Abstract
Chlorosomes of the green sulfur bacterium Chlorobium tepidum comprise mostly bacteriochlorophyll c (BChl c), small amounts of BChl a, carotenoids, and quinones surrounded by a lipid-protein envelope. These structures contain 10 different protein species (CsmA, CsmB, CsmC, CsmD, CsmE, CsmF, CsmH, CsmI, CsmJ, and CsmX) but contain relatively little total protein compared to other photosynthetic antenna complexes. Except for CsmA, which has been suggested to bind BChl a, the functions of the chlorosome proteins are not known. Nine mutants in which a single csm gene was inactivated were created; these mutants included genes encoding all chlorosome proteins except CsmA. All mutants had BChl c contents similar to that of the wild-type strain and had growth rates indistinguishable from or within ∼90% (CsmC− and CsmJ−) of those of the wild-type strain. Chlorosomes isolated from the mutants lacked only the protein whose gene had been inactivated and were generally similar to those from the wild-type strain with respect to size, shape, and BChl c, BChl a, and carotenoid contents. However, chlorosomes from the csmC mutant were about 25% shorter than those from the wild-type strain, and the BChl c absorbance maximum was blue-shifted about 8 nm, indicating that the structure of the BChl c aggregates in these chlorosomes is altered. The results of the present study establish that, except with CsmA, when the known chlorosome proteins are eliminated individually, none of them are essential for the biogenesis, light harvesting, or structural organization of BChl c and BChl a within the chlorosome. These results demonstrate that chlorosomes are remarkably robust structures that can tolerate considerable changes in protein composition.
Photosynthetic organisms have evolved a large variety of specialized light-harvesting structures that allow them to capture light energy. These antennae typically contain chlorophylls or linear tetrapyrroles as the chromophores. The molecular structures of many light-harvesting pigment-protein complexes are known in atomic detail, including those of light-harvesting complexes 1 and 2 from purple bacteria (10), light-harvesting complex II from chloroplasts (20), the Fenna-Matthews-Olson protein from green sulfur bacteria (2, 3, 21, 29), and various phycobiliproteins from cyanobacteria (27). A common feature of these structures is that the light-harvesting pigments (chlorophylls, carotenoids, and phycobilins) are organized and spatially arranged by the protein, although the pigments often interact both with one another and with the polypeptide. In addition, the components of these antenna structures are always present in a defined stoichiometric relationship.
The largest light-harvesting antenna structures known are the chlorosomes of green bacteria (2, 3, 17, 19, 24, 30). Chlorosomes are unique among light-harvesting antennae because they have the lowest protein-to-pigment ratio and because their major pigment, typically bacteriochlorophyll c (BChl c), is arranged in large aggregates whose structures do not involve protein molecules. Chlorosomes are also the only known antenna structures that do not have a stoichiometrically fixed ratio of their major pigment (BChl c) to a protein component (18, 31; this work). Additionally, chlorosomes do not have strictly defined dimensions, and isolated chlorosomes exhibit considerable variation in size and shape. For example, chlorosomes from Chlorobium tepidum are about 100 to 200 nm long and 40 to 60 nm wide (4). The BChl c aggregates are encapsulated by a lipid-protein envelope that harbors all of the proteins present in the chlorosomes (5, 8, 32). Chlorosomes also contain carotenoids, isoprenoid quinones, and small amounts of BChl a (14, 15). The BChl a in chlorosomes is almost certainly bound to the most abundant protein, CsmA, possibly in a one-to-one ratio (4, 23, 26). This CsmA-BChl a complex transfers the excitation energy away from the BChl c aggregates of the chlorosome to the BChl a-binding Fenna-Matthews-Olson protein and the reaction centers. The organization of the carotenoids and isoprenoid quinones is not well understood except that most of them interact with the BChl molecules and that most of them appear to be present in the interior of the chlorosome (15).
In addition to CsmA, chlorosomes from the green sulfur bacterium C. tepidum contain nine other proteins: CsmB, CsmC, CsmD, CsmE, CsmF, CsmH, CsmI, CsmJ, and CsmX (5-8, 32). The genes encoding all 10 proteins have been cloned, and antibodies to recombinant Csm proteins have been used to study the organization of these proteins, their subcellular distribution, and other aspects of chlorosome structure and biogenesis (4-8, 18, 31, 32). Those and other studies have shown that all 10 chlorosome proteins are exposed at the surface of the chlorosome and that some chlorosome proteins, notably CsmA, form oligomeric complexes, as revealed by chemical cross-linking (4). Apart from information generated by the studies concerning CsmA, it is not clear which functions the chlorosome proteins have or whether they are essential to the cells or to the integrity of the chlorosomes. In the present work, we have therefore systematically attempted to inactivate each of the 10 genes that encode chlorosome proteins and subsequently to identify phenotypic effects in the mutants.
MATERIALS AND METHODS
Organisms and growth conditions.
The strain of C. tepidum used and its cultivation and transformation have been described previously (13, 18). Growth rates in the absence of antibiotics were determined from the optical densities at 600 nm of cultures grown in 25-ml screw-cap tubes at 47°C as previously described (18). The growth rates were calculated as averages of four to six measurements, for which the standard deviation did not exceed 10% of the average. Various light intensities were obtained by adjusting the number of light bulbs and by using white paper or transparent plastic bubble wrap mounted on the glass door of the incubator for shading and insulation. Cell cultures for chlorosome preparations were grown in 2-liter flasks positioned in a water bath at 47°C and at a light intensity of approximately 120 μmol of photons m−2 s−1.
Escherichia coli strain DH5α (Bethesda Research Laboratories, Gaithersburg, Md.) was used for the propagation of all plasmids except pCT841 (Table 1), which was propagated in E. coli strain SCS110 (dam dcm; Stratagene, La Jolla, Calif.) to allow the digestion of the plasmid with the methylation-sensitive restriction enzyme BsaBI.
TABLE 1.
Plasmids used in this study
| Plasmid name | Size (kb) | Description | Phenotypea | Reference |
|---|---|---|---|---|
| Plasmids used as source of C. tepidum DNA | ||||
| pCT65r | 4.4 | pUC19 with a fragment containing csmED | Apr | 5 |
| pCT652 | 4.1 | pUC19 with a fragment containing csmED | Apr | 5 |
| pCT658 | 4.0 | pBluescript SK+ with a fragment containing csmE | Apr | 5 |
| pCT720 | 3.2 | pUC19 with a fragment containing csmC | Apr | 5 |
| pCT841 | 3.3 | pUC19 with a fragment containing csmB; propagated in E. coli SCS110 to allow digestion of BsaBI site | Apr | 5 |
| pET3d::csmI | 5.3 | CsmI protein expression construct | Apr | 32 |
| pET3d::csmJ | 5.3 | CsmJ protein expression construct | Apr | 32 |
| pET3d::csmH | 5.4 | CsmH protein expression construct | Apr | 32 |
| pET32a::csmA | 6.0 | CsmA protein expression construct | Apr | 32 |
| pET32a::csmF | 6.1 | CsmF protein expression construct | Apr | 32 |
| pET32a::csmX | 6.6 | CsmX protein expression construct | Apr | 32 |
| Gene inactivation constructs | ||||
| pCAS22 | 7.1 | pET32a::csmA in which csmA is interrupted at the NcoI site by the aadA cassette from pSRA2 | Apr Smr Spr | This work |
| pCBS11 | 4.3 | pCT841 in which csmB is interrupted at the BsaBI site by the aadA cassette from pSRA2 | Apr Smr Spr | This work |
| pCCS8 | 4.2 | pCT720 in which csmC is interrupted at the NsiI site by the aadA cassette from pSRA2 | Apr Smr Spr | This work |
| pCFS1 | 7.2 | pET32a::csmF in which csmF is interrupted at the NsiI site by the aadA cassette from pSRA2 | Apr Smr Spr | This work |
| pCHS2 | 6.4 | pET3d::csmH in which csmH is interrupted at the BstEII site by the aadA cassette from pSRA2 | Apr Smr Spr | This work |
| pCJ | 6.4 | pET3d::csmJ in which the EcoRI site is deleted and csmJ is interrupted at the ApoI site by the aacC1 cassette from pMS255 | Apr Gmr | This work |
| pCT652L2 | 5.1 | pCT652 in which csmD is interrupted at the HincII site by the aadA cassette from pSRA2 | Apr Smr Spr | This work |
| pCT658B1 | 5.1 | pCT658 in which csmE is interrupted at the StyI site by the aadA cassette from pSRA2 | Apr Smr Spr | This work |
| pCT65r1 | 4.8 | pCT65r in which csmED is interrupted between the StyI sites by the aadA cassette from pSRA2 | Apr Smr Spr | This work |
| pET3d::csmI::Ω | 7.3 | pET3d::csmI in which csmI is interrupted at the XcmI site by the aadA cassette from pHP45Ω | Apr Smr Spr | This work |
| pET32a::csmX::Ω | 8.6 | pET32a::csmX in which csmX is interrupted at the MfeI site by the aadA cassette from pHP45Ω | Apr Smr Spr | This work |
| Antibiotic resistance markers | ||||
| pHP45Ω | 4.4 | Contains a 2.1-kb aadA streptomycin and spectinomycin resistance cassette | Apr Smr Spr | 25 |
| pMS255 | 3.7 | Contains a 1.1-kb aacC1 gentamicin resistance cassette | Apr Gmr | 1 |
| pSRA2 | 3.7 | Contains a 1.1-kb aadA streptomycin and spectinomycin resistance cassette | Apr Smr Spr | This work |
Apr, ampicillin resistance; Gmr, gentamicin resistance; Smr, streptomycin resistance; Spr, spectinomycin resistance.
Antibiotic resistance markers.
Mutations in C. tepidum were created by using the gentamicin resistance marker aacC1 from plasmid pMS255 (1) or the streptomycin and spectinomycin resistance marker aadA from plasmid pHP45Ω (25) or pSRA2 (Table 1). Plasmid pSRA2 was derived from pHP45Ω (25) as follows. A 1,030-bp fragment containing the resistance-conferring gene aadA was amplified and modified to include HindIII restriction sites (underlined) by PCR with the primers aadAF (5′-ACTGGTCCAGAAGCTTGACCGA-3′) and aadAB (5′-AAGCGGCGTAAGCTTGAACGAA-3′) with pHP45Ω as the template. The resulting DNA fragment was digested with HindIII and cloned into the vector fragment of HindIII-digested pRL409 (12). A plasmid with the polylinkerconfiguration EcoRI-SacI-KpnI-BamHI-XbaI-SalI-HincII-PstI-SphI-HindIII-aadA-HindIII-SphI-PstI-HincII-SalI-XbaI-BamHI-KpnI-SacI-EcoRI was se-lected and named pSRA2. The EcoRI fragment from pSRA2 containing aadA contains 1,103 bp. Plasmid pSRA2 offers several advantages over pHP45Ω. It is a high-copy-number plasmid, the aadA cassette in pSRA2 has a more versatile polylinker, and the modified aadA gene is devoid of the transcription terminators, which are present in the cassette from pHP45Ω and which interfere with the PCR in vitro and gene transcription in vivo.
Construction of C. tepidum mutants.
Plasmids containing fragments of C. tepidum genomic DNA were obtained from an existing genomic library (5) and various protein expression constructs (32). The csm genes in these plasmids were insertionally inactivated by cloning the aadA cassette from either pSRA2 or pHP45Ω or by cloning the aacC1 cassette from pMS255 into the coding sequence. Table 1 and Fig. 1 contain details about the plasmids used and the production of the gene inactivation constructs. Natural transformation of C. tepidum was performed as described previously (13). Transformants were analyzed for segregation by PCR (csmB, csmC, csmD, csmE, csmED, csmF, and csmH mutants) or Southern blot hybridization analysis (csmI, csmJ, and csmX mutants) as described previously (13). The primers used for PCR analysis (Fig. 1) were derived from the genome sequence of C. tepidum (11) by using MacVector software (version 7.0; Genetics Computer Group, Madison, Wis.). Genomic DNA isolated from the csmI, csmJ, and csmX mutants for Southern blot hybridization analyses was digested with BsaBI, BlpI, and SacII, respectively. Hybridization probes for Southern analyses were obtained by excising the genes from their respective expression constructs (pET3d::csmI, pET3d::csmJ, and pET32a::csmX).
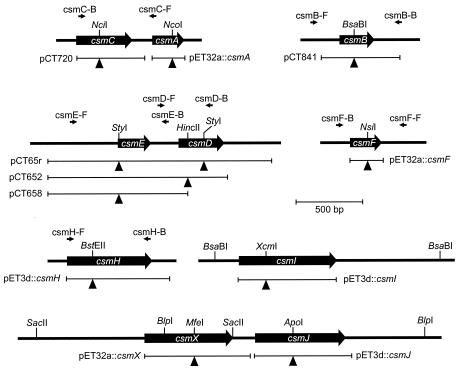
FIG. 1.
Restriction maps showing the regions contained in plasmids (Table 1) and the binding sites of primers used to verify the csm mutants of C. tepidum. Black triangles indicate the insertion sites of antibiotic resistance markers. See the text for details.
Chlorosome preparation and analysis.
Chlorosomes were prepared and their protein compositions were analyzed as described previously (32). Fluorescence emission spectra under oxidizing conditions were recorded after the incubation of the chlorosomes (absorption of 0.2 at the Qy peak around 745 nm) in aerobic buffer (10 mM KH2PO4, pH 7.0) for about 30 min and under reducing conditions after incubation with ∼10 mM sodium dithionite for about 2 h. Pigment analyses by absorption spectroscopy of methanol extracts and high-pressure liquid chromatography (HPLC) were performed as described previously (18), except that the HPLC pump and injector were replaced with an Agilent Technologies (Waldbronn, Germany) 1100 series binary pump (model G1312A), a vacuum degasser (model G1379A), and a manual injector (model G1328A).
Electron microscopy.
Chlorosomes were adsorbed on Formvar-coated copper grids, negatively stained with 2% (wt/vol) uranyl acetate, and visualized with a 1200EXII electron microscope (JEOL, Peabody, Mass.).
RESULTS
Construction of csm mutants.
Except for the transformants containing csmA::aadA, all csm transformants of C. tepidum segregated after two or three streakings on selective medium. Only merodiploid strains, which resisted segregation under the growth conditions used even after repeated streaking, were obtained from transformations with the csmA::aadA construct pCAS22 (Table 1). This result suggests that CsmA may play an essential role in light harvesting and that this is not the case for the other nine proteins of the chlorosome envelope. A csmE csmD double mutant was obtained by using the construct pCT65r1 (Table 1). The csmE csmD double mutant grew like the wild-type strain, produced chlorosomes, and did not exhibit any obvious growth or pigmentation phenotype (data not shown). Only the nine mutants lacking a single Csm protein were fully characterized in this work.
Characterization of cell cultures.
The following growth rates were measured for the wild-type strain (the light intensity, in micromoles of photons per square meter per second, is given in parentheses after the growth rate): 0.09 h−1 (8), 0.20 h−1 (30), 0.25 h−1 (120), 0.23 h−1 (210), and 0.21 h−1 (580). The csm mutants generally had growth rates that were indistinguishable from that of the wild-type strain at all light intensities tested. Exceptions to this situation were noted for the csmC and csmJ mutants. At a limiting light intensity (8 μmol of photons m−2 s−1), the csmC mutant consistently had a growth rate that was approximately 90% of that of the wild-type strain. In contrast, at saturating and inhibitory light intensities (≥120 μmol of photons m−2 s−1), the csmJ mutant had growth rates that were about 90% of the wild-type value.
Cells used for chlorosome preparations were grown in 2-liter batch cultures. The cellular BChl c contents of the wild-type strain and all mutant strains grown under these conditions, as judged from the ratios of BChl c absorption to the pigment-independent light scattering of the cells, were very similar for all cultures (data not shown). The absorbance properties of wild-type cells and all csm mutant cells were also generally very similar. The BChl c absorption maximum in the wild-type and mutant strains was about 750 nm, except in the csmC mutant, in which it was blue-shifted to 743 nm. The fluorescence emission spectra of all cells recorded under reducing conditions were also very similar. These spectra had emission maxima from BChl c at around 772 to 773 nm (769 nm for the csmC mutant) and emission peaks from BChl a at around 805 nm.
Characterization of isolated chlorosomes.
No obvious differences from wild-type cells were generally observed during the isolation of chlorosomes from the mutant cell cultures. The only exception to this generalization applies to the isolation of chlorosomes from the csmC mutant. For this mutant, a small amount of an orange-colored fraction banded at the top of the suspension after the ultracentrifugation of disrupted cells, whereas no such colored fraction appeared in material from wild-type cells. The absorption spectrum of this orange fraction was dominated by carotenoids in the 400-to-500-nm range, although smaller absorption maxima occurred at around 750 and 795 nm. This orange fraction also contained CsmA, as judged from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (data not shown). This absorption spectrum is very similar to the absorption spectrum of vestigial chlorosomes, designated carotenosomes, which can be isolated from a bchK mutant of C. tepidum that cannot synthesize BChl c (18; N.-U. Frigaard et al., unpublished data), except for the presence of a minor 750-nm peak. Thus, the orange fraction from the csmC mutant appears to represent a fraction of chlorosomes with a highly reduced BChl c content. This fraction represented only a small ratio of the total chlorosome fraction (roughly 5% based on carotenoids and BChl a), which was otherwise similar in quantity and density to the chlorosomes of the wild-type strain.
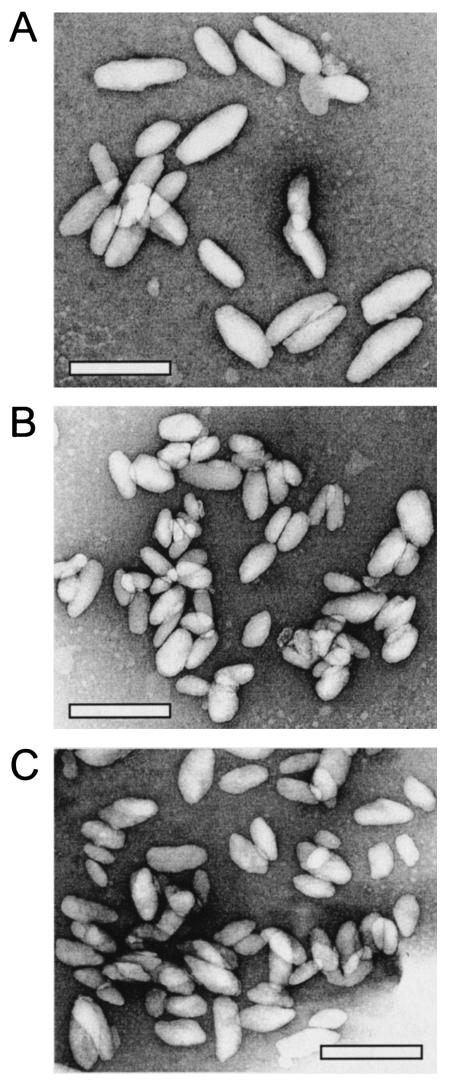
Electron microscopy of negatively stained chlorosomes revealed only minor differences among chlorosomes from the wild-type strain and the nine csm mutant strains. The measured dimensions of chlorosomes from the wild-type strain were as follows (sample size, 56): length, 142 ± 36 nm; width, 53 ± 10 nm; and length-to-width ratio, 2.7 ± 0.6 (Fig. 2A). Chlorosomes that had dimensions that deviated significantly were those from the csmC mutant (length, 95 ± 26 nm; width, 50 ± 12 nm; length-to-width ratio, 2.0 ± 0.5 [sample size, 44] [Fig. 2B]) and those from the csmH mutant (length, 112 ± 27 nm; width, 52 ± 10 nm; length-to-width ratio, 2.2 ± 0.4 [sample size, 63] [Fig. 2C]).
FIG. 2.
Transmission electron micrographs of chlorosomes isolated from the C. tepidum wild-type strain (A), the csmC mutant (B), and the csmH mutant (C). Chlorosomes were stained with 2% (wt/vol) uranyl acetate. Bars, 200 nm.
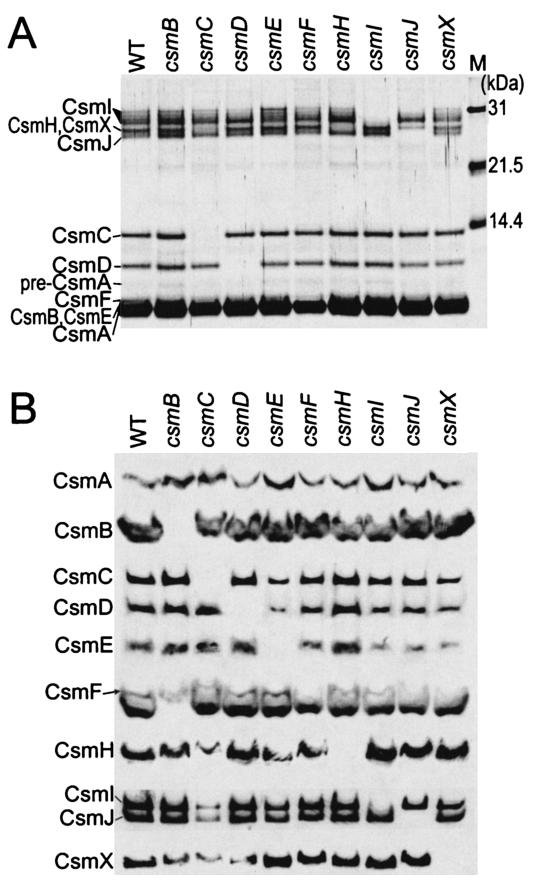
Figure 3A shows the results of an SDS-PAGE analysis of the chlorosomes isolated from the wild-type strain and the nine csm mutants. It is readily apparent from this silver-stained gel that chlorosomes from the csmC, csmD, csmI, and csmJ mutants lack the protein encoded by the inactivated gene and that the chlorosomes of all other mutants, in which these four genes are not inactivated, have normal amounts of the proteins encoded by these genes. Immunoblotting analyses (Fig. 3B) with antibodies raised against recombinant chlorosome proteins confirmed that the chlorosomes of each of the nine mutant strains lacked only the protein encoded by the gene inactivated in the particular mutant strain. Thus, it is clear from the results presented in Fig. 3 that the absence of any given chlorosome protein has little effect on the levels of the other chlorosome proteins. A minor exception to this generalization is that the chlorosomes of the csmC mutant appear to have a lower (Fig. 3A) but detectable (Fig. 3B) CsmH content.
FIG. 3.
Analysis of the protein compositions of chlorosomes isolated from the wild-type strain (WT) and csm mutants of C. tepidum. (A) SDS-PAGE gel stained with silver. Lane M, molecular mass markers. (B) Immunoblots probed with antibodies specific for recombinant chlorosome proteins. (Note that the antiserum to CsmF cross-reacts with CsmB, whose amino acid sequence is about 40% similar to that of CsmF. The antiserum to CsmB does not cross-react with CsmF.).
Table 2 summarizes the spectroscopic and biochemical properties of chlorosomes isolated from the wild-type and csm mutant strains. Wild-type chlorosomes contained about 0.27 g of protein per g of BChl c, a value that is in good agreement with previous measurements (0.23 g of protein per g of BChl c [32]). CsmA is the most abundant chlorosome protein and accounts for about one-third to one-half of the total chlorosome envelope protein (4). Thus, the absence of one of the other chlorosome proteins would not be expected to affect the total protein content significantly. The BChl a content of wild-type chlorosomes was about 12 mg per g of BChl c, and this value did not vary significantly in most of the mutants. However, chlorosomes from the csmC mutant contained roughly one-third less BChl a than wild-type chlorosomes did. The carotenoid contents of wild-type chlorosomes and all mutant chlorosomes were about 60 to 70 mg per g of BChl c, except for the csmB mutant, which produced chlorosomes containing significantly (∼25%) fewer carotenoids. HPLC analyses suggested that this decrease in the csmB mutant was due to a decrease in all carotenoid species rather than to a decrease in a specific carotenoid species. HPLC analyses of whole-cell extracts showed that roughly 50% of the cellular BChl a and about 80% of the cellular carotenoids were contained in the chlorosomes in all strains.
TABLE 2.
Characterization of chlorosomes isolated from the wild-type strain and csm mutants of C. tepiduma
| Strain | Absorption maximum (nm) | Fluorescence maximum (nm)b | Foxc | Fredd | Fred/Fox | Level (mg per g of BChl c) of:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein | BChl a | Carot- enoids | Mena- quinone-7 | Chlorobium- quinonese | Total quinonesf | ||||||
| Wild type | 745 ± 3 | 771 ± 1 | 1.8 ± 0.2 | 60 ± 2 | 31 ± 4 | 272 ± 74 | 12 ± 1 | 65 ± 2 | 10 ± 2 | 53 ± 4 | 63 ± 6 |
| csmB | 748 ± 2 | 772 ± 2 | 1.9 ± 0.1 | 52 ± 7 | 28 ± 2 | 206 ± 34 | 11 ± 1 | 48 ± 4 | 7 ± 2 | 41 ± 3 | 48 ± 5 |
| csmC | 737 ± 1 | 768 ± 0 | 1.7 ± 0.2 | 54 ± 5 | 26 ± 6 | 228 ± 37 | 8 ± 0 | 59 ± 8 | 12 ± 3 | 41 ± 2 | 54 ± 4 |
| csmD | 745 ± 1 | 771 ± 1 | 2.0 ± 0.1 | 55 ± 9 | 27 ± 6 | 252 ± 64 | 11 ± 1 | 59 ± 4 | 11 ± 1 | 33 ± 9 | 44 ± 10 |
| csmE | 746 ± 4 | 772 ± 2 | 2.3 ± 0.2 | 44 ± 8 | 18 ± 3 | 260 ± 93 | 13 ± 1 | 62 ± 1 | 16 ± 11 | 25 ± 2 | 41 ± 9 |
| csmF | 747 ± 1 | 771 ± 0 | 2.1 ± 0.3 | 46 ± 5 | 27 ± 1 | 251 ± 35 | 11 ± 2 | 68 ± 4 | 13 ± 2 | 34 ± 19 | 47 ± 17 |
| csmH | 744 ± 3 | 771 ± 1 | 1.8 ± 0.3 | 48 ± 11 | 23 ± 4 | 276 ± 25 | 13 ± 1 | 67 ± 4 | 13 ± 4 | 38 ± 6 | 51 ± 2 |
| csmI | 744 ± 3 | 770 ± 1 | 2.1 ± 0.2 | 48 ± 6 | 21 ± 3 | 278 ± 23 | 12 ± 0 | 70 ± 3 | 15 ± 4 | 32 ± 7 | 47 ± 11 |
| csmJ | 746 ± 1 | 771 ± 1 | 1.9 ± 0.1 | 46 ± 8 | 27 ± 5 | 306 ± 60 | 15 ± 4 | 64 ± 2 | 13 ± 2 | 43 ± 14 | 56 ± 16 |
| csmX | 747 ± 1 | 772 ± 1 | 2.0 ± 0.2 | 42 ± 9 | 23 ± 4 | 250 ± 30 | 12 ± 3 | 65 ± 0 | 11 ± 0 | 36 ± 5 | 47 ± 5 |
All values are the averages and standard deviations of at least two measurements for two separate chlorosome preparations from different cell cultures.
Fluorescence measured under reducing conditions.
Fluorescence intensity under oxidizing conditions (in arbitrary units normalized to the level of BChl c absorption).
Fluorescence intensity under reducing conditions (in arbitrary units normalized to the level of BChl c absorption).
Chlorobiumquinones include chlorobiumquinone and 1′-hydroxymenaquinone, which occurred in an approximate ratio of 5 to 1.
Total quinones is the sum of menaquinone-7 and chlorobiumquinones.
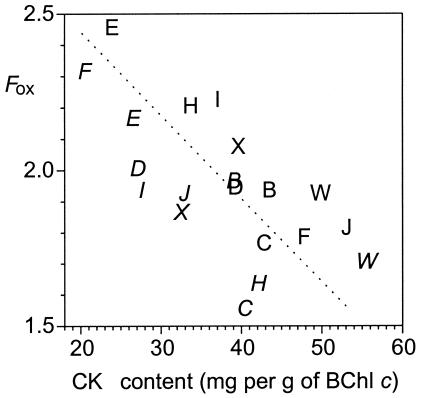
The quinone content of wild-type chlorosomes (10 mg of menaquinone and 53 mg of chlorobiumquinones [chlorobiumquinone and 1′-hydroxymenaquinone] per g of BChl c) is similar to previous measurements (14, 15). Chlorosomes of the nine mutant strains had similar menaquinone contents. However, the mutant chlorosomes consistently had significantly lower or, at most, similar chlorobiumquinone contents (Table 2 and Fig. 4). The total chlorobiumquinone content was varied among batches of chlorosomes from the mutants, and no other obvious pattern relating to the absence of a specific protein emerged from the data. The content of isoprenoid quinones in chlorosomes from the wild-type strain has also been observed to vary from batch to batch, and this variation may be due to subtle variations in growth conditions (e.g., small differences in levels of oxygen exposure in the cultures).
FIG. 4.
Correlation between the fluorescence emission intensity under oxidizing conditions (Fox) and the chlorobiumquinone (CK) contents in chlorosomes. Each letter refers to a separate chlorosome preparation (W, wild type; other letters refer to csm mutants).
The absorption maximum for BChl c in the wild-type chlorosomes was about 745 nm, and the BChl c absorption maxima were not significantly different from this value in the chlorosomes of most of the mutant strains. However, consistent with previous results (9), chlorosomes from the csmC mutant had an absorption maximum that was blue-shifted by about 8 nm (Table 2). HPLC analysis determined that the distributions of C-8 and C-12 methylation homologs of BChl c were not significantly different between the wild-type strain and any of the csm mutants. Thus, a difference in BChl c methylation cannot explain the altered absorbance properties of the chlorosomes of the csmC mutant. These results indicate that the organization of the BChl c aggregates was not significantly altered in the chlorosomes of any mutant strain except the one lacking CsmC.
In general, the amplitudes, shapes, and maxima of the fluorescence emission spectra were similar for chlorosomes from the wild-type strain and the various mutant strains (Table 2). The observation that both BChl c and BChl a fluorescence emissions were similar in the wild-type and mutant strains indicates that the organization of these pigments and the energy transfer between them were largely unaffected in the chlorosomes of the mutants. The only exception to this statement relates to the chlorosomes from the csmC mutant; the BChl c emission peak was blue-shifted about 3 nm for these chlorosomes (Table 2). In spite of the decreased level of BChl a in these chlorosomes (Table 2), no significant changes in the BChl a fluorescence of these chlorosomes or of the chlorosomes from any other mutant were detected (data not shown). Although the fluorescence emission intensities were similar under oxidizing conditions, one difference that was correlated with the total content of chlorobiumquinones was observed. Figure 4 shows the correlation between the relative fluorescence emission amplitude and the total contents of chlorobiumquinones from 20 different chlorosome preparations. In order to account for the natural variation in chlorobiumquinone contents, two samples for the wild-type strain and two samples for each mutant were analyzed. Previous observations have shown that chlorobiumquinones (and chemically related compounds) are better fluorescence quenchers than menaquinone (14, 16, 28), and thus the total chlorobiumquinone content should be inversely related to the fluorescence emission intensity under oxidizing conditions. As shown in Fig. 4, this is indeed the case. These observations strongly support the hypothesis that chlorobiumquinones are the components responsible for the redox regulation of energy transfer in chlorosomes in green sulfur bacteria (14-16, 28).
Estimation of protein numbers in wild-type chlorosomes.
In attempting to characterize the chlorosome proteins and their functions, it is useful to estimate the level of each protein in an average chlorosome and to compare that value to the amounts of other chlorosome components. A chlorosome from C. tepidum contains about 200,000 BChl c molecules (22) and, assuming a BChl a-to-BChl c ratio of about 0.012 (wt/wt) (Table 2), also contains roughly 2,200 BChl a molecules. The protein-to-BChl c ratio in chlorosomes is about 0.25 (wt/wt) (32) (Table 2). As estimated previously (4, 19), about one-third to one-half of this protein is CsmA (i.e., about 0.1 [wt/wt] on a BChl c basis), which corresponds to about 2,700 CsmA molecules per chlorosome. This number is in excellent agreement with the suggestion that CsmA binds BChl a at a 1:1 ratio and that there are about 2,700 CsmA-BChl a complexes per chlorosome (4, 23, 26). Based upon the staining intensities of the various constituent polypeptides on SDS-PAGE gels, a rough estimate of the stoichiometries of the remaining proteins on a weight basis is as follows. There are about 1,000 copies of CsmB; 300 copies of CsmC and CsmD; 200 copies of CsmE and CsmF; 100 copies of CsmH, CsmI, and CsmJ; and 20 copies of CsmX per chlorosome. Thus, an average chlorosome envelope contains about 5,000 protein molecules. The data in Table 2 indicate that the numbers of carotenoid, chlorobiumquinone, and menaquinone molecules per chlorosome are about 20,000, 15,000 and 3,000, respectively.
DISCUSSION
Only four of the csm mutants generated in this work (the csmB, csmC, csmH, and csmJ mutants) exhibited convincing phenotypes. Among the properties studied here, no characteristics were found in the csmD, csmE, csmF, csmI, and csmX mutants that distinguished them from the wild-type strain. The csmB mutant cells and chlorosomes contained about 25% less carotenoid than the wild-type strain (Table 2), which suggests that CsmB may play a role in organizing the carotenoids in chlorosomes. The only phenotype found in the csmH mutant was that the chlorosomes were about 20% shorter than those from the wild-type strain (Fig. 2), which suggests that CsmH, together with CsmC (see below), plays a role in determining the lengths of the BChl c aggregates. The csmJ mutant had slightly decreased growth rates at saturating and inhibitory light intensities. Because CsmJ is an iron-sulfur protein with a redox potential of about +90 mV (T. W. Johnson et al., unpublished data), it is possible that this protein directly or indirectly affects the turnover of photosynthetically generated reductants and that its absence may have an adverse effect on growth at high light intensities.
The only mutant with a more extensive phenotype was the csmC mutant. Compared to wild-type chlorosomes, chlorosomes isolated from this mutant were about 25% shorter (Fig. 2) and had blue-shifted BChl c absorption and fluorescence (Table 2). Thus, CsmC is the only protein whose absence seemed to affect BChl c organization. The small growth rate defect in this mutant at a limiting light intensity may be caused by the alteration in BChl c absorption, which might be correlated with a decreased light-harvesting efficiency. However, the reduced growth rate could also be due to the smaller size of the chlorosome. It is not clear how CsmC causes this effect. CsmC does not have a histidine residue, which is the most common ligand to chlorophylls in chlorophyll-binding proteins. It is noteworthy that the vestigial chlorosomes (designated carotenosomes) of a bchK mutant of C. tepidum that completely lacks BChl c also lack CsmC (18; Frigaard et al., unpublished). Observations for acetylene-treated Chlorobium vibrioforme cells also suggest a link between BChl c and CsmC. Acetylene specifically inhibits BChl c biosynthesis and causes the formation of abnormally small chlorosomes in green sulfur bacteria (31). However, the cellular levels of most chlorosome proteins are not significantly affected, and the chlorosome envelope seems to form normally even though the BChl c content decreases nearly 10-fold (31). A notable exception is that the cellular level of CsmC significantly increases in acetylene-treated cells. These results together suggest that CsmC and BChl c somehow interact and that changing the level of one affects the other.
How the small pool of BChl a in chlorosomes is organized is still not absolutely clear. However, since the levels and functional properties of BChl a in all of the csm mutants are similar to those in the wild-type strain, it is extremely unlikely that the nine eliminated Csm proteins bind BChl a or are necessary for its organization. Recent analyses of C. tepidum chlorosomes have shown that CsmA and BChl a are present in approximately equimolar amounts and that BChl a remains in the chlorosomes after the detergent-mediated extraction of all chlorosome proteins except CsmA (4). A subsequent treatment, which solubilizes CsmA from the chlorosome envelope without solubilizing the BChl c aggregates, also causes the release of BChl a. Since the spectroscopic properties of BChl a strongly suggest that it is protein bound, CsmA is the only candidate for the BChl a-binding protein. Montaño et al. (23) have recently shown CsmA to be a BChl a-binding protein in chlorosomes from the green filamentous bacterium Chloroflexus aurantiacus.
The absence of phenotypic effects in chlorosomes lacking a single protein could be due partly to the large number of protein types found in chlorosomes from C. tepidum. Although the chlorosome envelope contains 10 different proteins, these proteins can be classified into only four motif families (32). Thus, it is possible that some of the proteins can substitute for others and that obvious phenotypes will appear only if all proteins of a motif family are eliminated in the same cell. For example, CsmB and CsmF are about 40% similar in sequence, and CsmE is about 50% identical in sequence to CsmA and has a conserved histidine residue that could in principle serve as a ligand to BChl a (32). However, these proteins are present in very different amounts (about 5 times more CsmB than CsmF and about 14 times more CsmA than CsmE), and the level of one does not change when the other is eliminated (Fig. 3). In addition, since the inactivation of csmA appears to be lethal, or at least very difficult to achieve, CsmE can evidently not substitute for CsmA functionally or structurally. Nevertheless, due to the high sequence similarity, the possibility that CsmE binds a small, expendable pool of BChl a cannot be rigorously excluded. We are currently attempting to produce mutants of C. tepidum in which multiple genes have been inactivated. Technically, the maximum number of genes which can be inactivated in a given mutant strain is presently limited by the number of selective markers available (three) and by the degree of clustering of target genes (Fig. 1).
In spite of the phenotypes observed for four of the csm mutants, no csm mutant had a severe growth rate defect, and all mutants developed chlorosomes that were functionally and structurally indistinguishable from or similar to those from the wild-type strain. We therefore conclude that when they are eliminated individually, none of the nine chlorosome proteins whose genes were inactivated in this study are essential for the biogenesis, stability, or light-harvesting function of chlorosomes and their BChl c aggregates. This is a remarkable result, since the same suite of 10 envelope proteins are found in the chlorosomes from C. tepidum (containing BChl c), from C. vibrioforme (containing BChl d), and from Chlorobium phaeobacteroides (containing BChl e) (17, 31). These results demonstrate the remarkable robustness of the chlorosome design and are consistent with the fact that chlorosomes are an important determinant of the ability of these cells to grow at exceedingly low light intensities. The results of the present study are also consistent with those of previous studies that have concluded that pigment-pigment interactions, not pigment-protein interactions, are of principal importance in the structural and spectroscopic properties of the BChl c aggregates in chlorosomes (2, 3, 24). No other antenna structure in biology shows such a remarkable indifference towards the inactivation of the genes encoding its principal constituent protein species.
Acknowledgments
This work was supported by grant DE-FG02-94ER20137 to D.A.B. from the U.S. Department of Energy. N.-U.F. was supported by a grant from the Danish National Science Research Council.
We thank two anonymous reviewers for very helpful comments that improved the manuscript.
REFERENCES
- 1.Becker, A., M. Schmidt, W. Jäger, and A. Pühler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162:37-39. [DOI] [PubMed] [Google Scholar]
- 2.Blankenship, R. E., J. M. Olson, and M. Miller. 1995. Antenna complexes from green photosynthetic bacteria, p. 399-435. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Blankenship, R. E., and K. Matsuura. 2003. Antenna complexes from green photosynthetic bacteria, p. 195-217. In B. R. Green and W. W. Parson (ed.), Light-harvesting antennas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 4.Bryant, D. A., E. V. Vassilieva, N.-U. Frigaard, and H. Li. 2002. Selective protein extraction from Chlorobium tepidum chlorosomes using detergents. Evidence that CsmA forms multimers and binds bacteriochlorophyll a. Biochemistry 41:14403-14411. [DOI] [PubMed] [Google Scholar]
- 5.Chung, S. 1995. Characterization of chlorosomes in green sulfur bacteria. Ph.D. thesis. The Pennsylvania State University, University Park, Pa.
- 6.Chung, S., G. Frank, H. Zuber, and D. A. Bryant. 1994. Genes encoding two chlorosome proteins from the green sulfur bacteria Chlorobium vibrioforme strain 8327D and Chlorobium tepidum. Photosynth. Res. 41:261-275. [DOI] [PubMed] [Google Scholar]
- 7.Chung, S., and D. A. Bryant. 1996. Characterization of csmB genes from Chlorobium vibrioforme 8327D and Chlorobium tepidum and overproduction of the Chlorobium tepidum CsmB protein in Escherichia coli. Arch. Microbiol. 166:234-244. [DOI] [PubMed] [Google Scholar]
- 8.Chung, S., and D. A. Bryant. 1996. Characterization of the csmD and csmE genes from Chlorobium tepidum. The CsmA, CsmC, CsmD, and CsmE proteins are components of the chlorosome envelope. Photosynth. Res. 50:41-59. [DOI] [PubMed] [Google Scholar]
- 9.Chung, S., G. Shen, J. Ormerod, and D. A. Bryant. 1998. Insertional inactivation studies of the csmA and csmC genes of the green sulfur bacterium Chlorobium vibrioforme 8327: the chlorosome protein CsmA is required for viability but CsmC is dispensable. FEMS Microbiol. Lett. 164:353-361. [DOI] [PubMed] [Google Scholar]
- 10.Cogdell, R. J., N. W. Isaacs, T. D. Howard, K. McLuskey, N. J. Fraser, and S. M. Prince. 1999. How photosynthetic bacteria harvest solar energy. J. Bacteriol. 181:3869-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 99:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elhai, J., and C. P. Wolk. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119-138. [DOI] [PubMed] [Google Scholar]
- 13.Frigaard, N.-U., and D. A. Bryant. 2001. Chromosomal gene inactivation in the green sulfur bacterium Chlorobium tepidum by natural transformation. Appl. Environ. Microbiol. 67:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frigaard, N.-U., S. Takaichi, M. Hirota, K. Shimada, and K. Matsuura. 1997. Quinones in chlorosomes of green sulfur bacteria and their role in the redox-dependent fluorescence studied in chlorosome-like bacteriochlorophyll c aggregates. Arch. Microbiol. 167:343-349. [Google Scholar]
- 15.Frigaard, N.-U., K. Matsuura, M. Hirota, M. Miller, and R. P. Cox. 1998. Studies of the location and function of isoprenoid quinones in chlorosomes from green sulfur bacteria. Photosynth. Res. 58:81-90. [Google Scholar]
- 16.Frigaard, N.-U., S. Tokita, and K. Matsuura. 1999. Exogenous quinones inhibit photosynthetic electron transfer in Chloroflexus aurantiacus by specific quenching of the excited bacteriochlorophyll c antenna. Biochim. Biophys. Acta 1413:108-116. [DOI] [PubMed] [Google Scholar]
- 17.Frigaard, N.-U., E. V. Vassilieva, H. Li, K. J. Milks, J. Zhao, and D. A. Bryant. 2001. The remarkable chlorosome, article S1-003. In PS2001 proceedings of the 12th International Congress on Photosynthesis. CSIRO Publishing, Melbourne, Australia.
- 18.Frigaard, N.-U., G. D. Voigt, and D. A. Bryant. 2002. Chlorobium tepidum mutant lacking bacteriochlorophyll c made by inactivation of the bchK gene, encoding bacteriochlorophyll c synthase. J. Bacteriol. 184:3368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frigaard, N.-U., A. Gomez Maqueo Chew, H. Li, J. A. Maresca, and D. A. Bryant. 2003. Chlorobium tepidum: insights into the structure, physiology, and metabolism of a green sulfur bacterium derived from the complete genome sequence. Photosynth. Res. 78:93-117. [DOI] [PubMed]
- 20.Kühlbrandt, W., D. N. Wang, and Y. Fujiyoshi. 1994. Atomic model of plant light-harvesting complex by electron crystallography. Nature 367:614-621. [DOI] [PubMed] [Google Scholar]
- 21.Li, Y. F., W. L. Zhou, R. E. Blankenship, and J. P. Allen. 1997. Crystal structure of the bacteriochlorophyll a protein from Chlorobium tepidum. J. Mol. Biol. 271:456-471. [DOI] [PubMed] [Google Scholar]
- 22.Montaño, G. A., B. P. Bowen, J. T. LaBelle, N. W. Woodbury, V. B. Pizziconi, and R. E. Blankenship. 2001. Determination of the number of bacteriochlorophyll molecules per chlorosome light-harvesting complex in Chlorobium tepidum, article S1-020. In PS2001 proceedings of the 12th International Congress on Photosynthesis. CSIRO Publishing, Melbourne, Australia.
- 23.Montaño, G. A., H.-M. Wu, S. Lin, D. C. Brune, and R. E. Blankenship. 2003. Isolation and characterization of the B795 light-harvesting baseplate from the chlorosomes of Chloroflexus aurantiacus. Biochemistry 42:10246-10251. [DOI] [PubMed]
- 24.Olson, J. M. 1998. Chlorophyll organization and function in green photosynthetic bacteria. Photochem. Photobiol. 67:61-75. [Google Scholar]
- 25.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 26.Sakuragi, Y., N.-U. Frigaard, K. Shimada, and K. Matsuura. 1999. Association of bacteriochlorophyll a with the CsmA protein in chlorosomes of the photosynthetic green filamentous bacterium Chloroflexus aurantiacus. Biochim. Biophys. Acta 1413:172-180. [DOI] [PubMed] [Google Scholar]
- 27.Sidler, W. A. 1994. Phycobilisome and phycobiliprotein structures, p. 139-216. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 28.Tokita, S., N.-U. Frigaard, M. Hirota, K. Shimada, and K. Matsuura. 2000. Quenching of bacteriochlorophyll fluorescence in chlorosomes from Chloroflexus aurantiacus by exogenous quinones. Photochem. Photobiol. 72:345-350. [DOI] [PubMed] [Google Scholar]
- 29.Tronrud, D. E., M. F. Schmid, and B. W. Matthews. 1986. Structure and X-ray amino acid sequence of a bacteriochlorophyll a protein from Prosthecochloris aestuarii refined at 1.9 Å resolution. J. Mol. Biol. 188:443-454. [DOI] [PubMed] [Google Scholar]
- 30.Vassilieva, E. V., N.-U. Frigaard, and D. A. Bryant. 2000. Chlorosomes: the light-harvesting complexes of the green bacteria. Spectrum 13:7-13. [Google Scholar]
- 31.Vassilieva, E. V., J. G. Ormerod, and D. A. Bryant. 2002. Biosynthesis of chlorosome proteins is not inhibited in acetylene-treated cultures of Chlorobium vibrioforme. Photosynth. Res. 71:69-81. [DOI] [PubMed] [Google Scholar]
- 32.Vassilieva, E. V., V. L. Stirewalt, C. U. Jakobs, N.-U. Frigaard, K. Inoue-Sakamoto, M. A. Baker, A. Sotak, and D. A. Bryant. 2002. Subcellular localization of chlorosome proteins in Chlorobium tepidum and characterization of three new chlorosome proteins: CsmF, CsmH and CsmX. Biochemistry 41:4358-4370. [DOI] [PubMed] [Google Scholar]