Abstract
A major goal of plant biotechnology is the production of genetically engineered crops that express natural or foreign proteins at high levels. To enhance protein accumulation in transgenic plants, we developed a set of vectors that express proteins and peptides as C-terminal translational fusions with ubiquitin (UBQ). Studies of several proteins in tobacco (Nicotiana tabacum) showed that: (a) proteins can be readily expressed in plants as UBQ fusions; (b) by the action of endogenous UBQ-specific proteases (Ubps), these fusions are rapidly and precisely processed in vivo to release the fused protein moieties in free forms; (c) the synthesis of a protein as a UBQ fusion can significantly augment its accumulation; (d) proper processing and localization of a protein targeted to either the apoplast or the chloroplast is not affected by the N-terminal UBQ sequence; and (e) single amino acid substitutions surrounding the cleavage site can inhibit in vivo processing of the fusion by Ubps. Noncleavable UBQ fusions of β-glucuronidase became extensively modified, with additional UBQs in planta. Because multiubiquitinated proteins are the preferred substrates of the 26S proteasome, noncleavable fusions may be useful for decreasing protein half-life. Based on their ability to augment protein accumulation and the sequence specificity of Ubps, UBQ fusions offer a versatile way to express plant proteins.
Current biotechnological strategies for improving crop plants often require the high-level expression of natural and “foreign” proteins and peptides. Most of these polypeptides are intended to confer beneficial agronomic traits such as improved nutritional quality or resistance to herbicides, viruses, and insects (Shah, 1997; Yuan and Knauf, 1997). In addition, transgenic plants have also shown promise for the economic biosynthesis of pharmaceuticals, vaccines, and industrial proteins (Goddijn and Pen, 1995; Haq et al., 1995). A primary obstacle to protein expression in plants is low yields. Although the use of strong promoters has partially overcome transcriptional limitations to expression, barriers still exist with regard to the various posttranscriptional steps required to produce a fully active, mature protein. Numerous, largely anecdotal examples exist wherein proteins failed to accumulate to adequate levels in transgenic plants even though the introduced genes were actively transcribed (e.g. Odell et al., 1990; Ohtani et al., 1991; Cherry et al., 1993).
One strategy for augmenting protein expression involves the synthesis of the protein as a translational fusion to another (LaValle and McCoy, 1995). The fusion partner appears to boost expression by increasing translation of the mRNA and/or by enhancing solubility and folding of the protein, presumably by acting as a covalently linked chaperone. An additional proteolytic step is usually required after translation to release the protein from the fused partner.
A fusion partner that has received considerable attention in recent years is UBQ, a highly conserved, stable, and abundant protein in eukaryotes that functions in selective protein degradation (for review, see Vierstra, 1996). Its application stems from its unusual method of synthesis. Unlike most other eukaryotic proteins, UBQ is not synthesized as individual 76-amino-acid monomers but as fusions. The corresponding genes either encode a poly-UBQ precursor in which UBQ monomers are linked in tandem, or UBQ extension proteins in which a UBQ monomer is linked to the N terminus of an unrelated protein, some of which are ribosomal subunits. The initial translation products of these genes are accurately and rapidly cleaved in vivo by Ubps (UBQ C-terminal hydrolases or de-ubiquitinating enzymes), a family of novel, sequence-specific proteases that release the UBQ monomers (Wilkinson, 1997). Cleavage occurs irrespective of the amino acid immediately following UBQ, with the exception of P, which is processed inefficiently (Varshavsky, 1997).
When examined in microorganisms, fusion of UBQ to proteins has been shown to substantially enhance accumulation (Butt et al., 1989; Ecker et al., 1989; Baker, 1996) (see Fig. 1). This approach was especially beneficial for short peptides and gene products that were expressed poorly, if at all. For example, the levels of some recalcitrant proteins could be increased several hundred times in yeast and could account for up to 20% of the soluble protein in Escherichia coli simply by cotranslation with UBQ. In yeast, intact, nonfused proteins accumulate after cleavage by endogenous Ubp; the only exceptions are fused proteins beginning with a P residue. Because prokaryotes lack Ubps, unprocessed UBQ-fusion products accumulate in E. coli. These fusions can be processed in vivo by coexpression with Ubps or in vitro following the addition of purified Ubps. More recent studies suggest that the UBQ fusion approach could work in plants as well (Hondred and Vierstra, 1992; Garbino et al., 1995; Worley et al., 1998). Plants naturally express UBQ fusions (Callis et al., 1995) and have an array of Ubps, some of which can process UBQ fusions in vitro and in vivo (Sullivan et al., 1990; Chandler et al., 1997; N. Yan, T. Falbel, and R.D. Vierstra, unpublished data).
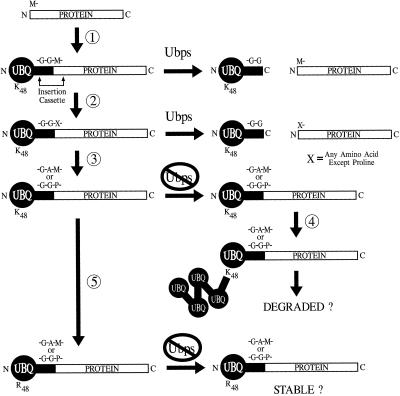
Figure 1.
Various strategies for expressing proteins as UBQ fusions in plants. 1, Genes are created that express chimeric UBQ fusion proteins in which the N terminus of the candidate protein is joined in-frame to the C-terminal G-76 of UBQ. Unique restriction sites in the fusion vector define a small insertional cassette in which codons at or near the UBQ-protein junction can be exchanged by swapping short oligonucleotide bridges. When expressed in plants, endogenous Ubps specifically cleave the UBQ-protein fusion after G-76 to generate both the candidate protein and UBQ in free forms. Processing is expected to occur regardless of the nature of the amino acid following the C-terminal G of UBQ; P is the only exception. 2, By altering the N-terminal codon(s) for the candidate protein, proteins with N-terminal residues other than M can be expressed and released in intact forms by Ubps. 3, By substituting G-76 of UBQ for A or by including P as the first amino acid of the candidate protein, processing of the UBQ fusion by Ubps can be effectively inhibited. 4, The UBQ conjugation pathway will assemble a chain of UBQs onto these noncleavable UBQ moieties provided that amino acid residue 48 is a K. Theoretically, these multiubiquitinated proteins can become substrates for degradation by the 26S proteasome. 5, If K-48 is replaced by R, the noncleavable UBQ moiety is protected from subsequent ubiquitination and the fusion protein may be stable. N, N terminus; C, C terminus.
Given the potential of UBQ fusions for enhancing protein production in transgenic plants (Hondred and Vierstra, 1992), we created a series of vectors for their expression. In this paper we show that chimeric UBQ-protein fusions can be synthesized in tobacco, accurately processed to yield unmodified active proteins, and correctly localized to their appropriate subcellular compartments. For two test proteins, GUS and LUC, we found that expression as a UBQ fusion can significantly increase accumulation. As a result, the UBQ fusion approach may have broad utility for enhancing protein production in plants.
MATERIALS AND METHODS
Construction of Chimeric UBQ Fusion Vectors
We generated the various constructions with standard cloning techniques, using replacement with appropriate oligonucleotide bridges and/or PCR with mutagenic primers to alter the DNA sequence. The design of each gene was verified by DNA sequence analysis. Diagrams of the completed vectors appear in Figures 1, 2, 5, and 6. More complete descriptions of the constructions are available upon request. Expression of all genes was directed by the CaMV 35S promoter obtained as a 645-bp XhoI/SpeI fragment from the plasmid pAMVBTS (Barton et al., 1987) and designed to contain ApaI, EagI, and NheI sites at the 5′ end. The promoter sequence was followed by the 39-bp 5′-UTR from AMV (Gehrke et al., 1983). Polyadenylation signals were obtained from the 3′ end of the Agrobacterium tumefaciens NOS gene (Bevan et al., 1983). The UBQ coding region was obtained from the third UBQ coding repeat within the Arabidopsis AtUBQ11 gene (Callis et al., 1995) and engineered to contain a translationally silent BglII site 10 bp downstream from the start codon, and a translationally silent SacII site 10 bp upstream from the terminal G codon. We used PCR to convert UBQ codon K-48 to that for R.
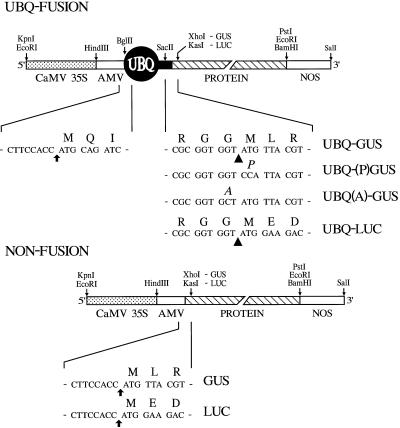
Figure 2.
Structures of the various gene constructions used for expressing GUS and LUC as UBQ fusions. The UBQ fusion vector was designed to contain the CaMV 35S promoter, the AMV 5′-UTR, and the sequence encoding UBQ. Appended to the UBQ sequence is DNA encoding GUS or LUC, followed by the polyadenylation signals from NOS. The nucleotide sequences surrounding the translational initiation site (arrows) and the UBQ/protein junction are shown. Convenient restriction sites used for assembly are indicated; the restriction sites within the 5′-coding region of GUS and LUC are XhoI and KasI, respectively. The structure and sequences of the respective nonfused genes that were used as controls are also shown. Arrowheads indicate the predicted cleavage site by Ubps. UBQ-(P)GUS contains GUS with its N-terminal Met changed to Pro; UBQ(A)-GUS contains UBQ with its C-terminal Gly changed to Ala.
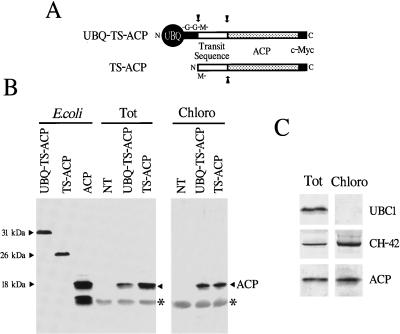
Figure 5.
Expression and chloroplastic localization of spinach ACPII after synthesis as a UBQ fusion in transgenic tobacco. A, Diagram of the ACP expression cassette. ACP bearing its N-terminal TS and a C-terminal c-Myc epitope tag was expressed either alone (TS-ACP) or as a UBQ fusion (UBQ-TS-ACP). Arrows indicate the predicted cleavage sites of the proteins after expression and import to the chloroplast. B, Immunodetection of ACP proteins expressed in tobacco. Protein was extracted and subjected to SDS-PAGE, and the ACP protein was visualized by immunoblot analysis with the c-Myc antibody 9E10. Tot represents 10 μg of total leaf protein. Chloro represents equal aliquots of protein extracted from the chloroplast stromal fraction following Percoll gradient centrifugation. Migration position of ACP alone (arrowhead), TS-ACP, and UBQ-TS-ACP was determined using the corresponding proteins expressed in E. coli. Lane NT, Sample from a nontransformed plant. (The approximately 15-kD protein [*] detected in the tobacco samples was the small subunit of Rubisco nonspecifically interacting with the c-Myc antibody. The approximately 15-kD protein in the E. coli-expressed ACP sample is a bacterial breakdown product of ACP.) C, Co-localization of ACP with the chloroplast protein CH-42 subunit of the Mg2+ protoporphyrin chelatase. Total leaf protein (Tot) and protein from the chloroplast stromal fraction (Chloro) were extracted from plants expressing UBQ-TS-ACP and subjected to SDS-PAGE and immunoblot analysis with antibodies against CH-42, c-Myc, or the cytosolic enzyme UBC1.
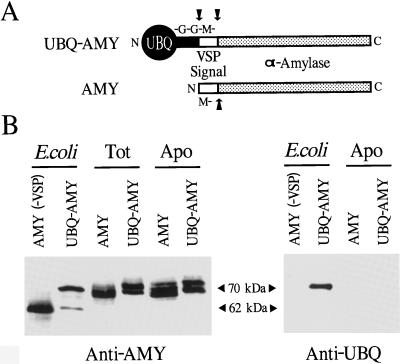
Figure 6.
Expression and apoplastic localization of B. licheniformis AMY after synthesis as a UBQ fusion in transgenic tobacco. A, Diagram of the AMY expression cassette. AMY bearing the N-terminal signal sequence from the soybean VSP was expressed either alone or as a UBQ fusion (UBQ-AMY). Arrows indicate the predicted cleavages sites of the proteins after expression and export to the apoplast. B, Immunodetection of AMY proteins expressed in tobacco. Protein was extracted and subjected to SDS-PAGE, and the AMY protein was visualized by immunoblot analysis with either AMY antibodies (left) or UBQ antibodies (right). Tot represents 10 μg of total soluble leaf proteins. Apo represents equal aliquots of protein extracted from the leaf apoplast. The migration positions of AMY (without the VSP signal sequence) and UBQ-AMY were determined using the corresponding proteins expressed in E. coli.
Using appropriate oligonucleotide bridges, GUS, LUC, AMY, and ACP coding regions were appended in-frame to the 35S/AMV/UBQ vector at the SacII site. DNA encoding GUS or LUC and the NOS 3′ end was isolated from pCMC1100 (McCabe et al., 1988) or pAB14016LBS (de Wet et al., 1987), respectively, and engineered to contain either a XhoI site (GUS) or a KasI site (LUC) at the 5′ end. The LUC-coding region retained its peroxisomal targeting sequence (Gould et al., 1990). We altered the amino acid sequence at the junction between UBQ and GUS by replacing the spanning DNA sequence between the SacII and XhoI sites with appropriate 31-bp oligonucleotide bridges. Coding DNA for the mature portion of Bacillis licheniformis AMY (minus the N-terminal 29-amino acid secretion peptide), obtained by PCR amplification of genomic DNA (Pen et al., 1992), was modified to include the coding sequence for a H-6 tag (RYLHHHHHH) appended in-frame to the 3′ end. The sequence encoding the N-terminal, 22-residue ER signal sequence from the soybean VSP β-subunit (Mason et al., 1988) was recreated by an oligonucleotide bridge and appended in-frame to the 5′ end of the AMY coding region. DNA containing the 48-amino acid TS and 131-amino acid coding region for spinach (Spinacia oleracea) root ACPII was obtained from the cDNA pKS21 (Schmid and Ohlrogge, 1990). We used PCR to add codons for the c-Myc epitope (EQKLISEEDL [Kolodzeij and Young, 1991]), followed by a stop codon in-frame to the 3′ end. We also generated a control plasmid for each gene that was identical in all aspects, except for the absence of the UBQ-coding region. The completed genes were inserted into the binary A. tumefaciens vector BIN19 at the KpnI and SalI sites (Bevan, 1984).
Plant Transformations
The completed BIN19 plasmids were introduced directly into A. tumefaciens strain LBA4404 to transform tobacco (Nicotiana tabacum cv Xanthi) leaf discs (Cherry et al., 1993). Stably transformed plants were selected by kanamycin resistance. Transgenic plants were transferred to soil and grown to maturity in a greenhouse. We performed transformations for each pair of UBQ fusion and control vectors simultaneously under identical conditions.
Enzyme Extraction and Analysis
Except as noted, proteins were isolated from the youngest fully expanded leaves of transformed plants. We measured the total soluble protein by the Bradford method (Bradford, 1976). For plants expressing GUS, leaf tissue was homogenized in 50 mm sodium phosphate (pH 7.0), 10 mm β-mercaptoethanol, 10 mm Na4EDTA, 20 mm sodium metabisulfite, 0.1% (w/v) sodium lauryl sarcosine, and 0.1% (v/v) Triton X-100. We used a fluorometric assay with 4-methylumbelliferyl β-d-glucuronide as the substrate (Gallagher, 1992) to determine the enzymatic activity from the clarified extracts. GUS protein was purified using ammonium sulfate precipitation (40%–50% saturation), followed by BioGel A1.5M size-exclusion chromatography (Bio-Rad), saccharo-1,4-lactone agarose (Sigma) affinity chromatography (Harris et al., 1973), and, finally, Mono-Q ion-exchange chromatography (Pharmacia). Purified GUS protein (50–100 μg) was subjected to N-terminal amino acid sequence analysis using an Applied Biosystems model 470A protein sequencer.
We homogenized leaf tissue expressing LUC in 25 mm Tris-phosphate (pH 7.8), 2 mm DTT, 2 mm 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% (v/v) glycerol, and 1% (v/v) Triton X-100. The amount of LUC protein was determined by a chemiluminescence assay (Promega) using purified LUC as the standard (Sigma).
The apoplastic fluid from young tobacco leaves was collected by centrifugal extraction of intercellular fluid (Pen et al., 1992). Tobacco leaf strips were washed, blotted dry, and vacuum infiltrated with 20 mm Hepes (pH 6.9), 3 mm MgCl2, and 3 mm DTT. Afterward, the strips were blotted dry, rolled between strips of Parafilm, and centrifuged at 4°C for 10 min at 200g. We isolated chloroplasts from 15-d-old plants by Percoll density-gradient centrifugation (Falbel and Staehelin, 1994), with the inclusion of 1 mm MgCl2 in the sorbitol buffer.
Immunoblot Analysis
Proteins were separated by SDS-PAGE, electrotransferred onto either nitrocellulose or Immobilon-P membranes (Millipore), and subjected to immunoblot analysis as previously described (van Nocker et al., 1993). Rabbit polyclonal antibodies were prepared against GUS (Clontech, Palo Alto, CA), LUC (Millar et al., 1992), plant UBQ (van Nocker et al., 1993), Arabidopsis CH42 (Guo et al., 1998), UBC1 (Sullivan et al., 1994), and B. licheniformis AMY (D. Mathews, unpublished data). The c-Myc epitope was detected with the monoclonal antibody 9E10 (Kolodziej and Young, 1991). We identified the immunoreactive proteins with the appropriate alkaline phosphatase-coupled goat anti-rabbit or goat anti-mouse IgGs (Kirkegaard & Perry Laboratories, Gaithersburg, MD). The UBQ-GUS conjugates were immunoprecipitated with GUS antibodies and Protein A Sepharose (Sigma).
To generate Escherichia coli-expressed versions of ACP and AMY, we amplified the coding regions by PCR from the appropriate BIN19 plasmids and inserted them into the pET28a or pET29a (Novagen, Madison, WI) vectors. Induced bacteria (strain BL21) were sonicated and the clarified cell lysates were used directly for the immunoblot analysis.
Nucleic Acid Extraction and Analysis
We extracted DNA from tobacco leaves using the hexadecyltrimethylammonium bromide method (Doyle and Doyle, 1987). The presence of introduced DNA sequences was confirmed by genomic DNA gel-blot analysis using 32P-labeled, sequence-specific DNA probes or by PCR of genomic DNA using primers specific for the coding sequence or the CaMV 35S promoter. Using the procedure of Wadsworth et al. (1988), we isolated total RNA from the youngest fully expanded leaves. RNA was electrophoresed under denaturing conditions in 2.2 m formaldehyde-containing agarose gels, transferred onto a Zetaprobe membrane (Bio-Rad), and subsequently hybridized with 32P-labeled RNA probes created using T3 or T7 RNA polymerase (Stratagene). The GUS-specific probe comprised the 2.3-kb XhoI-BamHI fragment from pCMC1100. The LUC-specific probe used an internal 1.2-kb EcoRI fragment from pAB14016LBS. We made a UBQ-specific probe from a 210-bp BglII-SacII DNA fragment from AtUBQ11.
RESULTS
Construction of the UBQ Fusion Vector
To examine the UBQ fusion approach in plants (Fig. 1), we created a cassette vector for expressing proteins/peptides as in-frame C-terminal fusions to UBQ (Hondred and Vierstra, 1992), a diagram of which appears in Figure 2. The coding sequence for UBQ was provided by the last UBQ repeat within the Arabidopsis UBQ11 poly-UBQ gene (Callis et al., 1995); its derived amino acid sequence is 100% identical to the canonical UBQ sequence present in higher plants. Two silent restriction sites (BglII and SacII) were introduced to facilitate the interchange of 5′-regulatory elements, 3′-coding regions, and the amino acid sequence at the UBQ/protein junction through replacement of short oligonucleotide bridges. Regulatory sequences for effective expression included the CaMV 35S promoter, the AMV 5′-UTR, and the NOS 3′-UTR. The nucleotide sequence immediately 5′ to the initiation codon conformed to the optimal Kozak sequence for mammalian translation initiation (CCACC ATG [Kozak, 1986]), although this sequence may be less than optimal in plants (Gallie, 1993). For comparison, a control vector was also created that was identical in all respects to the UBQ fusion vector, except that it was missing the UBQ-coding region (Fig. 2). We gave special attention to maintaining the same nucleotide sequence 5′ to the initiation codon to avoid any differences in translation efficiency not associated with the presence of the UBQ-coding sequence. We introduced all of the genes into tobacco and analyzed a number of independent, stable transformants for expression.
GUS Expression
We first compared the expression of GUS either alone or as a UBQ fusion. For tobacco expressing GUS alone, RNA gel-blot analysis with a GUS-specific RNA probe detected a 2.3-kb mRNA, consistent with the predicted size of the GUS transcript (Fig. 3A). In plants expressing the UBQ-GUS fusion (UBQ-GUS), a larger, 2.5-kb mRNA was present, in accord with the additional 228-bp coding region for UBQ. This 2.5-kb mRNA also hybridized with a UBQ-specific probe, confirming that the UBQ sequence was transcribed (Fig. 3A). mRNAs of 1.6 and 0.8 kb were also detected with the UBQ probe; they probably represent poly-UBQ and UBQ-extension genes endogenous to tobacco (Callis et al., 1995).
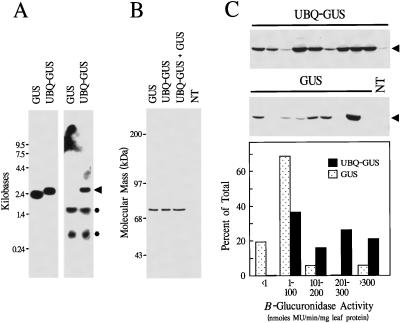
Figure 3.
Expression and processing of UBQ-GUS fusions in transgenic tobacco. A, RNA gel-blot analysis of 10 μg of total RNA isolated from young leaves expressing GUS alone or as a UBQ fusion (lanes UBQ-GUS). Left, RNA blot hybridized with a GUS-specific probe. Right, RNA blot hybridized with a UBQ-specific probe. The arrowhead indicates migration position of the UBQ-GUS transcript. RNAs of 1.6 and 0.8 kb (*) that also hybridize with the UBQ probe correspond in size to transcripts derived from endogenous tobacco UBQ genes. B, Immunoblot analysis of plants expressing GUS or UBQ-GUS. Soluble protein (10 μg) from young leaves was subjected to SDS-PAGE, and GUS protein was visualized by immunoblotting with GUS antibodies. Lane UBQ-GUS + GUS, Equal amounts of leaf extract from plants expressing UBQ-GUS and GUS mixed prior to electrophoresis. Lane NT, Sample from a nontransformed plant. C, Accumulation of GUS protein in plants transformed with either the GUS or the UBQ-GUS vector. Top and middle, Immunoblot analysis with GUS antibodies of soluble leaf protein extracted from randomly selected T0 plants independently transformed with either the UBQ-GUS or the GUS vector. Arrowheads indicate the position of mature GUS. NT, Sample from a nontransformed plant. Bottom, Distribution profile of GUS activity in a population of independently transformed tobacco: 19 UBQ-GUS and 16 GUS plants were analyzed. A fluorogenic assay determined GUS activity by measuring the conversion of the substrate 4-methylumbelliferyl β-d-glucuronide to the product 4-methylumbelliferone (MU).
Expression of the UBQ-GUS and GUS vectors resulted in the accumulation of active GUS protein. The GUS vector directed the synthesis of a 74-kD protein (Fig. 3B), in agreement with previous SDS-PAGE determinations of GUS (Jefferson et al., 1987). A protein of indistinguishable size was also synthesized from the UBQ-GUS vector, despite the additional sequence encoding UBQ (Fig. 3B). In fact, the products from the GUS and UBQ-GUS vectors comigrated as a single species during SDS-PAGE. The protein from UBQ-GUS was not recognized by UBQ antibodies, indicating that most, if not all, of the UBQ moiety was absent from the mature protein (Fig. 7A). Even after overloading the SDS-PAGE gels with protein and overdeveloping the blots, we were unable to detect the initial translation product of UBQ-GUS, suggesting that the UBQ moiety was rapidly and efficiently removed after synthesis (data not shown). It is possible that internal initiation of the UBQ-GUS mRNA generated a polypeptide lacking the UBQ sequence. However, subsequent analysis of noncleavable UBQ-GUS fusions demonstrated that the UBQ coding region was translated (see below).
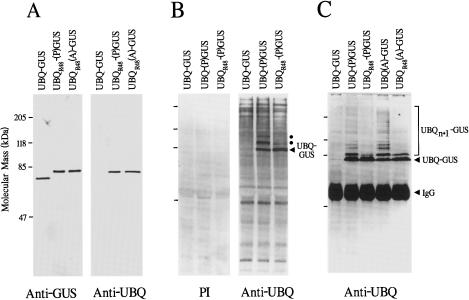
Figure 7.
Expression and ubiquitination of UBQ-GUS fusions containing mutations surrounding the Ubp cleavage site. Mutant versions of UBQ-GUS that contained the G-76-to-A substitution in UBQ (lanes UBQ[A]-GUS) or the M-1-to-P substitution in GUS (UBQ-[P]GUS), with or without K-48-to-A substitution in UBQ (UBQR48) were created (see Fig. 2). The proteins were expressed in tobacco and extracted from young leaves. Samples were subjected to SDS-PAGE and immunoblot analysis with UBQ preimmune serum (PI), GUS antibodies, or UBQ antibodies. A, Immunoblot analysis of the various GUS proteins purified from leaves by saccharo-1,4-lactone agarose-affinity chromatography. B, Immunoblot analysis of total leaf protein. Arrowhead indicates the migration position of nonprocessed UBQ-GUS. Dots show the migration positions of UBQ conjugates of UBQ-GUS formed in vivo. C, Immunoblot analysis of UBQ-GUS immunoprecipitated from total soluble protein with GUS antibodies and then subjected to SDS-PAGE and immunoblot analysis with UBQ antibodies. Arrowheads show unmodified UBQ-GUS and the heavy chain of the GUS IgG. The bracket indicates the UBQ conjugates of UBQ-GUS.
To determine the exact cleavage site(s) in the UBQ-GUS protein, we purified the GUS protein from tobacco expressing UBQ-GUS and GUS and subjected both preparations to N-terminal amino acid sequence analysis. Initial yields of phenylthiohydantoin amino acids were close to the expected values, indicating that each protein probably contained a free, nonacetylated N terminus (data not shown). The N-terminal sequence of the protein purified from the GUS-expressing plants was MLRPVETPTREIKKL for the first 15 cycles, which was identical to that predicted from the nucleotide sequence (Jefferson et al., 1987). The same sequence was obtained for UBQ-GUS. In the first cycle, Met was the only residue in significant quantity (>95% of total residues), with no evidence of other amino acids that would have resulted from imprecise cleavage at the UBQ/GUS junction (i.e. Gly, Leu, or Arg [Fig. 2]).
From a collection of independently transformed tobacco plants, we compared the amount of GUS protein generated from the GUS and UBQ-GUS vectors (Fig. 3C). All transgenic plants (19 plants for UBQ-GUS and 16 plants for GUS) that contained one or more nonrearranged copies of the appropriate gene (as determined by DNA gel-blot analysis) were included. As is commonly observed for transgene expression in plants (e.g. Martin et al., 1992; Cherry et al., 1993), the levels of GUS (determined either immunologically or enzymatically) varied widely among the independent transformants. However, when the expression levels were collectively assessed, significantly greater amounts of GUS protein were generated on average from the UBQ-GUS vector than from the GUS vector (Fig. 3C). By enzymatic assay, 4.1 times more GUS activity was synthesized in the UBQ-GUS plants (Table I). Furthermore, the percentage of the transgenic plants that accumulated GUS to high levels (>200 nmol 4-methylumbelliferone min−1 mg−1 leaf protein) was 10-fold greater in the UBQ-GUS population (Fig. 3C).
Table I.
Accumulation of GUS and LUC expressed alone or as a UBQ fusion
| Construction | Activitya | UBQ-Protein/Protein |
|---|---|---|
| UBQ-GUS (n = 19) | 166 ± 133 | 4.1 (P < 0.001)b |
| GUS (n = 16) | 41 ± 68 | |
| UBQ-LUC (n = 64) | 101 ± 99 | 2.2 (p < 0.0002)b |
| LUC (n = 43) | 47 ± 40 | P |
Protein content was assayed in young, mature-green leaves of independently transformed T0 plants. GUS levels were determined by fluorogenic assay that measured the conversion of the substrate 4-methylumbelliferyl β-d-glucuronide to the product 4-methylumbelliferone. LUC levels were determined by chemiluminescence assay using the activity of the purified LUC as the standard. n, Number of independent transformants analyzed for each vector.
Activity units are nanomoles 4-methylumbelliferone per minute per milligram of leaf protein for GUS and picograms of LUC protein per microgram of leaf protein for LUC. Activity is expressed as ±sd.
Student's t test of sample means.
LUC Expression
In a manner similar to the procedure for GUS, we examined the potential benefits of UBQ fusion for expressing LUC. As can be seen in Figure 4, a collection of independently transformed tobacco was generated that expressed LUC or UBQ-LUC. For the LUC vector, an mRNA of the appropriate size (2.0 kb) was transcribed and translated into an active LUC protein of 62 kD (Fig. 4, A and B), in agreement with the apparent molecular mass of the LUC protein reported previously (de Wet et al., 1987). For the UBQ-LUC vector, a slightly larger mRNA of 2.3 kb that contained both LUC- and UBQ-coding sequences was evident (Fig. 4A). This mRNA generated a protein indistinguishable in size to that expressed from the LUC vector (Fig. 4B). Similar to our observations with UBQ-GUS, the UBQ-LUC protein was easily recognized by LUC antibodies but not by UBQ antibodies, indicating that the UBQ moiety was removed posttranslationally. Moreover, the 62-kD LUC protein was the only species detected even when the SDS-PAGE gels were overloaded with protein and the blots were overdeveloped (data not shown), suggesting that cleavage was rapid and quantitative.
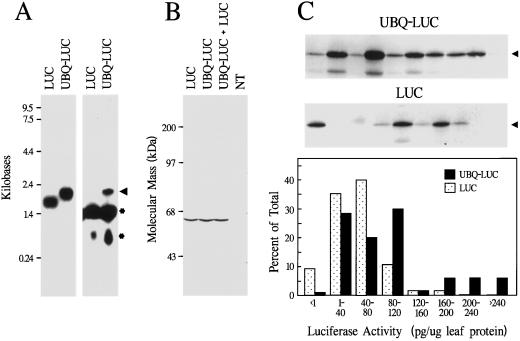
Figure 4.
Expression and processing of UBQ-LUC fusions in transgenic tobacco. A, RNA gel-blot analysis of 10 μg of total RNA isolated from young leaves expressing LUC alone or as a UBQ fusion (lanes UBQ-LUC). Left, RNA blot hybridized with a LUC-specific probe. Right , RNA blot hybridized with a UBQ-specific probe. The arrowhead indicates migration position of the UBQ-LUC transcript. RNAs of 1.6 and 0.8 kb (*) that also hybridize with the UBQ probe correspond in size to transcripts derived from endogenous tobacco UBQ genes. B, Immunoblot analysis of plants expressing LUC or UBQ-LUC. Soluble protein (10 μg) from young leaves was subjected to SDS-PAGE and LUC protein was visualized by immunoblotting with LUC antibodies. Lane UBQ-LUC + LUC, Equal amounts of leaf extract from plants expressing UBQ-LUC and LUC mixed prior to electrophoresis. Lane NT, Sample from a nontransformed plant. C, Accumulation of LUC protein in plants transformed with either the LUC or the UBQ-LUC vector. Top and middle, Immunoblot analysis with LUC antibodies of soluble leaf protein extracted from randomly selected T0 plants independently transformed with either the UBQ-LUC or the LUC vector. Arrowheads indicate the position of mature LUC. Bottom, Distribution profile of LUC activity in a population of independently transformed tobacco: 64 UBQ-LUC and 43 LUC plants were analyzed. LUC activity was determined by chemiluminescence assay using the activity of the purified protein as a standard.
From enzymatic and immunoblot analyses of a collection of transgenic plants harboring either the LUC or the UBQ-LUC vectors (43 and 64 independent transformants, respectively), we found that the addition of the UBQ moiety also increased the level of LUC protein (Fig. 4C). Using a chemiluminescence assay, an average of 2.2 times more active LUC protein accumulated (Table I). Furthermore, the percentage of the transgenic plants that had high levels of LUC protein (>160 pg mg−1 leaf protein) was 8-fold greater in the population expressing UBQ-LUC (Fig. 4C).
Expression of Compartmentalized Proteins
Although it is apparent that cytoplasmic proteins can be readily synthesized as UBQ fusions (Garbino et al., 1995; Worley et al., 1998; the present study), this approach may not be successful for many compartmentalized proteins in which the UBQ moiety could interfere with proper localization. This is especially true for chloroplastic and ER/secretory system-resident proteins because the UBQ moiety would immediately precede the requisite N-terminal transport sequences. The added UBQ moiety may be less intrusive for chloroplast proteins because their import occurs posttranslationally (Cline and Henry, 1996), thus allowing time for UBQ removal by Ubps. However, ER-directed protein is imported cotranslationally (Walter and Johnson, 1994). Johnson and Varshavsky (1995) showed previously that inserting the UBQ moiety following the signal sequence did not impair ER localization in yeast. However, if UBQ preceded the signal sequence, it was unclear whether removal of the UBQ moiety would be sufficiently rapid to allow import.
As an example of a chloroplast-localized protein, we tested spinach ACPII bearing its own TS (Schmid and Ohlrogge, 1990). ACP was modified at the C terminus to contain the 10-amino acid c-Myc epitope (Kolodziej and Young, 1991) to help discriminate the spinach protein from the tobacco ACPs (see Fig. 5A for a description). When TS-ACP was expressed in transgenic tobacco, the tagged 17.5-kD ACP protein was easily detected in crude leaf extracts (Fig. 5B). It was indistinguishable in size from mature ACP and was substantially smaller than ACP bearing the TS (both expressed in E. coli), indicating that proper removal of the TS had probably occurred in tobacco. We also detected a similarly sized protein of 17.5 kD in plants synthesizing ACP as a UBQ fusion (UBQ-TS-ACP) (Fig. 5B). The tobacco-expressed UBQ-TS-ACP was easily recognized by the c-Myc antibody, but unlike an E. coli-derived version, it was not detected with UBQ antibodies (data not shown). Both the antigenicity and the size of the product from the UBQ-TS-ACP vector indicated that the UBQ moiety and the TS had been removed in planta.
We addressed the proper localization of ACP from the analysis of chloroplasts isolated from young tobacco leaves. Following the isolation by Percoll gradient centrifugation, intact chloroplasts were lysed to release the stromal fraction. Enrichment of the stromal fraction and the lack of cytosolic contamination were confirmed by the presence of the stromal enzyme CH-42 subunit of the Mg2+ protoporphyrin chelatase (Guo et al., 1998) and the absence of the cytosolic protein UBC1 (Sullivan et al., 1994) in the final preparations, as determined by immunoblot analysis (Fig. 5C). As can be seen in Figure 5, B and C, the ACP protein expressed from either the TS-ACP or UBQ-TS-ACP vectors properly localized to chloroplasts. Using transgenic plants expressing near-equivalent levels of ACP, we found a similar level of enrichment of ACP protein in the chloroplast stromal fraction.
As an example of an ER-targeted protein, we used an AMY from B. licheniformis (Pen et al., 1992), which was directed to the ER and ultimately secreted to the apoplast by the N-terminal signal sequence from the soybean VSP (Mason et al., 1988) (see Fig. 6A for a description). Previous studies showed that the AMY protein expressed in tobacco by this strategy had an apparent molecular mass of 62 kD after removal of the signal sequence, and that this polypeptide had become glycosylated and exported to the apoplast as a mixture of approximately 70-kD enzymatically active proteins (Pen et al., 1992; D.E. Mathews, unpublished data). We confirmed these observations with the AMY vector described here. In total leaf homogenates from AMY plants, we detected immunologically a mixture of four to five approximately 70-kD proteins with molecular masses higher than the single 62-kD AMY protein expressed in E. coli without the VSP signal sequence (Fig. 6B). These species were present in the apoplastic fraction.
A similar processing and localization was observed for AMY expressed as a UBQ fusion. In total leaf homogenates from tobacco expressing UBQ-AMY, a comparable mixture of approximately 70-kD AMY proteins was detected that could be extracted with the apoplast fluid (Fig. 6B). In fact, when AMY and UBQ-AMY plants expressing equivalent levels of protein were compared, similar levels of AMY protein were present in the apoplast fraction. The only notable difference between the proteins expressed from the AMY and the UBQ-AMY vectors was an enrichment for the forms of higher molecular mass in the UBQ-AMY plants (Fig. 6B). These higher-mass forms could be created by various mechanisms, including differences in glycosylation, improper cleavage of the signal sequence, and/or retention of the N-terminal UBQ moiety. From immunoblot analyses with UBQ antibodies, we eliminated the possibility that much of the UBQ moiety remained attached. Whereas UBQ-AMY expressed in E. coli was easily detected, an equivalent amount of its counterpart collected from tobacco apoplastic fluid was not (Fig. 6B).
Expression of Noncleavable UBQ Fusions
Processing of UBQ fusions by Ubps is highly sensitive to the amino acid sequence immediately surrounding the cleavage site. In particular, substituting other amino acids for the C-terminal Gly of UBQ or introducing Pro as the first residue of the adjacent polypeptide can effectively inhibit processing (Baker, 1996; Varshavsky, 1997). The retained N-terminal UBQ moiety in turn can destabilize the fusion by serving as an acceptor site for linking the multiubiquitin chain, which then targets the proteins for breakdown by the 26S proteasome (see Fig. 1). In yeast this conjugation is accomplished by the UBQ-fusion degradation subpathway and probably uses the free ε-amino group in Lys-48 in the fused UBQ for binding additional UBQs (Johnson et al., 1995). By various combinations of UBQ mutations at the Ubp cleavage site and at Lys-48, noncleavable UBQ fusions can be created that either destabilize or stabilize proteins in yeast. A noncleavable fusion with wild-type UBQ (K-48) can be a target for further ubiquitination and thus may confer a shorter half-life, whereas one bearing a UBQ mutant containing R at position 48 (R-48) could be resistant to multiubiquitination and consequently may confer a longer half-life (Fig. 1). Worley et al. (1998) recently provided evidence that a similar affect may occur in plants. From studies in which LUC was transiently expressed in tobacco, they found that appending a truncated and theoretically noncleavable version of UBQ (residues 1–72) significantly reduced the levels of active LUC, suggesting that the half-life of the modified LUC was substantially reduced.
To further investigate the use of noncleavable UBQ fusions in plants, we generated two sequence variants of the UBQ-GUS vector (see Fig. 1) that substituted either A for the C-terminal G-76 of UBQ (UBQ[A]-GUS) or P for the N-terminal M-1 of GUS (UBQ-[P]GUS). These mutations were combined with either wild-type UBQ or the R mutant (UBQR-48). Both the UBQ(A)-GUS and UBQ-(P)GUS mutants were readily expressed in tobacco, producing protein that was enzymatically active and easily detected by GUS antibodies (Fig. 7A and data not shown). Consistent with the retention of the UBQ moiety, each had an apparent molecular mass of 80 kD, which was approximately 6 kD larger than that of GUS (Fig. 7A). The presence of the UBQ moiety was confirmed subsequently by the recognition of both proteins with UBQ antibodies (Fig. 7A). In a similar fashion, we expressed in tobacco a LUC mutant, bearing a P for M-1 substitution (UBQ-[P]LUC), and found that this variant also retained the UBQ moiety (data not shown).
From immunoblot analysis with UBQ antibodies, it became apparent that the retained UBQ moiety in UBQ(A)-GUS and UBQ-(P)GUS stimulated further ubiquitination of the fusion, provided wild-type UBQ (K-48) was used. When total tobacco leaf homogenates were subjected to immunoblot analysis with UBQ antibodies, we observed a heterogeneous smear of UBQ conjugates (Fig. 7B) that represented a wide range of polypeptides modified with multiple UBQs in vivo (van Nocker et al., 1993). In addition to these, the 80-kD, noncleaved UBQ fusion was evident in plants expressing UBQ(A)-GUS or UBQ-(P)GUS (Fig. 7B and data not shown). In fact, these species were the most abundant UBQ conjugates in the extracts. Above the 80-kD UBQ-GUS fusion, we detected additional UBQ-immunoreactive proteins in the UBQ(A)-GUS and UBQ-(P)GUS plants but not in plants expressing UBQR-48(A)-GUS or UBQR-48-(P)GUS. Their size increments were consistent with the posttranslational addition of one or more UBQs, suggesting that the UBQ(A)-GUS and UBQ-(P)GUS proteins became targets for further ubiquitination in a process that required K-48.
To confirm the identity of the UBQ conjugates, GUS protein was immunoprecipitated from the extracts with GUS antibodies and the immunoprecipitates were then subjected to immunoblot analysis with UBQ antibodies. As can be seen in Figure 7C, we detected a heterogeneous array of multiubiquitinated GUS proteins from the noncleavable UBQ-GUS proteins containing wild-type UBQ (i.e. K-48 (UBQ[A]-GUS or UBQ-[P]GUS). This array was substantially reduced in plants expressing the noncleavable fusions bearing the R-48 UBQ mutation (UBQR-48[A]-GUS or UBQR-48-[P]GUS). Given the possibility that multiubiquitination of UBQ-(P)GUS destabilizes the protein, we expected that the half-life of UBQ-(P)GUS would be substantially shorter. However, using pulse-chase analysis of leaf discs, we found that the half-lives of UBQ-(P)GUS and UBQR-48-(P)GUS, and GUS were indistinguishable (about 70 h [data not shown]).
DISCUSSION
In both yeast and E. coli, UBQ fusions offer a versatile method to manipulate protein expression (Baker, 1996; Varshavsky, 1997). By appropriate combinations of amino acids within or adjacent to the UBQ moiety, accumulation of protein can be dramatically enhanced or, alternatively, the stability of the protein can be substantially reduced (Fig. 1). Here we show that UBQ fusions may provide similar benefits in plants. We found using transgenic tobacco that: (a) protein expression as a UBQ fusion can augment protein accumulation; (b) the N-terminal UBQ moiety does not interfere with proper localization of chloroplast- or ER-targeted proteins; (c) through the use of wild-type or modified forms of UBQ, the UBQ moiety can be rapidly released or remain stably attached to the protein; and (d) that noncleavable variants of UBQ-protein fusions can become substrates for further ubiquitination. In yeast multiubiquitinated forms of these noncleavable fusions are rapidly degraded (Varshavksy, 1997). We observed in all cases that UBQ fusion expression did not detectably alter function of the gene or activity of the fused protein. In addition to GUS, LUC, ACP, and AMY, synthesis of active oat phytochrome A, B. thuringiensis δendotoxin, and Aequoria victoria green fluorescent protein was possible (data not shown).
For all proteins tested, we found that the UBQ moiety was rapidly removed from the initial translation product to release the fused protein in an unmodified form. In fact, this processing was so efficient that we were unable to detect the unprocessed form of any of the four proteins tested. This processing is not restricted to tobacco; in transgenic potato and rice, the initial translation product of UBQ-GUS was also rapidly cleaved, releasing quantitatively the 74-kD GUS protein (Garbino et al., 1995; P. Christou, D. Hondred, and R.D. Vierstra, unpublished data). It is remotely possible that internal initiation ignores the UBQ coding sequence in the fusion-vector mRNAs. However, results from noncleavable UBQ fusions bearing amino acid substitutions 76 or 77 codons downstream of the UBQ initiation codon strongly suggest that the UBQ sequence was translated and removed posttranslationally.
Although the plant enzyme(s) involved in UBQ processing have not yet been identified, the nature of the cleavage site (between G-76 of UBQ and M-1 of GUS) and the sensitivity of cleavage to amino acid substitutions at this site (G-76 to A or M-1 to P) are consistent with the action of Ubps (Varshavsky, 1997; Wilkinson, 1997). Plants contain a number of distinct Ubps that are possible candidates (16 Ubps identified so far in Arabidopsis), some of which can process UBQ fusions in vitro and/or in vivo (Sullivan et al., 1990; Chandler et al., 1997; N. Yan, T. Falbel, and R.D. Vierstra, unpublished data). In yeast, processing of UBQ fusions by Ubps appears to be cotranslational (Johnson and Varshavsky, 1995; Varshavsky, 1997). A similar timing is likely in plants given our observations that an N-terminal UBQ moiety does not block protein import into the ER. Such import would probably require removal of the UBQ moiety before docking of the signal sequence with the ER transport machinery.
As is observed in microorganisms, synthesis of proteins as UBQ fusions can significantly augment accumulation in plants. From analysis of a population of transgenic tobacco, we found that the presence of the N-terminal UBQ moiety increased the enzymatically detectable levels of GUS and LUC. A similar benefit was suggested by Garbino et al. (1995) from expression of a UBQ-GUS fusion in transgenic potato. However, because their GUS and UBQ-GUS vectors differed at the Kozak sequence and because their UBQ-GUS vector also contained an intron within the 5′-UTR, it was not possible to differentiate the effects caused by the UBQ coding region from those caused by the intron or sequence differences surrounding the translation initiation codon. The enhancements observed here are more modest than some of those reported using E. coli or yeast (Butt et al., 1989; Ecker et al., 1989; Baker, 1996). However, we should note that both the GUS and LUC proteins are easily expressed and stable in plants. As a result, greater benefits may be possible for proteins more recalcitrant to high-level expression. It remains to be determined whether the UBQ-fusion approach can increase expression of compartmentalized proteins. Although we showed that AMY and ACP can be synthesized as UBQ fusions and correctly localized, an insufficient number of transformants was available to compare expression levels with confidence. Preliminary studies on a small group of transgenic plants certainly indicated that the UBQ moiety was not detrimental to ACP and AMY accumulation (data not shown).
The mechanism(s) whereby an N-terminal UBQ moiety can augment protein accumulation remain unclear. One possibility is that the favorable codon bias of UBQ enhances translation of the appended coding region. However, this possibility is diminished by the fact that yeast UBQ works in E. coli despite substantial differences in codon bias (Butt et al., 1989). Our analysis of individual GUS and UBQ-GUS plants showed that the amount of GUS protein synthesized per mRNA was indistinguishable for the fusion and control vectors (J.M. Walker and R.D. Vierstra, unpublished data). Still, it remains possible that enhanced translation stabilizes the mRNA by association with polysomes, so that the levels of the protein and the mRNA rise concomitantly. Another possibility is that the UBQ moiety facilitates folding and/or stability of the nascent polypeptide or shields the protein from cotranslational degradation. This is supported by the observations that UBQ is an extremely stable and proteolytically resistant protein that folds rapidly (Briggs and Roder, 1992). Given the likelihood that the UBQ sequence is removed quickly, its benefits to protein accumulation would have to occur before translation is complete.
As shown previously in yeast, we were able to express noncleaved UBQ fusions simply by substituting single amino acids at the cleavage site. Changes in either residue 76 of UBQ or introduction of P as the first residue of the adjacent fusion were effective. Whereas most Ubp are unable to process UBQ fusions linked via a P residue, Gilchrist et al. (1997) recently reported on two from mammalian cells that are able to break this linkage. The stability of UBQ-P fusions in tobacco suggests that homologs of these Ubps are not present in plants.
The UBQ fusion vector offers two simple ways to confer a shorter half-life to transgenic proteins (Fig. 1). The use of cleavable UBQ fusions allows the production of proteins with N-terminal residues other than M, after processing of the fusions by Ubps (all residues are possible except P). By exposing specific “destabilizing” amino acids at the N terminus, the protein may be rapidly degraded by the N-end rule pathway, a subpathway within the UBQ system (Varshavsky, 1997). Although the N-end rule pathway is not yet fully described in plants, recent studies suggest that the same hierarchy of stabilizing and destabilizing amino acids exists (Potuschak et al., 1998; Worley et al., 1998).
Another way to confer a short half-life is to express the protein as a noncleavable fusion with wild-type UBQ. As shown in yeast, these fusions became targets for further ubiquitination through the K-48 residue, which enhanced their degradation by the UBQ pathway (Johnson et al., 1995). A similar situation may exist in plants as well but it appears to be target specific. Worley et al. (1998) reported that the addition of a noncleavable version of UBQ to LUC dramatically impaired LUC expression in tobacco, suggesting that the half-life of the protein was decreased. However, that study and the present study observed no such “destabilizing” effect for a similar fusion with GUS. Here we found that a noncleavable version of UBQ-GUS became extensively ubiquitinated in a reaction that required K-48. Taken together, the data suggest that the presence of the noncleavable UBQ and subsequent linkage of additional UBQs are insufficient for rapid turnover and that other properties (e.g. structure, conformational stability) of the protein may be involved as well.
Conversely, because noncleavable UBQ fusions bearing the R-48 mutation appeared to be immune to further ubiquitination, they may be stable in plants. If so, such fusions may provide a way to stabilize unstable proteins and could be especially useful for short peptides or protein subdomains that are typically degraded rapidly if expressed by themselves.
In conclusion, we found that expressing proteins as UBQ fusions offers a number of ways to manipulate protein expression in transgenic plants. Most important is the observation that synthesis of a protein as a UBQ fusion can augment accumulation. Conveniently, the UBQ moiety is subsequently removed so that only the unmodified protein accumulates in vivo. Of immediate interest is a new set of stronger GUS and LUC reporter vectors that should help in expression analysis of weak promoters (Gallagher, 1992). Given that the enhancement probably occurs posttranscriptionally, the UBQ-fusion strategy can be combined with enhancements in other aspects of transgene expression to elevate protein accumulation. Clearly, additional examples (especially of those proteins difficult to express) will be required to determine the extent to which these vectors will benefit protein production in plants.
ACKNOWLEDGMENTS
We thank Agracetus, Mycogen, E.I. DuPont Nemours, and Proctor & Gamble for supplying the oligonucleotides used to assemble the various constructions. We are grateful to Dr. Steve Kay for providing LUC antibodies; to Drs. David Russell and Mike Miller at Agracetus for the plasmid pCMC1100 (GUS); to Drs. Anthony Cashmore and William B. Terzaghi for plasmid pAB14016LBS (LUC); to Clara Kielkopf for technical assistance; and to Dr. Jane Wallent (University of Wisconsin Biotech Center) for help with protein sequence analysis.
Abbreviations:
- ACP
acyl carrier protein
- AMV
alfalfa mosaic virus
- AMY
α-amylase
- CaMV
cauliflower mosaic virus
- LUC
luciferase
- NOS
nopaline synthase
- TS
transit sequence
- Ubp
UBQ-specific protease
- UBQ
ubiquitin
- UTR
untranslated region VSP, vegetative storage protein
Footnotes
This work was supported the U.S. Department of Agriculture funded through the Consortium for Plant Biotechnology Research (grant no. 92-34190-6941) and the North Central Biotechnology Initiative (grant no. 94-34190-1204), Pioneer Hi-Bred International, Proctor & Gamble, ICI Seeds, Rhône Poulenc S.A., Agrigenetics, Dow Elanco, Northrup King, the Graduate School of the University of Wisconsin, and the Research Division of the University of Wisconsin, College of Agriculture and Life Sciences, Madison.
LITERATURE CITED
- Baker RT. Protein expression using ubiquitin fusion and cleavage. Curr Opin Biotechnol. 1996;7:541–546. doi: 10.1016/s0958-1669(96)80059-0. [DOI] [PubMed] [Google Scholar]
- Barton KA, Whiteley HR, Yang N-S. Bacillus thuringiensis delta-endotoxin expressed in transgenic Nicotiana tabacum provides resistance to Lepidopteran insects. Plant Physiol. 1987;85:1103–1109. doi: 10.1104/pp.85.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M, Barnes WM, Chilton M-D. Structure and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acid Res. 1983;11:369–385. doi: 10.1093/nar/11.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford HM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briggs MS, Roder H. Early hydrogen-bonding events in the folding reaction of ubiquitin. Proc Natl Acad Sci USA. 1992;89:2017–2021. doi: 10.1073/pnas.89.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt TR, Jannalagadda S, Monia B, Sternberg E, Marsh JA, Stadel JM, Ecker DJ, Crooke ST. Ubiquitin fusion augments the yield of cloned gene products in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:2540–2544. doi: 10.1073/pnas.86.8.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis JA, Carpenter TB, Sun CW, Vierstra RD. Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics. 1995;139:921–939. doi: 10.1093/genetics/139.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JS, McArdle B, Callis J. AtUBP3 and AtUBP4 are two closely related Arabidopsis thaliana ubiquitin-specific proteases present in the nucleus. Mol Gen Genet. 1997;255:302–310. doi: 10.1007/s004380050501. [DOI] [PubMed] [Google Scholar]
- Cherry JR, Hondred D, Walker JM, Keller JM, Hershey HP, Vierstra RD. Carboxy-terminal deletion analysis of oat phytochrome A reveals the presence of separate domains required for structure and biological activity. Plant Cell. 1993;5:565–575. doi: 10.1105/tpc.5.5.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Henry R. Import and routing of nucleus-encoded chloroplast proteins. Annu Rev Cell Dev Biol. 1996;12:1–26. doi: 10.1146/annurev.cellbio.12.1.1. [DOI] [PubMed] [Google Scholar]
- de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid RNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Ecker DJ, Stadel JM, Butt TR, Marsh JA, Monia BP, Powers DA, Gorman JA, Clark PE, Warren F, Shatzman A and others. Increasing gene expression in yeast by fusion to ubiquitin. J Biol Chem. 1989;264:7715–7719. [PubMed] [Google Scholar]
- Falbel TG, Staehelin LA. Characterization of a family of chlorophyll-deficient wheat (Triticum) and barley (Hordeum vulgare) mutants with defects in the magnesium-insertion step of chlorophyll biosynthesis. Plant Physiol. 1994;104:639–648. doi: 10.1104/pp.104.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR. Quantitation of GUS activity by fluorometry. In: Gallagher SR, editor. GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. Boston: Academic Press; 1992. pp. 47–59. [Google Scholar]
- Gallie DR. Posttranslational regulation of gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:77–105. [Google Scholar]
- Garbino JE, Oosumi T, Belknap WR. Isolation of a polyubiquitin promoter and its expression in transgenic potato plants. Plant Physiol. 1995;109:1371–1378. doi: 10.1104/pp.109.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke L, Auron PE, Quigley GJ, Rich A, Sonenberg N. 5′-Conformation of capped alfalfa mosaic virus ribonucleic acid 4 may reflect its independence of the cap structure or of cap-binding protein for efficient translation. Biochemistry. 1983;22:5157–5164. doi: 10.1021/bi00291a015. [DOI] [PubMed] [Google Scholar]
- Gilchrist CA, Gray DA, Baker RT. A ubiquitin-specific protease that efficiently cleaves the ubiquitin-proline bond. J Biol Chem. 1997;272:32280–32285. doi: 10.1074/jbc.272.51.32280. [DOI] [PubMed] [Google Scholar]
- Goddijn OJM, Pen J. Plants as bioreactors. Trends Biotechnol. 1995;13:379–387. [Google Scholar]
- Gould SJ, Keller GA, Schneider M, Howell SH, Garrard LJ, Goodman JM, Distel B, Tabak H, Subramani S. Peroxisomal protein import is conserved between yeast, plants, insects, and mammals. EMBO J. 1990;9:85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Luo M, Weinstein JD. Magnesium-chelatase from developing pea leaves. Plant Physiol. 1998;116:605–615. [Google Scholar]
- Haq TA, Mason HS, Clements JD, Arntzen CJ. Oral immunization with a recombinant bacterial antigen produced in transgenic plants. Science. 1995;268:714–719. doi: 10.1126/science.7732379. [DOI] [PubMed] [Google Scholar]
- Harris RG, Rowe JJM, Stewart PS, Williams DC. Affinity chromatography of β-glucuronidase. FEBS Lett. 1973;29:189–192. doi: 10.1016/0014-5793(73)80558-7. [DOI] [PubMed] [Google Scholar]
- Hondred D, Vierstra RD. Novel applications of the ubiquitin-dependent proteolytic pathway in plant genetic engineering. Curr Opin Biotechnol. 1992;3:147–151. doi: 10.1016/0958-1669(92)90144-8. [DOI] [PubMed] [Google Scholar]
- Jefferson R, Kavanagh T, Bevans M. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Johnson N, Varshavsky A. Ubiquitin-assisted dissection of protein transport across membranes. EMBO J. 1995;13:2686–2698. doi: 10.1002/j.1460-2075.1994.tb06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P, Young RA. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- LaValle ER, McCoy JM. Gene fusion expression systems in Escherichia coli. Curr Opin Biotechnol. 1995;6:501–506. doi: 10.1016/0958-1669(95)80083-2. [DOI] [PubMed] [Google Scholar]
- Martin T, Wohner R-V, Hummel S, Willmitzer L, Frommer WB. The GUS reporter system as a tool for the study of plant gene expression. In: Gallagher SR, editor. Gus Protocols: Using the GUS Gene as a Reporter of Gene Expression. San Diego, CA: Academic Press; 1992. pp. 23–43. [Google Scholar]
- Mason HS, Guerrero FD, Boyer JS, Mullet JE. Proteins homologous to leaf glycoproteins are abundant in stems of dark-grown soybean seedlings: analysis of proteins and cDNAs. Plant Mol Biol. 1988;11:845–856. doi: 10.1007/BF00019524. [DOI] [PubMed] [Google Scholar]
- McCabe DE, Swain WF, Martinell BJ, Christou P. Stable transformation of soybean (Glycine max) by particle acceleration. Biotechnology. 1988;6:923–926. [Google Scholar]
- Millar AJ, Short SR, Chua N-H, Kay S. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell. 1992;4:1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Caimi PG, Yadav NS, Mauvais CJ. Comparison of increased expression of wild-type and herbicide-resistant acetolactate synthase genes in transgenic plants, and implications of posttranscriptional limitations on enzyme activity. Plant Physiol. 1990;94:1647–1654. doi: 10.1104/pp.94.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua N-H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Ohtani T, Galili G, Wallace JC, Thompson GS, Larkins B. Normal and lysine-containing zeins are unstable in transgenic tobacco. Plant Mol Biol. 1991;16:117–128. doi: 10.1007/BF00017922. [DOI] [PubMed] [Google Scholar]
- Pen J, Molendijk L, Quax WJ, Sijmons PC, van Ooyen AJJ, van den Elzen PJM, Rietveld K, Hoekema A. Production of active Bacillus licheniformis alpha-amylase in tobacco and its application in starch liquefaction. Biotechnology. 1992;10:292–296. doi: 10.1038/nbt0392-292. [DOI] [PubMed] [Google Scholar]
- Potuschak T, Stary S, Schlögelhofer P, Becker F, Nejinskaia V, Bachmair A. PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc Natl Acad Sci USA. 1998;95:7904–7908. doi: 10.1073/pnas.95.14.7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KM, Ohlrogge JB. A root acyl carrier protein-II from spinach is also expressed in leaves and seeds. Plant Mol Biol. 1990;15:765–778. doi: 10.1007/BF00016126. [DOI] [PubMed] [Google Scholar]
- Shah DM. Genetic engineering for fungal and bacterial diseases. Curr Opin Biotechnol. 1997;8:208–214. doi: 10.1016/s0958-1669(97)80104-8. [DOI] [PubMed] [Google Scholar]
- Sullivan ML, Callis JA, Vierstra RD. HPLC resolution of ubiquitin pathway enzymes from wheat germ. Plant Physiol. 1990;94:710–716. doi: 10.1104/pp.94.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan ML, Carpenter TB, Vierstra RD. Wheat ubiquitin-conjugating enzymes, TaUBC1 and TaUBC4, are encoded by small multigene families in Arabidopsis thaliana. Plant Mol Biol. 1994;24:651–661. doi: 10.1007/BF00023561. [DOI] [PubMed] [Google Scholar]
- van Nocker S, Vierstra RD. Multiubiquitin chains linked through lysine-48 are abundant in vivo and competent intermediates in the ubiquitin-dependent proteolytic pathway. J Biol Chem. 1993;268:24766–24773. [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1997;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. Proteolysis in plants: mechanisms and functions. Plant Mol Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- Wadsworth GJ, Redinbaugh MG, Scandalios JG. A procedure for the small-scale isolation of plant RNA suitable for RNA blot analysis. Anal Biochem. 1988;172:279–283. doi: 10.1016/0003-2697(88)90443-5. [DOI] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Wilkinson K. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- Worley CK, Ling R, Callis J. Engineering in vivo instability of firefly luciferase and Escherichia coli β-glucuronidase in higher plants using recognition elements from the ubiquitin pathway. Plant Mol Biol. 1998;37:337–347. doi: 10.1023/a:1006089924093. [DOI] [PubMed] [Google Scholar]
- Yuan L, Knauf VC. Modification of plant components. Curr Opin Biotechnol. 1997;8:227–233. doi: 10.1016/s0958-1669(97)80107-3. [DOI] [PubMed] [Google Scholar]