0.2% glycerol, 8 μM N-α-benzyol-l-arginine-p-aminobenzoic acid (Bachem, Torrance, Calif.), 0.2% arabinose, and 30 μg of chloramphenicol per ml. A total of 2,500 colonies obtained through mutagenesis were patched on M9 medium plates with or without N-α-benzyol-l-arginine-p-aminobenzoic acid to eliminate false-positive colonies. Colonies that grew only in the presence of the p-aminobenzoic acid (PABA) adduct were selected, and the production of active tissue plasminogen activator (tPA) in these organisms was examined by the fibrin plate assay as described previously (18). Of 120 colonies examined in this manner, 8 showed enhanced fibrin clearance and were studied further.
Mutation mapping.
A Tn5 transposon containing a dihyrofolate reductase (DHFR) resistance marker was inserted into E. coli mutant strains by in vitro transposition by using a EZ::TN <DHFR-1> Tnp Transposome kit (Epicentre, Madison, Wis.). About 4,000 colonies selected for resistance to 10 μg of trimethoprim per ml were pooled and used to generate a P1 lysate which was used to transduce E. coli MCZ4(pBAD33-vtPA). Transductants were selected for growth on plates containing N-α-benzyol-l-arginine-p-aminobenzoic acid as described above. The insertion sites of the transposon in linked transductants were determined by inverse PCR (14). Standard linkage analysis was employed to estimate the physical distance between the position of the transposon insertion and the mutant allele. Subsequently, several kilobases of DNA flanking the transposon insertion sites was sequenced to determine the precise location of the mutations.
Enzymatic activity assays.
tPA activity was determined either by the fibrin plate assay (18) or by an indirect chromogenic assay (American Diagnostica, Stamford, Conn.) as previously described (3). β-Galactosidase activity was determined with cells grown in M9 medium as previously described by using o-nitrophenyl-β-d-galactoside (Sigma) as the substrate (12). Production of correctly folded bovine pancreatic trypsin inhibitor (BPTI) was assayed by an enzyme-linked immunosorbent assay as previously described (16).
RNA preparation and RNase protection assays.
Overnight cultures grown in LB medium were diluted 1:100 in fresh prewarmed LB medium. At an A600 of ∼0.8 an aliquot of cells was harvested to measure the steady-state level of dsbC transcripts. Rifampin was added to the remaining culture at a final concentration of 200 μg/ml to block transcription. Subsequently, cells were harvested at different times and rapidly chilled in an ethanol-dry ice bath, and RNA was isolated with an RNeasy kit (Qiagen, Valencia, Calif.). Total RNA was quantified spectrophotometrically at 260 nm. RNase protection assays were carried out with an RPA III kit (Ambion, Austin, Tex.) by following the manufacturer's protocol. The probe used in the RNase protection assay was transcribed from the first 250 bp of the dsbC gene, which was amplified by PCR by using primers dsbC-Up and dsbC-Dn and subsequently cloned in the TOPO vector (Invitrogen, Carlsbad, Calif.).
RESULTS
Genetic selection for E. coli mutants with enhanced disulfide isomerization activity.
In the bacterial periplasm, the rearrangement of nonnative protein disulfide bonds is catalyzed primarily by the disulfide isomerase DsbC (20, 27) and to a lesser degree by DsbG. We sought to develop a genetic system for isolation of mutations that enhance disulfide isomerization. The development of an appropriate selection technique was complicated because (i) E. coli dsbC or dsbG mutants do not exhibit any obvious phenotype (2, 13) and (ii) no E. coli proteins whose folding is absolutely dependent on disulfide bond isomerization are known. It has been shown previously that the folding in E. coli of eukaryotic proteins with multiple disulfide bonds is critically dependent on DsbC (16, 20). In particular, the expression of proteolytically active human tPA (h-tPA), a complex protein with a total of 17 disulfide bonds, is absolutely dependent on disulfide bond isomerization (18). We found that cells expressing active h-tPA or a variant of tPA (v-tPA) (a truncated version of h-tPA consisting of the kringle 2 and protease domains of the intact protein and containing nine disulfide bonds) can specifically cleave the adduct N-α-benzoyl-l-arginine-p-aminobenzoic acid to release PABA. In turn, the released PABA allowed growth of the PABA auxotrophic strain MCZ4 (DH5α pabB::Tn5Kn) in minimal media supplemented with the appropriate concentration of N-α-benzyol-l-arginine-p-aminobenzoic acid.
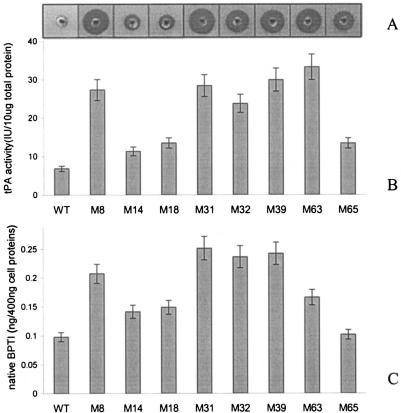
MCZ4 containing the v-tPA expression vector pBAD33-vtPA was subjected to chemical mutagenesis with MNNG as the mutagen. After mutagenesis, cells were plated on M9 minimal medium containing 8 μM N-α-benzoyl-l-arginine-p-aminobenzoic acid and 0.2% arabinose to induce the synthesis of v-tPA. A total of 2,500 colonies obtained after mutagenesis were patched on M9 minimal medium with or without N-α-benzoyl-l-arginine-p-aminobenzoic acid and an arabinose supplement to eliminate suppressor mutations that restored growth in the absence of an exogenous source of PABA. Approximately 120 colonies that failed to grow in the absence of N-α-benzoyl-l-arginine-p-aminobenzoic acid were cultured in rich medium with arabinose, and the v-tPA activities in cell lysates were measured by the fibrin plate assay (18). Eight mutants that exhibited the highest v-tPA activities in this assay were selected for further study (Fig. 1A).
FIG. 1.
Disulfide isomerase activities in mutant strains. Cells were grown in rich medium as described in Materials and Methods, harvested by centrifugation, and lysed with a French pressure cell. Equal amounts of cell lysate protein were used to determine formation of active tPA, as measured by the fibrin clearance assay (A), tPA activity, as determined by a coupled chromogenic assay that monitored the rate of conversion of plasminogen to plasmin (B), and accumulation of folded BPTI, as monitored by an enzyme-linked immunosorbent assay (C). In panels B and C the data are averages of three experiments. WT, wild type.
Quantitative activity assays revealed that the mutants isolated exhibited two- to fivefold-higher yields of active v-tPA than the parental wild-type strain (Fig. 1B). The expression of correctly folded BPTI, a 56-amino-acid protein with three disulfide bonds, was tested in the mutants as well. Like folding of v-tPA, folding of BPTI in the E. coli periplasm is completely dependent on disulfide bond isomerization by DsbC (16, 19). A similar increase in the yield of correctly folded BPTI protein was obtained (Fig. 1C). Finally, a similar effect was also seen with a third protein substrate, mouse urokinase, which contains nine disulfide bonds in its native, catalytically active conformation (data not shown).
Increased yield of multiple disulfide proteins is due to upregulation of DsbC expression.
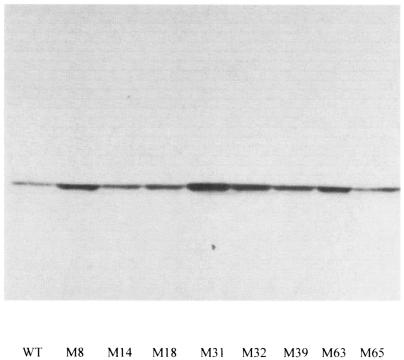
Since the folding of both v-tPA and BPTI is dependent on the disulfide isomerase activity in the periplasmic space, we examined whether the expression of proteins involved in disulfide bond formation had been altered in the mutant strains. Western blot analysis revealed that expression of the E. coli disulfide isomerase DsbC increased significantly in the mutants. The DsbC levels correlated with v-tPA activity, and the strongest mutants (M32, M39, and M63) exhibited much higher steady-state levels of the protein (Fig. 2). In contrast, no change was detected in the steady-state level of the periplasmic thiol oxidase DsbA or the minor disulfide isomerase DsbG (data not shown).
FIG. 2.
DsbC expression in mutant strains. Equal amounts of cells from cultures grown in rich medium as described in Materials and Methods were used to determine the abundance of the DsbC protein by Western blotting. All mutants showed increased DsbC protein expression. WT, wild type.
Mutations that increase dsbC expression map in rne.
To identify the molecular lesion in mutant M32 responsible for the increased active v-tPA production, genetic mutations were identified by a linkage mapping technique by using a Tn5 transposon carrying a DHFR marker. It was first determined that transposon mutagenesis of wild-type strain MCZ4(pBAD33-vtPA) does not confer increased v-tPA activity (data not shown). Subsequently, a Tn5::DHFR transposon was randomly inserted into the chromosome of M32 by in vitro transposition. A P1 lysate, generated from the pool of trimethoprim-resistant (Tmr) cells, was used to transduce wild-type strain MCZ4(pBAD33-vtPA), and colonies were scored for v-tPA activity and trimethoprim resistance. Three linked transductants (25, 5, and 90% linkage) were isolated with the DHFR marker inserted into the ycfX, ycfD, and yceC genes located in the region between 24.5 and 25 min on the E. coli chromosome, respectively. On the basis of the most closely linked marker insertion, the chromosomal region encompassing the rne gene to the plsX gene was sequenced. Two transition mutations were identified in the rne gene encoding endonuclease RNase E, which resulted in missense mutations R15H and A651V (Table 2).
TABLE 2.
Summary of mutants isolated in this study
| Mutant strain | rne allele(s)a |
|---|---|
| M8 | A319V, R368C |
| M14 | WTb |
| M18 | WT |
| M31 | S52N, G584D, A920T, A943T |
| M32 | R15H, A651V |
| M39 | A8T |
| M63 | Q390STOPUAG |
| M65 | WT |
The rne gene in all mutants was amplified by PCR. PCR products were subjected to sequencing. No mutations were found in the rne allele in M14, M18, and M65.
WT, wild type.
Mutations in two other strong mutants, M39 and M63, were also linked to marker insertions in the same chromosomal region as M32. DNA sequencing revealed that both strains had transition mutations in the rne gene (Table 2). The entire rne gene, including the promoter region and coding sequence, in the rest of the mutants was also sequenced. All but three mutants (M14, M18, and M65) were found to have mutations in the rne gene, which resulted in missense mutations in the RNase E protein (Table 2).
RNase E autoregulates its expression by modulating the rate of degradation of its own mRNA (7). Since the rne 5′ untranslated region is a substrate for RNase E, the β-galacatosidase activity expressed from a translational rne::lacZ fusion increases when the catalytic activity of RNase E decreases (7). The relative enzymatic activity of the RNase E protein in isolated mutants was analyzed quantitatively by using such an rne::lacZ fusion. The eight mutant strains isolated in this study were transformed with pEZ201, a low-copy-number plasmid encoding the rne::lacZ fusion. Cells were grown in shake flask cultures, and β-galactosidase activity was determined in the mid-exponential phase. All but one mutant (M14) exhibited significantly increased β-galactosidase activity (Table 3). The rest of the mutants exhibited between 50 and 400% higher β-galacatosidase levels than the wild-type strain, indicating that the RNase E activity in the mutants was reduced to various degrees. It is interesting that RNase E activity was reduced in mutants M18 and M65, even though rne alleles in these two mutants have no known mutation.
TABLE 3.
RNase E activities in isolated mutants
| Strain | β-Galactosidase activity (Miller units)a |
|---|---|
| M8 | 315 ± 21.1 |
| M14 | 108 ± 1.6 |
| M18 | 163 ± 5.5 |
| M31 | 484 ± 13.6 |
| M32 | 414 ± 6.1 |
| M39 | 233 ± 1.7 |
| M63 | 440 ± 12.6 |
| M65 | 228 ± 1.3 |
| MCZ4 (wild type) | 112 ± 3.1 |
Plasmid pEZ201, which contained an rne::lacZ translational fusion, was transformed into all eight mutant strains and the parental strain MCZ4. β-Galactosidase activity was measured as previously described (16). The values are the averages of three independent experiments.
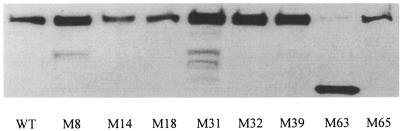
Western blot analysis revealed that the rne mutant alleles increased the levels of the RNase E protein in strains M8, M31, M32, and M39 (Fig. 3). In addition, an intense band corresponding to the molecular weight of RNase E (amino acids 1 to 390) cross-reacted with anti-RNase E sera in M63. The increased levels of RNase E polypeptide in these strains are consistent with the β-galactosidase activity data obtained from the rne::lacZ fusions. Interestingly, the three mutations that were not allelic to rne did not have a noticeable effect on the steady-state level of the RNase E polypeptide. This was surprising given that two of the mutants, M18 and M65, exhibited significantly increased activity from the rne::lacZ fusion, indicating that the autoprocessing of the rne transcript had been impaired. It appears that the lesions in these strains exerted a compensatory effect that allowed the RNase E levels to remain at nearly wild-type values even though the processing of the 5′ untranslated region of rne had been reduced.
FIG. 3.
Western blot analysis of RNase E level. Cells were grown in rich medium to the mid-exponential phase and harvested by centrifugation, and the level of RNase E was analyzed by Western blotting. As a result of RNase E self-regulation, expression of RNase E protein increased in selected mutants. WT, wild type.
rne mutations increase the half-life of dsbC mRNA.
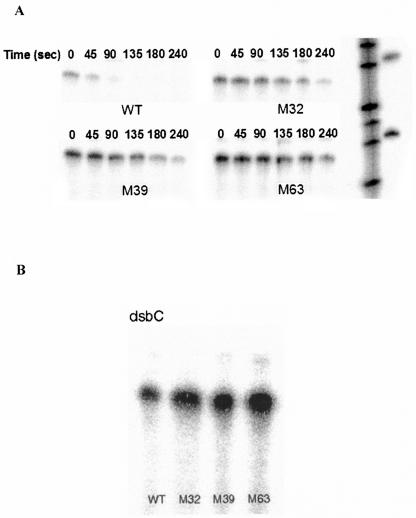
RNase E is an essential E. coli endoribonuclease that degrades mRNAs and assists in the maturation of a variety of rRNAs and tRNAs (4, 24). We tested whether the attenuation in RNase E activity contributes to the increased expression level of DsbC by slowing down the degradation of its transcript. The half-lives of dsbC mRNA in the wild-type and mutant strains were determined quantitatively by RNase protection assays following inhibition of transcription with rifampin. The half-life of the dsbC message at 37°C increased significantly from 0.8 min in the wild-type strain to 2.1, 1.6, and 2.5 min in mutant strains M32, M39, and M63, respectively (Fig. 4A). The steady-state levels of dsbC transcripts were also shown to increase in these three mutants by an RNase protection assay (Fig. 4B).
FIG. 4.
dsbC mRNA half-life. Cells were grown to the late exponential phase, and then rifampin was added to inhibit transcription. Samples were collected at different times, and RNA was extracted and quantified spectrophotometrically. Ten micrograms of total RNA was used for the RNase protection assay, in which RNA transcribed from a 270-bp fragment of the dsbC coding sequence was used as the probe. (A) dsbC mRNA half-life increased from 0.82 ± 0.05 min in wild-type cells to 2.1 ± 0.3, 1.6 ± 0.3, and 2.5 ± 0.3 min in M32, M39 and M63, respectively. (B) The mutants accumulated dsbC mRNA at a steady-state level. WT, wild type.
Classical rne mutation upregulates DsbC expression.
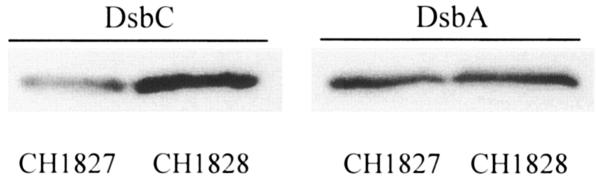
Finally, we examined whether the well-characterized rne mutant allele ams-1 (1, 15), which was not selected on the basis on its effect on disulfide isomerization, also affects DsbC expression. As shown by Western blotting, the expression level of the DsbC protein was markedly higher in the ams-1 mutant strain CH1828 than in its isogenic parent, CH1827 (Fig. 5). Under these conditions the abundance of DsbA was not affected. This result indicates that upregulation of DsbC is a general property of mutants with reduced RNase E activity.
FIG. 5.
Upregulation of DsbC is a general property of mutants with impaired RNase E activity. The DsbC expression level was studied by Western blotting by using anti-DsbC antibody in ams-1 mutant CH1828 and in the isogenic wild-type strain CH1827.
DISCUSSION
Even though disulfide isomerases were discovered by Anfinsen 40 years ago and have been the subject of extensive studies ever since, genetic analyses of the disulfide isomerization pathway have so far relied on indirect assays for phenotypes unrelated to the rearrangement of protein disulfide bonds per se. For example, dsbC was originally isolated as a suppressor of dsbA mutants deficient in protein thiol oxidation and as a gene that confers dithiothreitol sensitivity (13, 22). It took several years of subsequent biochemical studies to establish the true physiological role of DsbC as the major disulfide isomerase in the E. coli periplasm (19, 23, 27). In this work we designed a strategy to render growth of E. coli dependent on disulfide bond isomerization. We took advantage of (i) the requirement for disulfide bond rearrangement in the folding of an h-tPA variant that contains nine disulfide bonds, (ii) the fact that correctly folded v-tPA is biologically active, (iii) the observation that active v-tPA expressed in the bacterial periplasmic space can specifically hydrolyze micromolar concentrations of the PABA adduct N-α-benzoyl-l-arginine-p-aminobenzoic acid, which is not a substrate for native E. coli amidases, and (iv) the observation that generation of free PABA by v-tPA is sufficient to complement PABA auxotrophy and thus support colony formation on selective plates. In other words, disulfide bond isomerization activity determines the yield of active v-tPA, which in turn is required for cell growth. We found that following a step to eliminate revertants that can synthesize PABA endogenously, selection was quite efficient and resulted in a high yield of mutants having elevated tPA activity. To our knowledge, the present study represents the first genetic analysis specifically aimed at identification of lesions that directly affect disulfide bond isomerization.
Eight mutants that conferred two- to fivefold-higher active v-tPA production than the production by the wild-type strain were isolated from a chemically mutagenized library and studied in detail. We found that curing the mutant strains of the v-tPA expression plasmid completely abolished fibrin-cleaving activity. However, retransforming the cured cells with the pBAD33-vtPA plasmid restored v-tPA activity to the original level of the mutants isolated. These observations indicate that the increased active v-tPA yield is dependent on expression of v-tPA from a plasmid and is due to chromosomally encoded mutations. Furthermore, the mutants exhibited a similar effect on the folding of BPTI, although the increase in the yield of native protein was smaller than that observed with v-tPA. These results indicate that the lesions in the eight mutants confer a general increase in disulfide bond isomerization activity that is not restricted to one particular substrate. No mutations that affect the synthesis of v-tPA were found, which is consistent with previous observations that disulfide bond isomerization is the rate-limiting step in the expression of functional protein and that the yield of active v-tPA cannot be enhanced by increasing the rate of its synthesis (18).
All the chromosomal lesions which we isolated resulted in an increase in the abundance of DsbC. It has been shown previously that overexpression of DsbC, but not overexpression of DsbA, can dramatically increase the expression of active tPA in the E. coli periplasm (18). In general, the yield of active v-tPA in the mutants tracked, but did not vary linearly, with the expression level of DsbC in the cells. The lack of linearity was probably a consequence of the complex kinetics of disulfide bond formation and isomerization in a protein such as v-tPA that has a very complex folding pathway. Taken together, our results demonstrate that (i) as anticipated, the genetic selection for mutants that enhance the folding of v-tPA led to isolation of mutant strains with increased disulfide isomerization activity; and (ii) increased disulfide isomerization activity resulted mainly from substantially higher level of DsbC protein in the periplasm. The fact that DsbC expression was increased in all isolated mutants again demonstrates the important role of DsbC as the major E. coli disulfide isomerase. It also suggests that the most practical way to increase disulfide isomerization activity in E. coli is to increase DsbC expression.
The molecular lesions responsible for the increased DsbC expression were analyzed in detail. Linkage mapping analysis revealed that mutations in five of the eight mutants were allelic to the rne gene, which encodes E. coli endonuclease RNase E. These five mutants each had at least one amino acid substitution within the N-terminal catalytic domain of RNase E, the segment comprising at least the first 417 amino acids (17) of the 1,061 amino acids of the intact protein. The catalytic domain is responsible for the endonuclease and 3′-polyadenylase activities of the protein and is essential for growth (7, 9, 25). The isolation of mutants containing multiple DNA substitutions in rne was probably due to the mutagenesis conditions used in this study. Interestingly, mutant M63 contained a nonsense amber mutation at amino acid 390 which gave rise to an RNase E fragment that should have been devoid of catalytic activity and unable to allow cell growth (11). The viability of this strain must have been due to the suppression of the amber codon by the supE allele in MCZ4 which resulted in the accumulation of a small amount of intact RNase E. Consistent with this explanation, a weak band corresponding to full-length RNase E protein was detected by Western blotting (Fig. 3 and data not shown). In addition, efforts to transduce a marked rne allele from M63 into strains lacking a supE suppressor were unsuccessful.
The rne gene in another two mutants, M18 and M65, contained no mutations, as judged by the sequencing results. Transduction of wild-type rne gene from strain CH1827 did not alter the β-galactosidase level from the rne::lacZ fusion, nor did it mitigate the upregulation of DsbC (data not shown). Both of these lesions conferred high β-galactosidase levels from the rne::lacZ fusion, indicating that RNase E activity had been impaired. However, we did not observe a noticeable increase in the amount of RNase E polypeptide, as would be expected from a reduction in the processing of the rne transcript. One possibility is that the lesions in M18 and M65 affect the transcription of rne in a manner that compensates for the increased stability of the transcript and thus results in no net change in protein synthesis. Characterization of these mutations and their mechanism of action may thus provide more insight into the regulation of RNase E activity. However, regardless of the precise mechanism, the increased levels of rne::lacZ activity indicate that RNA processing is affected to some degree in these strains.
One initial goal of this investigation was to identify genes involved in the disulfide isomerization pathway in the periplasmic space. We expected to identify mutations in the promoter region and in the coding sequence of the Dsb protein(s) that could alter the transcription level or redox properties and contribute to an increase in disulfide isomerization activity. Surprisingly, seven of the eight mutations that caused increased disulfide isomerization activity affected the enzymatic activity of RNase E, the major endonuclease involved in the RNA decay pathway. In a separate search for genes that enhance disulfide isomerization when multiple copies are expressed, one of the genes which we isolated, apart from dsbC itself, upregulated the synthesis of DsbC by stabilizing its mRNA via a mechanism that involves interactions of the corresponding polypeptides (which are not among the known degradosome components) with RNase E (10). Taken together, these results support the notion that the expression of DsbC is controlled by the stability of its mRNA, which has a short half-life, and the conclusion that the DsbC expression level is critical for expression of complicated multiple disulfide proteins in the E. coli periplasm.
Acknowledgments
We are grateful to J. Beckwith, J. Belasco, C. Gross, S. Kushner, and T. J. Silhavy for providing E. coli strains and plasmids and to Nancy McFarland for initial studies related to the PABA auxotrophy selection. We especially thank S. Kushner for providing anti-RNase E antibodies.
This work was supported by NSF grant BES 963406 and by NIH grant GM 55090.
REFERENCES
- 1.Babitzke, P., and S. R. Kushner. 1991. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 88:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessette, P. H., J. J. Cotto, H. F. Gilbert, and G. Georgiou. 1999. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J. Biol. Chem. 274:7784-7792. [DOI] [PubMed] [Google Scholar]
- 3.Bessette, P. H., J. Qiu, J. C. Bardwell, J. R. Swartz, and G. Georgiou. 2001. Effect of sequences of the active-site dipeptides of DsbA and DsbC on in vivo folding of multidisulfide proteins in Escherichia coli. J. Bacteriol. 183:980-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 62:55-108. [DOI] [PubMed] [Google Scholar]
- 5.Collet, J. F., and J. C. Bardwell. 2002. Oxidative protein folding in bacteria. Mol. Microbiol. 44:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Freedman, R. B., P. Klappa, and L. W. Ruddock. 2002. Model peptide substrates and ligands in analysis of action of mammalian protein disulfide-isomerase. Methods Enzymol. 348:342-354. [DOI] [PubMed] [Google Scholar]
- 7.Jain, C., and J. G. Belasco. 1995. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes Dev. 9:84-96. [DOI] [PubMed] [Google Scholar]
- 8.Katzen, F., and J. Beckwith. 2000. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell 103:769-779. [DOI] [PubMed] [Google Scholar]
- 9.Kido, M., K. Yamanaka, T. Mitani, H. Niki, T. Ogura, and S. Hiraga. 1996. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J. Bacteriol. 178:3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, K., X. Zhan, J. Gao, J. Qiu, Y. Feng, R. Meganathan, S. N. Cohen, and G. Georgiou. 2003. RraA: a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell 114:623-634. [PubMed] [Google Scholar]
- 11.McDowall, K. J., and S. N. Cohen. 1996. The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding site. J. Mol. Biol. 255:349-355. [DOI] [PubMed] [Google Scholar]
- 12.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Missiakas, D., C. Georgopoulos, and S. Raina. 1994. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 13:2013-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ochman, H., J. W. Ajioka, D. Garza, and D. L. Hartl. 1990. Inverse polymerase chain reaction. Bio/Technology 8:759-760. [DOI] [PubMed] [Google Scholar]
- 15.Ono, M., and M. Kuwano. 1980. Chromosomal location of a gene for chemical longevity of messenger ribonculeic acid in a temperature-sensitive mutant of Escherichia coli. J. Bacteriol. 142:325-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostermeier, M., K. De Sutter, and G. Georgiou. 1996. Eukaryotic protein disulfide isomerase complements Escherichia coli dsbA mutants and increases the yield of a heterologous secreted protein with disulfide bonds. J. Biol. Chem. 271:10616-10622. [DOI] [PubMed] [Google Scholar]
- 17.Ow, M. C., and S. R. Kushner. 2002. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 16:1102-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu, J., J. R. Swartz, and G. Georgiou. 1998. Expression of active human tissue-type plasminogen activator in Escherichia coli. Appl. Environ. Microbiol. 64:4891-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rietsch, A., D. Belin, N. Martin, and J. Beckwith. 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:13048-13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rietsch, A., P. Bessette, G. Georgiou, and J. Beckwith. 1997. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol. 179:6602-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritz, D., and J. Beckwith. 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 55:21-48. [DOI] [PubMed] [Google Scholar]
- 22.Shevchik, V. E., G. Condemine, and J. Robert-Baudouy. 1994. Characterization of DsbC, a periplasmic protein of Erwinia chrysanthemi and Escherichia coli with disulfide isomerase activity. EMBO J. 13:2007-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sone, M., Y. Akiyama, and K. Ito. 1997. Differential in vivo roles played by DsbA and DsbC in the formation of protein disulfide bonds. J. Biol. Chem. 272:10349-10352. [DOI] [PubMed] [Google Scholar]
- 24.Steege, D. A. 2000. Emerging features of mRNA decay in bacteria. RNA 6:1079-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taraseviciene, L., G. R. Bjork, and B. E. Uhlin. 1995. Evidence for an RNA binding region in the Escherichia coli processing endoribonuclease RNase E. J. Biol. Chem. 270:26391-26398. [DOI] [PubMed] [Google Scholar]
- 26.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 27.Zapun, A., D. Missiakas, S. Raina, and T. E. Creighton. 1995. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry 34:5075-5089. [DOI] [PubMed] [Google Scholar]