Abstract
DNA microarrays were used to compare gene expression in dividing and nondividing (filamentous) cultures of Escherichia coli. Although cells from these cultures differed profoundly in morphology, their gene expression profiles were nearly identical. These results extend previous evidence that there is no division checkpoint in E. coli, and progression through the cell cycle is not regulated by the transcription of different genes during different parts of the cell cycle.
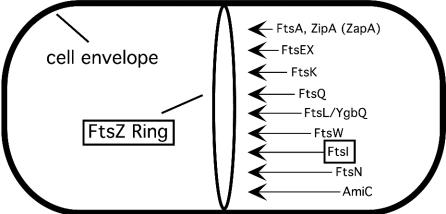
Cell division in Escherichia coli requires about a dozen proteins, all of which localize to the midcell, where they direct assembly of the division septum (Fig. 1) (for recent reviews, see references 12, 24, and 30). Most of these proteins are named Fts, for filamentation temperature sensitive. If any of these proteins is rendered nonfunctional, as occurs when an appropriate temperature-sensitive mutant is shifted to the nonpermissive temperature, the cells grow into long, aseptate filaments with regularly spaced nucleoids. The filaments ultimately lyse, so blocking cell division is lethal. Nevertheless, for several generations after the inhibition of cell division, the only obvious effects are morphological-growth rate (as assessed by mass increase), DNA replication, and chromosome segregation all appear to continue as if nothing has happened.
FIG. 1.
Recruitment of proteins to the septal ring of E. coli. Assembly of the septal ring starts with formation of the FtsZ ring at the midcell. The remaining proteins are then recruited in the order indicated, with AmiC being the last. Proteins targeted for inhibition in this study are boxed.
Studies of protein localization in filaments formed after the inactivation of the various division proteins indicate that these proteins localize to the midcell in a defined order (Fig. 1). The first is FtsZ, an abundant tubulin-like GTPase that forms a contractile ring at the inner face of the cytoplasmic membrane. FtsA, ZipA, and ZapA (14) bind directly to the Z ring and probably localize as the ring is assembling. Subsequently, FtsEX (31a), FtsK, FtsQ, FtsL/YbgQ (a probable heterodimer), FtsW, FtsI, FtsN, and AmiC (3) join the septal ring in that order.
The objectives of the studies described in this report were (i) to determine whether E. coli can sense a failure to divide and respond to the problem by changing gene expression, (ii) to identify new division genes, and (iii) to identify cell cycle-regulated genes, if any. We approached these objectives by blocking cell division at an early (FtsZ) and a late (FtsI) step in septal ring assembly. DNA microarrays were then used to obtain transcriptional profiles of the resulting nondividing (filamentous) populations and of control populations that were dividing normally. Interestingly, only a few changes in gene expression were observed. These changes were small and affected characterized genes, none of which are related to cell division. These findings argue against the existence of a division checkpoint or the cell cycle regulation of gene expression in E. coli.
Transcriptional response to inhibition of septal ring constriction.
In our initial experiments, we blocked cell division by inhibiting FtsI (also called penicillin-binding protein 3, or PBP3), a transpeptidase required for cross-linking of the peptidoglycan cell wall during division (see reference 1 and references therein). We inhibited FtsI by treating growing cells with aztreonam, an FtsI-selective β-lactam antibiotic. Aztreonam forms a covalent adduct with a serine residue in the transpeptidase catalytic site and thus prevents synthesis of septal peptidoglycan (20, 28). Aztreonam does not prevent localization of FtsI to the septal ring (37), nor does it prevent subsequent recruitment of FtsN (M. Wissel and D. Weiss, unpublished data). Thus, so far as is known, filamentous cells obtained by aztreonam treatment contain assembled septal rings that are unable to constrict, owing to the inactivation of FtsI. Whether AmiC localizes under these conditions is not known and probably does not matter in the context of our experiment because AmiC is not needed for constriction. Rather, AmiC is a peptidoglycan hydrolase needed for the separation of daughter cells after the division septum has formed (16).
The procedure for obtaining and analyzing “aztreonam filaments” (i.e., filamentous cells obtained by aztreonam treatment) was as follows. Our laboratory strain of E. coli MG1655, called strain EC251, was inoculated into Luria-Bertani medium and grown for about 6 h. This culture was then diluted serially into culture tubes containing 5 ml of N−C− minimal medium (21) with 0.4% glucose as a carbon source and 10 mM NH4Cl as a nitrogen source and grown overnight at 37°C. The next morning, a culture in exponential growth (optical density at 600 nm [OD600], ∼0.5) was used to inoculate two 250-ml baffle flasks, each containing 30 ml of prewarmed N−C− medium, to an initial OD600 of 0.02. Cultures were grown at 37°C and 210 rpm to an OD600 of 0.13, at which time aztreonam was added to one of the flasks to a final concentration of 1 μg/ml. To maintain the cells in exponential growth, cultures were diluted 1:6 into 60 ml of prewarmed medium when the OD600 reached 0.4, and growth was continued until the OD600 returned to 0.3. This density level corresponds to three to four mass doublings after the addition of the aztreonam and was as long as we could grow cells without observing lysis, which occurred after four to five mass doublings in the aztreonam-treated culture (Fig. 2A). The inhibition of cell division had no effect on the growth rate during the course of the experiment (doubling time, approximately 60 min), but it had a profound effect on cell morphology. The length of the aztreonam-treated cells was 43 ± 4 μm (mean ± standard deviation; n = 145), compared to 2.7 ± 0.1 μm (n = 150) for cells from the control culture.
FIG. 2.
Effect of aztreonam (A) and sulA induction (B) on growth and division. Insets show phase contrast micrographs of untreated and treated cells. Cultures were maintained in exponential growth by dilution at the time indicated. OD600 values were adjusted to account for this dilution and are therefore plotted as a continuous growth curve.
Cells were harvested and RNA was extracted as described previously (42). Transcription profiles were obtained by using the GeneChip E. coli Antisense Genome Array (Affymetrix, Santa Clara, Calif.). The synthesis of cDNA, fragmentation, labeling, and hybridization were done according to the GeneChip expression analysis protocol from Affymetrix, except that the amount of input RNA was increased to 15 μg. Hybridizations were performed overnight at 45°C with an Affymetrix fluidic station. Arrays were scanned on a GeneArray Scanner (Affymetrix) at 570 nm and a resolution of 3 μm.
We performed three biological replicates of this experiment. Comprehensive transcript profiles are available at http://www.medicine.uiowa.edu/WeissLab. We used software from Affymetrix (Microarray Suite 5.0) to scale and normalize the signal intensities and to calculate a signal log ratio for each gene. This ratio is the increase (or decrease) in amount of transcript relative to a baseline value, expressed as the log2 ratio. The baseline value in this case is the signal intensity for each gene in untreated cells. The software also calculates change calls and change P values (statistical significance for change calls). Change calls indicate whether a gene exhibits increased, decreased, or unchanged expression. They are based on statistical criteria (17, 22). We used default parameters for these calculations. To keep the list of genes whose expression responds to blocking division with aztreonam from being artificially long, we considered only genes that had a statistically significant change call in at least two of the three replicates. These genes were then subjected to a Student's t test (Affymetrix Data Mining Tools, version 3.0; default parameters) to identify those with a statistically significant difference (P ≤ 0.05) for dividing versus filamentous cells over all the three replicates. Table 1 lists the genes that passed these statistical tests and had an average log2 ratio greater than 1.32 or less than −1.32 (i.e., ≥ 2.5-fold change). Genes that passed the statistical tests but whose change in expression was <2.5-fold are listed in the supplemental table at http://www.medicine.uiowa.edu/WeissLab.
TABLE 1.
Genes induced or repressed in filamentous cells
| b no. | Gene | Descriptiona | Log2 ratiob |
|---|---|---|---|
| Induced upon inhibition of FtsI function with aztreonam: b2005 | wcaE | Colanic acid biosynthesis | 3.43 ± 2.02 |
| Repressed upon inhibition of FtsI function with aztreonam | |||
| b3517 | gadAc | Glutamate decarboxylase isozyme | −2.47 ± 0.29 |
| b1493 | gadBc | Glutamate decarboxylase isozyme | −2.13 ± 0.23 |
| b3512 | yhiE | Transcriptional activator | −2.13 ± 0.64 |
| b3508 | yhid | Putative transport ATPase | −1.43 ± 0.40 |
| Induced upon inhibition of FtsZ function via expression of sulA: b3561 | yiaH | Putative membrane protein | 1.77 ± 0.81 |
| Repressed upon inhibition of FtsZ function via expression of sulA | |||
| b3512 | yhiE | Transcriptional activator | −2.23 ± 0.81 |
| b3511 | hdeD | Putative membrane protein | −2.23 ± 0.38 |
| b3506 | slp | Outer membrane protein | −1.87 ± 0.49 |
| b3508 | yhiD | Putative transport ATPase | −1.50 ± 0.66 |
Gene descriptions are from the Affymetrix sequence information database and were updated with information from the Colibri and EcoGene databases (as of 15 August 2003).
Mean ± standard deviation for three independent experiments.
Although gadA and gadB are 97% identical at the DNA level, Affymetrix asserts that their probe pairs distinguish between these genes.
Only one gene was induced when cell division was blocked with aztreonam: the gene wcaE was induced about 10-fold. This gene is involved in capsule synthesis and resides in an operon with several other capsule synthesis genes. Inspection of the primary data revealed that expression of the other genes in this operon also increased, although none achieved the 2.5-fold cutoff, implying that the induction observed for wcaE is not an artifact. Because capsule synthesis is not required for cell division, we doubt that induction of the wca operon is a response to the failure to divide per se. Rather, capsule synthesis is induced in response to desiccation, osmotic shock, and a variety of lesions in the cell envelope (8, 9, 32). Since the aztreonam filaments started to lyse shortly after they were harvested (Fig. 2A), it seems likely that they already had envelope defects at the time of harvest.
Four genes were repressed by aztreonam treatment: gadA, gadB, yhiE, and yhiD. All of these genes are associated with the acid response (36). The gadA and gadB genes encode glutamate decarboxylases (33), yhiE encodes a transcriptional activator that responds to low pH (26, 36), and yhiD encodes a membrane protein of unknown function whose expression is activated by yhiE. Inspection of the primary data revealed that numerous other acid response genes were also down-regulated but not sufficiently or consistently enough to satisfy all the criteria for inclusion in Table 1. Why acid response genes were slightly repressed in filamentous cells is a matter of conjecture. We doubt that it has anything to do with pH, since the medium was pH 7.1 at the time of harvest with and without aztreonam treatment. A direct involvement in cell division seems unlikely because null mutants have been characterized for each of these genes (7, 25, 36), and none has been reported to be essential for viability or to have a division phenotype. Curiously, changes in expression (mostly induction) for these genes have been observed in a bewildering array of transcriptional profiling experiments, including studies of the responses to pH, acetate, oxidative stress, nitrogen starvation, an antibiotic, trimethylamine N-oxide, and growth as a biofilm (2, 4, 25, 26, 29, 39, 42). Moreover, the acid response genes reside at the bottom of a complex regulatory hierarchy that places them under the direct or indirect control of HN-S, CAP, RpoS, EvgA, YdeO, YhiE, and GadX (6, 26). We infer that expression of the acid response genes is exquisitely sensitive to changes in cell physiology, but these genes are not involved in cell division.
Transcriptional response to inhibition of septal ring assembly.
The first step in assembly of the septal ring is formation of the FtsZ ring at the midcell. FtsZ ring assembly is highly regulated. The MinCDE proteins along with nucleoid occlusion direct the FtsZ ring to the midcell and control the timing of FtsZ ring assembly (recently reviewed in reference 12). The SulA protein also regulates FtsZ ring assembly. Ordinarily, sulA is under lexA control and is induced in response to DNA damage as part of the SOS response (18, 19). SulA binds to FtsZ and prevents FtsZ ring assembly (10, 27, 35), thus stalling division until DNA has been repaired. We used induction of sulA to study the transcriptional response to an early defect in assembly of the septal ring. To avoid complications associated with the SOS response, we placed sulA under the control of an arabinose-inducible promoter, PBAD (15). This procedure was accomplished in the following steps. First, we deleted the araBAD operon from strain EC251 so that arabinose would behave as a gratuitous inducer rather than as a carbon and energy source. The araBAD deletion was constructed by allele replacement with a Kanr cassette from plasmid pKD11 (11) that had been amplified by PCR with primers P498 (5′-ATTGGCTGTGGTTTTATACAGTCATTACTGCCCGTAATATGCCTTCGCGtgtgtaggctggagctgcttc-3′) and P499 (5′-TACCCGTTTTTTTGGATGGAGTGAAACGATGGCGATTGCAATTGGCCTCGCATATGAATATCCTCCTTAG). In the primer sequences, uppercase letters denote homology to araB and araD, respectively. Kanr derivatives of MG1655 were confirmed by PCR. The Kanr cassette was then excised as previously described (11) to create a markerless deletion. An isolate was designated strain EC1097, which was confirmed by PCR and by a failure to grow on minimal medium with arabinose as a carbon and energy source. Separately, sulA was cloned into plasmid pBAD18-Kan (15) and integrated into the chromosome of strain EC251 at the λ att site by λInCh (5). This arabinose-inducible sulA allele was moved into EC1097 by P1-mediated transduction to create strain EC1098.
To obtain SulA filaments for DNA microarray studies, strain EC1098 was grown in parallel in two flasks in N−C− minimal medium as described above except that the carbon source was 0.4% glycerol. Expression of sulA was induced by the addition of arabinose (0.2% final concentration), and growth was continued for three to four mass doublings (Fig. 2B), at which time the cells were harvested and processed for DNA microarray analysis. Induction of sulA had no discernible effect on the growth rate (doubling time, approximately 90 min), but at the time of harvest, the arabinose-treated cells were 40 ± 6 μm long (n = 152), while the untreated cells were 2.7 ± 0.1 μm long (n = 186).
Blocking cell division by using arabinose to induce sulA appeared to result in the induction of about 15 genes, most of which clearly have nothing to do with cell division. For instance, the most highly induced genes are known to be involved in arabinose metabolism, as follows (fold inductions in parentheses): araE (60-fold), araF (50-fold), araH1 (27-fold), and araH2 (25-fold). In addition, sulA was induced 30-fold. These inductions make sense and validate our experimental procedures. Some of the induced genes, such as ylcC (now cusC), which was induced about 13-fold and is a component of a copper and silver ion efflux system (13), could not be dismissed so easily. To distinguish between genes that responded to arabinose (or contaminants in the arabinose) and those that responded to the division block imposed by SulA, we performed an arabinose-induction experiment with strain EC1097. This strain is the parent of EC1098; it has the same araBAD deletion but not a chromosomal copy of sulA under PBAD control. Genes that responded to SulA were defined by the same criteria used for the aztreonam-responsive genes above and, in addition, had to show no response to arabinose alone (i.e., no change in EC1097). Table 1 summarizes these genes. Genes that did not pass all of the tests, including genes that appeared to be induced or repressed by arabinose, are listed in the supplemental table (http://www.medicine.uiowa.edu/WeissLab).
Only one gene was induced upon inhibition of FtsZ ring assembly by SulA: yiaH was induced threefold (Table 1). Even this induction is somewhat suspect because yiaH was induced 1.6-fold in strain EC1097. The yiaH gene appears to encode a 331-amino-acid membrane protein of unknown function, although amino-acids 1 to 167 of YiaH show 26% identity and 48% similarity to the VanT amino acid racemase from several Enterococcus species.
Only four genes appeared to be specifically repressed by SulA: yhiE, hdeD, slp, and yhiD. All of these genes are associated with the acid response, and two made the list of aztreonam-repressed genes, yhiE and yhiD. Many other acid response genes were also repressed but failed to meet all the criteria for inclusion in Table 1. Several of these genes (e.g., gadB, xasA, and hdeA) appeared to respond to arabinose rather than SulA, because they were repressed more than twofold in strain EC1097. Null mutants of yhiE, hdeD, slp, and yhiD have been studied in two laboratories, neither of which reported a division phenotype (25, 36). As discussed above in connection with the aztreonam experiment, repression of the acid response genes more likely reflects their extreme sensitivity to physiological perturbations than their having a heretofore-unappreciated role in cell division.
Conclusions.
We have used DNA microarrays to compare the gene expression profiles of dividing and nondividing (filamentous) E. coli cells. Despite a profound difference in morphology (∼3 versus ∼40 μm long), gene expression was nearly identical in these two cell types; only eight genes exhibited changes that were ≥2.5-fold and that met statistical criteria for reproducibility. We doubt that any of these genes are truly connected to cell division. The reasons differ from gene to gene and are discussed in detail above but reflect the following considerations: mutants are available and do not have a division phenotype (wcaE and the acid response genes), the change in expression was seen with only one method of inhibiting division (wcaE and yiaH), plausible alternative explanations can account for the change in expression (wcaE and the acid response genes), and similar changes were observed with a treatment that did not inhibit division (arabinose induction of yiaH and repression of several acid response genes in strain EC1097). We did not attempt to confirm any of the observed changes by an independent method such as real-time PCR, because none of the changes in expression are likely to be relevant to cell division.
What is the likelihood that we have overlooked important changes in gene expression? In this context it is worth noting that we readily detected the induction of genes in the araC regulon when we used arabinose induction of sulA to prevent cell division. We also consistently detected all of the known division genes in these experiments, even though their expression did not change when cell division was inhibited. All of the experiments were also performed with glass slide microarrays (42), with essentially the same results, although the fold induction obtained from glass slides was about half that observed with the Affymetrix chip. The Affymetrix software consistently scored about 25% of the genes on the DNA chip as not present, meaning the fluorescence signal was not above background. Presumably, these genes were not expressed in either dividing or filamentous cells and, thus, had no change in expression, but we cannot exclude the possibility of changes from one low level of expression to another. Finally, changes in protein levels due to translational regulation or differential proteolysis would not have been detected by DNA microarrays.
It has long been apparent that E. coli does not have a division checkpoint that prevents the initiation of a new round of chromosome replication until cytokinesis has occurred. Indeed, the absence of such a checkpoint explains why inhibiting division leads to the formation of long filaments with regularly spaced nucleoids. Our study extends these observations by showing that such filamentous cells fail to mount a subtler response when division is blocked; E. coli neither elevates the expression of genes that are needed for cell division nor lowers the expression of genes needed for a subsequent cell cycle event.
Our results also imply that no E. coli genes are expressed in a division cycle-dependent manner. Otherwise, blocking cell division should have trapped the population in a physiological state where these genes were over- or underexpressed relative to a population of cells that are cycling normally. Because the cells employed in our studies were not synchronized for DNA replication, the results say nothing about whether any genes are transcribed periodically with respect to chromosome replication. Nevertheless, our findings are consistent with previous evidence that orderly progression through the cell cycle appears to be achieved primarily by governing the activity rather than the abundance of the relevant proteins (23, 34, 38, 40, 41). In this regard, cell cycle regulation is strikingly different in E. coli than in model eukaryotes and bacteria such as Caulobacter crescentus (31), which not only regulate the activity of key proteins but also have cell cycle-regulated gene expression and checkpoint controls. These differences enable E. coli to engage in multiple rounds of DNA replication simultaneously in the same cell, which in turn confers a competitive advantage by allowing for a faster growth rate.
Acknowledgments
We thank Sydney Kustu for introducing us to microarray methods and the DNA core facility at the University of Iowa for help with analysis of the Affymetrix chips.
This work was supported by a grant from the National Institutes of Health (GM59893). S.J.R.A. was supported by a National Science Foundation Research Training Grant (DBI9602247) and by an NIH Training Grant in Biotechnology (T32 GM08365-13).
REFERENCES
- 1.Adam, M., C. Fraipont, N. Rhazi, M. Nguyen-Distèche, B. Lakaye, J. M. Frere, B. Devreese, J. Van Beeumen, Y. van Heijenoort, J. van Heijenoort, and J. M. Ghuysen. 1997. The bimodular G57-V577 polypeptide chain of the class B penicillin-binding protein 3 of Escherichia coli catalyzes peptide bond formation from thiolesters and does not catalyze glycan chain polymerization from the lipid II intermediate. J. Bacteriol. 179:6005-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhardt, T. G., and P. A. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordi, C., L. Theraulaz, V. Mejean, and C. Jourlin-Castelli. 2003. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol. Microbiol. 48:211-223. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castanie-Cornet, M. P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 7.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, M. H., S. Takeda, H. Yamada, Y. Ishii, T. Yamashino, and T. Mizuno. 2001. Characterization of the RcsC→YojN→RcsB phosphorelay signaling pathway involved in capsular synthesis in Escherichia coli. Biosci. Biotechnol. Biochem. 65:2364-2367. [DOI] [PubMed] [Google Scholar]
- 9.Clavel, T., J. C. Lazzaroni, A. Vianney, and R. Portalier. 1996. Expression of the tolQRA genes of Escherichia coli K-12 is controlled by the RcsC sensor protein involved in capsule synthesis. Mol. Microbiol. 19:19-25. [DOI] [PubMed] [Google Scholar]
- 10.Cordell, S. C., E. J. Robinson, and J. Löwe. 2003. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc. Natl. Acad. Sci. USA 100:7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke, S., G. Grass, C. Rensing, and D. H. Nies. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidrich, C., M. F. Templin, A. Ursinus, M. Merdanovic, J. Berger, H. Schwarz, M. A. de Pedro, and J. V. Höltje. 2001. Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167-178. [DOI] [PubMed] [Google Scholar]
- 17.Hubbell, E., W. M. Liu, and R. Mei. 2002. Robust estimators for expression analysis. Bioinformatics 18:1585-1592. [DOI] [PubMed] [Google Scholar]
- 18.Huisman, O., and R. D'Ari. 1981. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature 290:797-799. [DOI] [PubMed] [Google Scholar]
- 19.Huisman, O., R. D'Ari, and S. Gottesman. 1984. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc. Natl. Acad. Sci. USA 81:4490-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keck, W., B. Glauner, U. Schwarz, J. K. Broome-Smith, and B. G. Spratt. 1985. Sequences of the active-site peptides of three of the high-Mr penicillin-binding proteins of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 82:1999-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kustu, S. G., N. C. McFarland, S. P. Hui, B. Esmon, and G. F. Ames. 1979. Nitrogen control of Salmonella typhimurium: co-regulation of synthesis of glutamine synthetase and amino acid transport systems. J. Bacteriol. 138:218-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, W. M., R. Mei, X. Di, T. B. Ryder, E. Hubbell, S. Dee, T. A. Webster, C. A. Harrington, M. H. Ho, J. Baid, and S. P. Smeekens. 2002. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics 18:1593-1599. [DOI] [PubMed] [Google Scholar]
- 23.Lutkenhaus, J. F., B. A. Moore, M. Masters, and W. D. Donachie. 1979. Individual proteins are synthesized continuously throughout the Escherichia coli cell cycle. J. Bacteriol. 138:352-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 25.Masuda, N., and G. M. Church. 2002. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 184:6225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee, A., C. Cao, and J. Lutkenhaus. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholas, R. A., J. L. Strominger, H. Suzuki, and Y. Hirota. 1985. Identification of the active site in penicillin-binding protein 3 of Escherichia coli. J. Bacteriol. 164:456-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phadtare, S., I. Kato, and M. Inouye. 2002. DNA microarray analysis of the expression profile of Escherichia coli in response to treatment with 4,5-dihydroxy-2-cyclopenten-1-one. J. Bacteriol. 184:6725-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothfield, L., S. Justice, and J. Garcia-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 31.Ryan, K. R., and L. Shapiro. 2003. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 19:19. [DOI] [PubMed] [Google Scholar]
- 31a.Schmidt, K. L., N. D. Peterson, R. J. Kustusch, M. C. Wissel, B. Graham, G. J. Phillips, and D. S. Weiss. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186:785-793. [DOI] [PMC free article] [PubMed]
- 32.Sledjeski, D. D., and S. Gottesman. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, D. K., T. Kassam, B. Singh, and J. F. Elliott. 1992. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol. 174:5820-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Theisen, P. W., J. E. Grimwade, A. C. Leonard, J. A. Bogan, and C. E. Helmstetter. 1993. Correlation of gene transcription with the time of initiation of chromosome replication in Escherichia coli. Mol. Microbiol. 10:575-584. [DOI] [PubMed] [Google Scholar]
- 35.Trusca, D., S. Scott, C. Thompson, and D. Bramhill. 1998. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J. Bacteriol. 180:3946-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 184:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wientjes, F. B., T. J. Olijhoek, U. Schwarz, and N. Nanninga. 1983. Labeling pattern of major penicillin-binding proteins of Escherichia coli during the division cycle. J. Bacteriol. 153:1287-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, P., J. A. Bogan, K. Welch, S. R. Pickett, H. J. Wang, A. Zaritsky, and C. E. Helmstetter. 1997. Gene transcription and chromosome replication in Escherichia coli. J. Bacteriol. 179:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, P., and C. E. Helmstetter. 1994. Relationship between ftsZ gene expression and chromosome replication in Escherichia coli. J. Bacteriol. 176:6100-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]