Abstract
Histone deacetylase (HDAC) inhibitors are a class of promising anticancer reagents. They are able to induce apoptosis in embryonic carcinoma (EC) cells. However, the underlying mechanism remains poorly understood. Here we show that increased expression of zinc-finger protein regulator of apoptosis and cell-cycle arrest (Zac1) is implicated in HDAC inhibitor-induced apoptosis in F9 and P19 EC cells. By chromatin immunoprecipitation analysis we identified that increased Zac1 expression is mediated by histone acetylation of the Zac1 promoter region. Knockdown of Zac1 inhibited HDAC inhibitor-induced cell apoptosis. Moreover, HDAC inhibitors repressed nuclear factor-κB (NF-κB) activity, and this effect is abrogated by Zac1 knockdown. Consistently, Zac1 overexpression suppressed cellular NF-κB activity. Further investigation showed that Zac1 inhibits NF-κB activity by interacting with the C-terminus of the p65 subunit, which suppresses the phosphorylation of p65 at Ser468 and Ser536 residues. These results indicate that Zac1 is a histone acetylation-regulated suppressor of NF-κB, which is induced and implicated in HDAC inhibitor-mediated EC cell apoptosis.
Keywords: Zac1, NF-κB, apoptosis, histone acetylation, epigenetic regulation
In multi-cellular organisms, apoptotic cell death is a conserved self-destruct and disposal mechanism evolved to eliminate useless or potentially harmful cells. The balance between cell death and survival is regulated by various proapoptotic stimuli1, 2 and survival signals. Nuclear factor-κB (NF-κB) is a family of widely expressed transcription factors composed of a group of evolutionally conserved and structurally related homo- or heterodimeric DNA-binding proteins. It is an important survival signal, which is frequently observed to be constitutively activated in tumor cells.3 The p50/p65 heterodimer is the most abundant form of the NF-κB family. In normal cells, its activity is tightly controlled by an array of highly orchestrated events, including subcellular translocation and post-translational modifications.4 It has been known that NF-κB increases cell viability through activation of the expression of a number of antiapoptotic genes.5
Histone acetylation is one of the most important epigenetic mechanisms controlling gene expression. Increased histone acetylation is generally associated with a higher gene transcription activity. Aberrant histone acetylation has been found to be implicated in cancer development.6, 7 The cellular histone acetylation status is tightly regulated by the reciprocal activities of acetyltransferases and histone deacetylases (HDACs). Abnormal HDAC activity or overexpression has been observed in a variety of cancers.8 HDAC inhibitors include a series of structurally unrelated compounds that can induce apoptosis in tumor cells from diverse origins. Among them, SAHA has been approved for the treatment of cutaneous T-cell lymphoma, and some other HDAC inhibitors are undergoing phase-I or -II clinical trials in malignant diseases.9 The molecular mechanisms underlying HDAC inhibitor-induced apoptosis of cancer cells are still not well understood. It has been reported that HDAC inhibitor-induced apoptosis is accompanied with the repression of NF-κB activity.10, 11 These evidences suggest that suppression of NF-κB activity is a critical event in the HDAC inhibitor-induced apoptosis of cancer cells. However, how HDAC inhibitors suppress cellular NF-κB activity remains unclear.
Teratocarcinoma is a cancer that mainly affects young adults. It originates from abnormal embryonic cells or transformed totipotent germinal stem cells. Teratocarcinoma is a heterogeneous tissue composed of differentiated teratomas and highly aggressive embryonic carcinoma (EC) cells that account for the cancer metastasis or relapse after surgery remove. A recent study showed that treatment of EC cells with the HDAC inhibitor apicidin induced apoptosis,12 suggesting that HDAC inhibitors can be potentially used in the treatment of teratocarcinoma. Thus, investigation of the underlying mechanism through which HDAC inhibitors induce the apoptosis of EC cells can be important for developing the mechanism-based therapeutic strategy.
Zac1 was originally indentified as a zinc-finger protein that induces cell apoptosis and cell-cycle arrest.13 Zac1 functions as a transcription factor that binds to a specific DNA sequence14 or as a transcription cofactor for certain nuclear receptors.15 Zac1 is also a transcriptional coactivator that can enhance p53 transcriptional activation.16 Although a crosstalk between the p53 and NF-κB pathways has been reported in recent years,17 the role of Zac1 in NF-κB signaling pathways remains unidentified.
Epigenetic alterations, such as unusual DNA methylation and chromatin histone acetylation, have important roles in the transcriptional silencing of certain antiproliferative genes and in promoting tumorigenesis.18 It has been reported that Zac1 expression is silenced through DNA methylation in cancer cells.19 However, whether histone acetylation is involved in regulating Zac1 gene expression needs to be established.
In this study we aimed to investigate the mechanism of HDAC inhibitor-induced apoptosis in F9 and P19 EC cells. We observed that induction of Zac1 by HDAC inhibitors is mediated by histone acetylation of the Zac1 promoter region and associated with the suppression of NF-κB activity. Further experiments showed that Zac1 mediates the suppression of NF-κB activity in HDAC inhibitor-induced apoptosis. In addition, the interaction between Zac1 and the C-terminal region of the NF-κB p65 subunit was also characterized. To explore how Zac1 suppresses NF-κB activity, we examined the subcellular distribution of p65 and its post-translational modifications in response to HDAC inhibitors. We found that phosphorylation at Ser468 and Ser536 of p65 is strongly regulated by HDAC inhibitors. This effect is abrogated by Zac1 knockdown and Zac1 overexpression suppresses p65 phosphorylation at these two sites. Moreover, we found that binding of Zac1 to the NF-κB p65 subunit is required for Zac1-mediated p65 phosphorylation inhibition and suppression of NF-κB activity. These data provide a novel understanding on the mechanism underlying the suppression of NF-κB activity in HDAC inhibitor-induced apoptosis.
Results
HDAC inhibitors activate Zac1 transcription by increasing its promoter histone acetylation
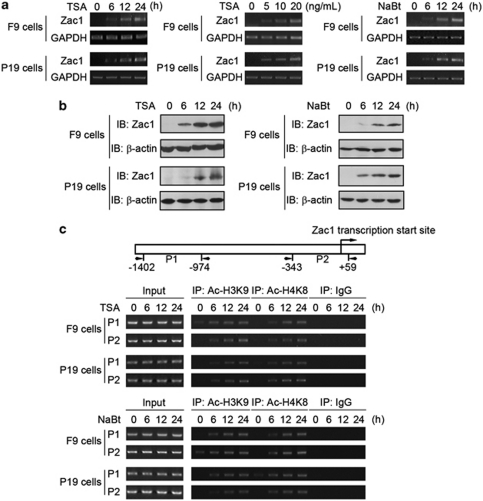
F9 and P19 are EC cells established from testis and embryo tissue, respectively. In response to treatment with the HDAC inhibitors trichostatin-A (TSA) and sodium butyrate (NaBt), the cells underwent apoptosis dose dependently, as shown by both flow cytometry analysis (Supplementary Figure S1a) and caspase-3 activity assay (Supplementary Figure S1b). It has been shown recently that HDAC inhibitor-induced cancer cell apoptosis is initiated primarily in a transcription-dependent manner.20, 21 To explore the underlying mechanisms, we examined the transcription levels of a series of genes involved in the regulation of apoptosis before and after HDAC inhibitor treatment. Among these genes, transcription of Zac1 was markedly increased by TSA (Figure 1a, left and middle panels). Similarly, increased Zac1 mRNA levels were also induced by NaBt (Figure 1a, right panel). Western blot analysis further verified the upregulation of Zac1 by TSA (Figure 1b, left panel) and NaBt (Figure 1b, right panel). To determine whether HDAC inhibitors induce Zac1 transcription by increasing chromatin acetylation in the Zac1 promoter, we performed chromatin immunoprecipitation (ChIP) assays. HDAC inhibitors treatment time dependently increased acetylated histone H3 (Lys9) and histone H4 (Lys8) in the Zac1 promoter in both F9 and P19 cells (Figure 1c). The above data indicate that HDAC inhibitor treatment upregulates histone acetylation in the Zac1 promoter region and activates the transcription of Zac1.
Figure 1.
HDAC inhibitors induce Zac1 transcription by increasing histone acetylation in its promoter. (a) F9 and P19 cells were treated with TSA and NaBt for the indicated time periods and concentrations. Left: Time-dependent effect of TSA (20 ng/ml); middle: dose–effect of TSA (24 h); right: effect of NaBt (1.5 mM). Zac1 mRNA levels were determined by RT-PCR. GAPDH was used as loading control. (b) Cells were treated with 20 ng/ml TSA (left) or 1.5 mM NaBt (right) for the indicated time periods. Zac1 protein levels were examined by western blotting. (c) Upper: A schematic illustration of a Zac1 gene genomic DNA fragment showing the relative location of the transcription start site and the two amplicons corresponding to two pairs of primers indicated by P1 and P2 used in the ChIP assay. Middle and lower: Cells were treated with 20 ng/ml TSA (middle) or 1.5 mM NaBt (lower) for the indicated time period. ChIP assay was performed using antibodies against the indicated acetylated histones. Rabbit IgG was used as negative control. The precipitated DNA was analyzed by semi-quantitative PCR
Zac1 is involved in HDAC inhibitor-induced EC cell apoptosis
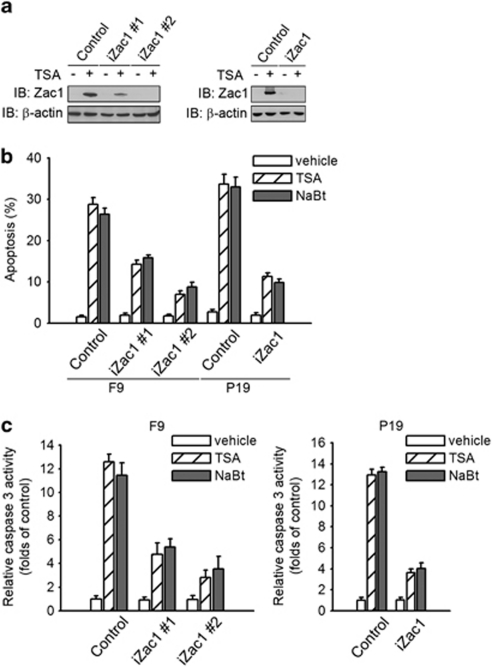
To determine whether Zac1 upregulation is implicated in cell apoptosis induced by HDAC inhibitors, we designed a small-hairpin RNA (shRNA) targeting Zac1, using a scrambled sequence as the negative control, and obtained two F9 (iZac1#1 and iZac1#2) and one P19 cell clones (iZac1) stably expressing Zac1-shRNA. Zac1 expression levels are strongly reduced after TSA treatment in Zac1-shRNA clones compared with that in the control clones (Figure 2a). In these cells, TSA- and NaBt-induced apoptosis was significantly inhibited as indicated by FACS (Figure 2b) and caspase-3 activity (Figure 2c) assays. These findings indicate that Zac1 induction is critical for HDAC inhibitor-induced apoptosis.
Figure 2.
Zac1 upregulation and HDAC inhibitor-induced cell apoptosis. (a) F9 and P19 cell clones were treated with 20 ng/ml TSA for 24 h and Zac1 protein levels were examined by western blotting. (b) F9 and P19 cell clones were treated with 20 ng/ml TSA or 1.5 mM NaBt for 48 h as indicated. Cell apoptosis was examined by FACS assay. (c) F9 and P19 cell clones were treated with 20 ng/ml TSA or 1.5 mM NaBt for 48 h, and the cell apoptosis was examined by caspase-3 activity assay
Zac1 upregulation is required for HDAC inhibitor-induced suppression of NF-κB activity
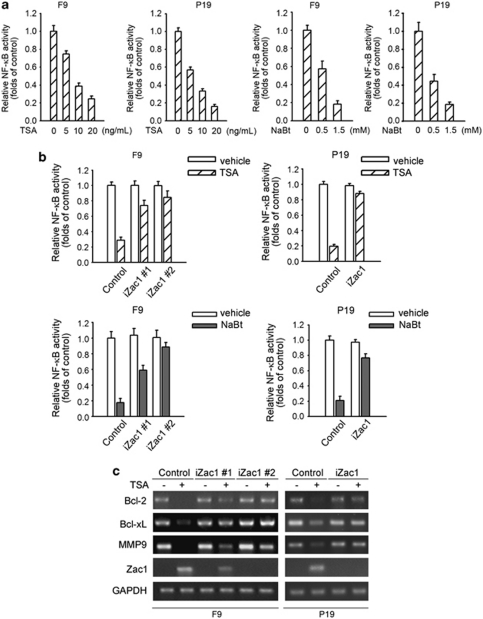
As it has been shown previously that Zac1 functions as a p53 transcription cofactor,16 we examined whether the p53 pathway is involved in the apoptosis. When F9 cells were treated with TSA for 12 h, Zac1 level was significantly upregulated, without any change in cell viability. At this time point, p53 activity was examined using a p53 luciferase reporter system. As shown in Supplementary Figure S2a, TSA treatment showed no significant effect on p53 activity. BAX and p21 are two typical target genes of p53. We also examined the effect of TSA on the protein expression levels of these genes. However, no increase in the level of these proteins was detected (Supplementary Figure S2b). A possible explanation of this fact is that basal p53 activity is very high in these cells as compared with that in non-transformed murine AML-12 hepatocytes (Supplementary Figure S2c). NF-κB is an important cellular survival factor. Changes in NF-κB activity have been reportedly associated with HDAC inhibitor-induced cell apoptosis.10, 11 Therefore, we are interested in determining whether Zac1 upregulation-mediated apoptosis is related with the suppression of NF-κB activity. We examined NF-κB-dependent gene transcription in response to HDAC inhibitor. As shown in Figure 3a, TSA and NaBt treatment (12 h) caused a dramatical decrease in the basal level of NF-κB activity in both F9 and P19 cells, as measured using an NF-κB luciferase reporter. We then performed NF-κB luciferase reporter assay in Zac1-shRNA cell clones and found that HDAC inhibitor-induced suppression of NF-κB activity was significantly reduced in these cell clones (Figure 3b). To further confirm the role of Zac1 in HDAC inhibitor-induced NF-κB repression, we examined the transcription levels of a subset of NF-κB-responsive genes such as Bcl-2, Bcl-xL, and MMP-9. In control cell clones, transcription of these NF-κB-responsive genes was markedly downregulated after treatment with the HDAC inhibitor TSA. However, this effect was significantly attenuated in cells stably expressing Zac1-shRNA (Figure 3c).
Figure 3.
Zac1 is required for HDAC inhibitor-induced NF-κB activity repression. (a) Cells were co-transfected with 0.5 μg of 3 × κB-Luc reporters and the same amount of Renilla luciferase reporter plasmid. Twenty-four hours after transfection, cells were stimulated with the indicated concentration of TSA or NaBt. Luciferase activity was measured 12 h after treatment. Luciferase activity was presented relative to Renilla luciferase activity. (b) 3 × κB-Luc and Renilla luciferase reporters were co-transfected into different F9 and P19 cell clones as indicated. Twenty-four hours later, cells were treated with 20 ng/ml TSA or 1.5 mM NaBt for 12 h. Vehicle-treated cells were used as controls. Cells were subjected to luciferase activity assay. (c) F9 and P19 cell clones were treated with 20 ng/ml TSA for 24 h. Then the mRNA levels of the indicated genes were analyzed by RT-PCR
Zac1 represses NF-κB-dependent gene transcription
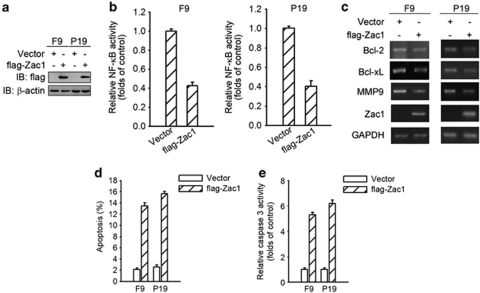
The fact that Zac1 downregulation significantly reduces HDAC inhibitor-mediated inhibition of NF-κB activity suggests that Zac1 overexpression may suppress the NF-κB activity. To test this possibility, we transfected flag-tagged Zac1 into F9 and P19 cells (Figure 4a). Indeed, Zac1 overexpression caused a significant decrease in constitutive NF-κB transactivity (Figure 4b). It also attenuated the transcription of certain genes downstream from NF-κB (Figure 4c). As examined by FACS and caspase-3 activity assays (Figures 4d and e), Zac1 overexpression induced cell apoptosis. These data show that Zac1 acts as a suppressor of NF-κB activity, which contributed to its proapoptotic function.
Figure 4.
Zac1 inhibits NF-κB signaling. (a) Empty or flag-tagged Zac1 (flag-Zac1) expression vectors were transfected into F9 and P19 cells. Twenty-four hours later, exogenous Zac1 levels were determined by western blotting using anti-flag antibody. (b) F9 and P19 cells were co-transfected with 3 × κB-Luc, Renilla luciferase reporter, and flag-Zac1 expression vectors. Cells were subjected to luciferase activity assay 24 h after transfection. (c) F9 and P19 cells were transfected with flag-Zac1 expression plasmids or empty vectors; forty-eight hours later, total RNA was extracted from the cells and the mRNA levels of the indicated genes were measured by RT-PCR. (d) F9 and P19 cells were transfected with plasmids expressing flag-Zac1. Forty-eight hours after transfection, cell apoptosis was measured by FACS assay. (e) Empty or Zac1-expressing vectors were transfected into F9 and P19 cells. Cell apoptosis was examined by caspase-3 activity assay after 48 h
Zac1 interacts with NF-κB p65 subunit
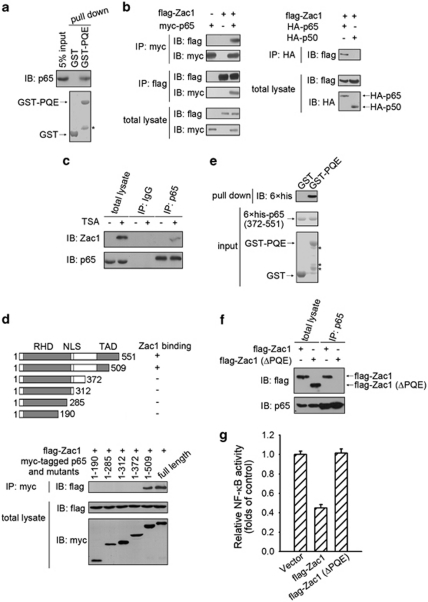
Multiple sequence alignment of Zac1 proteins from 10 species showed that there are two phylogenetically conserved regions in the primary sequence of the Zac1 protein. One is the C2H2 zinc finger containing a DNA-binding region and the other is a Pro-Gln-Glu (PQE)-rich region (data not shown). This observation suggests that the PQE-rich region of Zac1 may have an important role in its biological functions. Using GST-fusion protein chromatography, interaction between the NF-κB p65 subunit and the PQE-rich region of Zac1 was determined (Figure 5a) and was further confirmed by co-immunoprecipitation experiments using either anti-myc or the anti-flag antibodies in 293T cells co-expressing myc-tagged p65 and flag-tagged Zac1 (Figure 5b, left panel). By contrast, co-immunoprecipitation of flag-tagged Zac1 and HA-tagged NF-κB p50 subunits showed no detectable interaction (Figure 5b, right panel). Moreover, interactions between endogenous p65 and Zac1 were detected by immunoprecipitation and western blotting using anti-p65 and anti-Zac1 antibodies (Figures 5c and 6b). RelB and cRel are another two members of the NF-κB transcription factor family. We also examined whether Zac1 binds to RelB and cRel, but no binding was observed (Supplementary Figure S3). To identify which portion of the p65 protein is required for its interaction with Zac1, we constructed several truncated p65 expression plasmids (Figure 5d, upper panel). Among these p65 mutants, only the one containing the 372- to 509-amino-acid sequence can interacts with Zac1 protein molecules (Figure 5d, lower paenl), indicating that the C-terminal region of p65 is required for Zac1 binding. To determine whether the PQE-rich motif of Zac1 directly interacts with the p65 C-terminal region, GST pull-down assays were performed using recombinant GST-PQE and a 6 × his-tagged 372- to 551-amino-acid sequence region of p65 (6 × his-p65372–551). As shown in Figure 5e, GST-PQE, but not GST, was able to pull down purified 6 × his-p65372–551. In purified GST-PQE proteins some degradation bands were observed. The degradation of these fusion proteins is probably because of their fast turnover rate in bacterial cells. To determine whether the Zac1/p65 interaction is required for the NF-κB-inhibitory role of Zac1, a mutated Zac1 with its PQE-rich motif deleted (Zac1 ΔPQE) was constructed. Owing to this mutation Zac1 lost its ability to bind to p65 (Figure 5f) and its inhibitory effect on NF-κB activity was abolished (Figure 5g). Thus, the interaction between the PQE-rich motif of Zac1 protein and the C-terminal region of the NF-κB p65 subunit is critical for inhibition of NF-κB activity by Zac1.
Figure 5.
Zac1 interacts with the NF-κB p65 subunit. (a) GST pull-down assay was performed using GST fused with the PQE-rich motif of Zac1 (GST-PQE) and F9 cell total lysates. Associated proteins were eluted and analyzed by western blotting with anti-p65 antibody. The purified proteins used in this assay were visualized by Coomassie blue staining. (b) 293T cells were transfected with the plasmids as indicated. Twenty-four hours later, cell extracts were subjected to immunoprecipitation using the indicated antibodies and then analyzed by western blotting. (c) F9 cells were treated with vehicle or 20 ng/ml TSA for 24 h. Then cell lysates were prepared. Immunoprecipitation and western blotting were performed using the indicated antibodies. (d) Upper: A schematic illustration of p65 and its mutants (RHD, Rel homology domain; NLS, nuclear localization signal; TAD, transactivation domain). Lower: 293T cells were co-transfected with flag-Zac1 expression plasmids and myc-p65, or its truncations vectors. Cell lysates were immunoprecipitated using anti-myc antibody and the bound proteins were analyzed by western blotting using anti-flag antibody. (e) The GST pull-down assay was performed using GST-PQE and bacterially expressed 6 × his-p65372–551. Associated 6 × his-p65372–551 was detected by western blotting using anti-6 × his antibody. The purified proteins used in this assay were visualized by Coomassie blue staining. (f) Flag-tagged wild-type Zac1 or mutated Zac1 were transfected into F9 cells. Forty-eight hours later, cell extracts were subjected to immunoprecipitation using the indicated antibodies and then analyzed by western blotting. (g) F9 cells were co-transfected with 3 × κB-Luc, Renilla luciferase reporter, and the indicated Zac1 expression vectors. Cells were subjected to luciferase activity assay 24 h after transfection. The asterisks in this figure denote the degraded bands of the GST-PQE fusion protein
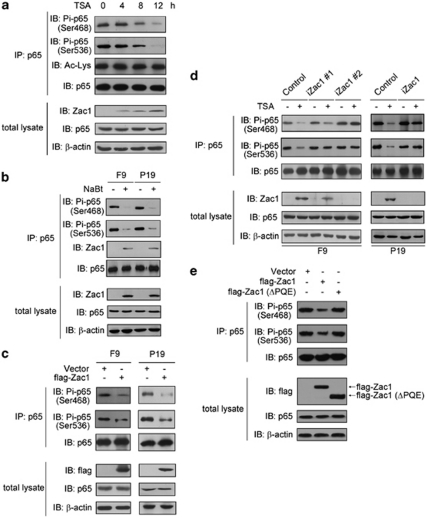
Figure 6.
Zac1 inhibits NF-κB p65 subunit phosphorylation at Ser468 and 536 residues. (a) F9 cells were exposed to 20 ng/ml TSA for the indicated time periods and then p65 protein was immunoprecipitated using an anti-p65 antibody. Total p65 and p65 phosphorylation at Ser468 and Ser536 were visualized using corresponding antibodies. Acetylation of p65 was detected using an antibody specific to acetylated lysine. Zac1 and p65 protein levels in the total lysate were also shown. (b) F9 and P19 cells were treated with 1.5 mM NaBt for 12 h, and then p65 phosphorylation levels at Ser468 and Ser536 were detected as described in panel a. The Zac1 proteins interacting with p65 are also shown. (c) F9 and P19 cells were transfected with plasmids encoding flag-Zac1. Forty-eight hours after transfection, p65 phosphorylation levels at Ser468 and Ser536 were detected. The level of transfected Zac1 (detected by anti-flag tag antibody) and the total p65 level (detected by anti-p65 antibody) in the total lysate were also shown. (d) F9 and P19 cell clones were treated with 20 ng/ml TSA (12 h). Cellular Zac1, p65, Ser468-, and Ser536-phosphorylated p65 were examined as described above. (e) F9 cells were transfected with the indicated plasmids. Forty-eight hours later, p65 phosphorylation levels at Ser468 and Ser536 were detected. The level of transfected Zac1 and total p65 level in cell lysate were also shown
Zac1 impairs p65 phosphorylation at Ser468 and Ser536
p65 nuclear translocation is important for NF-κB signaling. To further understand how Zac1 inhibits NF-κB activity, we examined the subcellular location of the p65 subunit before and after TSA treatment, and observed no change (Supplementary Figure S4). NF-κB activity is also regulated by acetylation of its p65 subunit. However, no detectable change in p65 acetylation was observed (Figure 6a). Phosphorylation of the p65 protein at its serine residues is another important regulatory mechanism of NF-κB activity. Ser468 and Ser536 are two important phosphorylation sites locate in the C-terminal of p65. Because binding of Zac1 to the C-terminal region of p65 was observed (Figures 5d and e), we tested p65 phosphorylation levels at these two sites before and after HDAC inhibitor treatment. TSA treatment strongly decreased the phosphorylation levels of Ser468 and Ser536 residues, which correlated with an increase in the level of Zac1 (Figure 6a). Similarly, NaBt treatment decreased p65 phosphorylation at these two sites in both F9 and P19 cells (Figure 6b). To test whether Zac1 overexpression inhibits the phosphorylation of p65 at these sites, flag-tagged Zac1 was transfected into F9 and P19 cells. Zac1 overexpression decreased the levels of p65 Ser468 and Ser536 phosphorylation in both F9 and P19 cells, whereas p65 expression level in the total cell lysate remained unchanged (Figure 6c). This effect was further confirmed in F9 and P19 Zac1-shRNA cell clones. In control cells, besides induction of Zac1 by TSA, a corresponding decrease in the level of Ser468- and Ser536-phosphorylated p65 was induced. However, expression of Zac1-shRNA abolished this effect (Figure 6d), and in consistent with this observation, mutated Zac1 without its PQE-rich motif that does not bind to p65 cannot inhibit p65 phosphorylation at these two sites (Figure 6e). The above results indicate that Zac1/p65 interaction inhibits p65 phosphorylation at Ser468 and Ser536, which is implicated in the inhibitory effect of Zac1 on NF-κB activity.
The expression levels of Zac1 and cellular responses to HDAC inhibitors
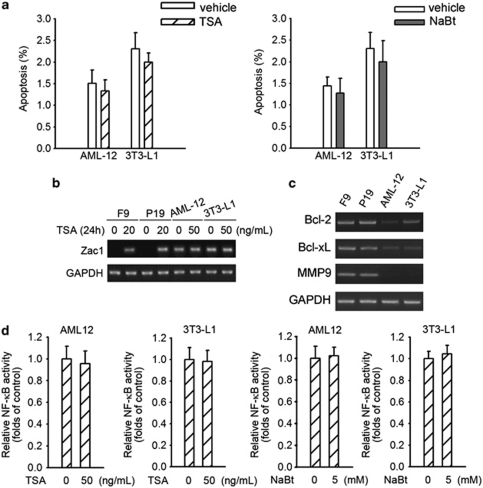
Although HDAC inhibitors are potent inducers of apoptosis in EC cells, they do not have the same effect on non-cancer cells such as AML-12 murine hepatocytes and 3T3-L1 fibroblasts (Figure 7a). To get an insight into this phenomenon, we examined the mRNA levels of Zac1 before and after HDAC inhibitor treatment in these cell lines. In sharp contrast to F9 and P19 EC cells, the basal levels of Zac1 in AML-12 and 3T3-L1 cells are very high but cannot be further upregulated by TSA (Figure 7b). Low Zac1 levels in F9 and P19 cells correlated with high basal transcription levels of NF-κB-responsive genes, and high Zac1 levels correlated with low basal transcription levels of NF-κB-responsive genes in AML-12 and 3T3-L1 cells (Figure 7c). In addition, HDAC inhibitors showed no inhibitory effect on basal NF-κB activity in these non-cancer cells (Figure 7d). These data indicate a relationship between the different cellular responses and the basal levels of NF-κB activities.
Figure 7.
The effect of HDAC inhibitors on several non-cancer cell lines. (a) Cells were treated with 50 ng/ml TSA (left) or 5 mM NaBt (right) for 48 h. Cell apoptosis was determined by FACS assay. (b) Cells were treated with the indicated concentration of TSA for 24 h. Then the Zac1 transcriptional level was examined by RT-PCR. (c) Total RNA was extracted from the cells, and then the mRNA levels of the indicated genes were detected by RT-PCR. (d) Renilla luciferase and 3 × κB-Luc reporters were co-transfected into cells as indicated. Twenty-four hours later, cells were treated with TSA (left two panels) or NaBt (right two panels) for 12 h, followed by luciferase activity assay
Discussion
Relatively little is known about the effects of HDAC inhibitors on EC cells. Further studies in this field are important for increasing our understanding and for developing potential mechanism-based molecular targets. In this presented study, we showed that upregulation of histone acetylation in the Zac1 promoter induces Zac1 gene expression and suppression of NF-κB activity, which account for HDAC inhibitor-induced apoptosis. These data indicate that epigenetic regulation of Zac1 expression modulates NF-κB activity and the balance between survival and death of EC cells. Because Zac1 is widely expressed in normal tissues, but its expression is undetectable or highly reduced in a variety of tumors,22, 23, 24 the above findings provide a new understanding of the potential mechanisms of cancer formation.
To determine whether HDAC inhibitor-induced histone acetylation in the Zac1 promoter is simply due to an increase in the globe histone acetylation, the effect of TSA on cellular globe histone acetylation was examined (Supplementary Figure S5). TSA-induced increase in globe acetylation occurs later and is weaker than that in the Zac1 promoter (Supplementary Figure S5 and Figure 1c), indicating that upregulation of histone acetylation in the Zac1 promoter appears to be an early cellular event independent of increment of globe histone acetylation. It is likely that a relatively high level of HDAC activity is required for maintaining the Zac1 promoter at a hypo-acetylated state.
Zac1 knockdown markedly decreases HDAC inhibitor-induced apoptosis, suggesting that the Zac1/NF-κB pathway has a major role in the HDAC inhibitor-induced apoptosis of EC cells. Because it appeared that a relatively complete inhibition of Zac1 expression by Zac1-shRNA cannot totally abolish HDAC inhibitor-induced apoptosis, it is possible that other unidentified mechanisms may also participate in HDAC inhibitor-induced apoptosis in EC cells. For instance, acetylation of non-histone proteins and changes in the expression levels of other proteins may also contribute to the apoptosis induction effect of HDAC inhibitors.25, 26, 27
HDAC inhibitors suppress HDAC activity in both normal and tumor cells. However, the cytotoxic effect of HDAC inhibitors in tumor cells is fundamentally much stronger than that in non-cancer cells.28 Our present study provides a possible explanation for these phenomena. As both NF-κB activity and proapoptotic p53 signal levels are high in EC cells but relatively much lower in normal cells, a high level of NF-κB activity constitutes an important factor for the survival of F9 and P19 cells. Thus, HDAC inhibitors selectively exerted their proapoptotic effect in EC cells through inhibition of NF-κB activity. This possibility was further supported by the observation that the expression level of Zac1 is very low but can be strongly induced by HDAC inhibitors in EC cells. In non-cancer cells, the fact that a high Zac1 basal level or treatment with HDAC inhibitors does not result in cell apoptosis suggests that low NF-κB activity alone is not sufficient to cause cell death, because the basal proapoptotic p53 signal is correspondingly very low in these cells.
Zac1 can interact with the p65 but not the p50, RelB or cRel subunit of NF-κB, and the PQE-rich motif of Zac1 interacts with the C-terminal region of the NF-κB p65 subunit. These two observations are compatible with each other because there are no homolog sequences among the C-termini of p65 and the other three subunits. Phosphorylation of the p65 subunit of NF-κB is a major post-translational modification that regulates NF-κB activity. A series of phosphorylation sites on the NF-κB p65 subunit has been identified. Ser468 and Ser536 are two typical phosphorylation sites in the C-terminal region of p65. The phosphorylation levels of these two sites are dramatically increased upon stimulation of NF-κB activity.29, 30 Moreover, studies show that tumor cell apoptosis induced by anticancer reagents is associated with decreased p65 phosphorylation at Ser536 residue,31, 32, 33 suggesting that p65 Ser536 phosphorylation is important for maintaining cancer cell survival and can be inhibited by anticancer reagents. In this study we showed that not only phosphorylation of Ser536 but also phosphorylation of Ser468 is downregulated during HDAC inhibitor-induced apoptosis of EC cells. Although a negative regulatory role of Ser468 phosphorylation in NF-κB activation has been observed,34 some later reports showed that phosphorylation of Ser468 serves to enhance NF-κB-dependent gene transcription in different systems.29, 35 Our findings support a positive role of p65 Ser468 phosphorylation in NF-κB activity, and suggest that repression of p65 phosphorylation at both Ser468 and Ser536 residues is involved in the inhibition of NF-κB activity and cell apoptosis.
Phosphorylation of p65 Ser468 and Ser536 residues can be induced by certain kinases.29, 35, 36, 37 However, very little is known about factors that inhibit the phosphorylation of these residues. In this study, we showed that Zac1 inhibits the phosphorylation of p65 Ser468 and Ser536. The basal level of Zac1 is very low in untreated EC cells. Zac1 upregulation through increased acetylation in its gene promoter by HDAC inhibitors inhibits the phosphorylation of p65 Ser468 and Ser536, and causes cell apoptosis, which suggests that repression of Zac1 expression by promoter hypo-acetylation accounts for the high phosphorylation levels in Ser468 and Ser536 residues, and the high NF-κB activity.
Consistent with previous reports,10, 11 we observed that HDAC inhibitors repress NF-κB activity in EC cells. On the other hand, Chen et al.38 reported that HDAC inhibitors are able to induce NF-κB activity through directly increasing the acetylation of the NF-κB p65 subunit. In our study we did not find changes in the acetylation of p65 protein by HDAC inhibitor treatment. HDAC inhibitor represses NF-κB activity through increasing histone acetylation in the Zac1 promoter, which activates Zac1 gene transcription. A possible explanation for this discrepancy is that acetylation of the histones in the Zac1 promoter is more sensitive to inhibition of HDAC activity than p65 protein. In addition, the concentration of TSA we used is much lower than that used in their study, which may not be high enough to increase p65 acetylation. We cannot exclude the possibility that complex mechanisms are involved in cellular NF-κB regulation in response to HDAC inhibitors.
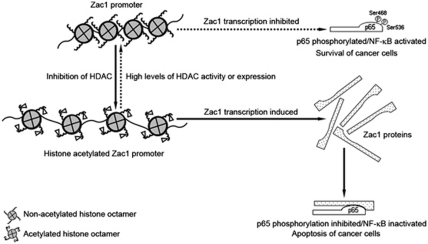
In brief summary, our results show that Zac1 has a critical role in HDAC inhibitor-mediated apoptosis in F9 and P19 EC cells. HDAC inhibitors induce the histone acetylation of the Zac1 promoter and thereby increase its expression. Zac1 interacts with and inhibits the phosphorylation of the Ser468 and Ser536 residues of the p65 subunit, leading to suppression of NF-κB activity and induction of apoptosis. The results indicate that an epigenetic regulation of Zac1 expression and suppression of NF-κB activity by Zac1 are implicated in the HDAC inhibitor-induced apoptosis of F9 and P19 EC cells. The above results are schematically illustrated in Figure 8.
Figure 8.
Summary by schematic illustration. The histone of the Zac1 promoter in untreated EC cells was hypo-acetylated, which repressed Zac1 gene transcription. HDAC inhibitors induced histone acetylation and increased Zac1 expression. Zac1 protein bound to the NF-κB p65 subunit and inhibited its phosphorylation at Ser468 and Ser536, leading to suppression of NF-κB activity and induction of cell apoptosis. The reverse process, as indicated by a dashed arrow, shows a potential pathway that maintains NF-κB activity and is important for cancer cell survival. High levels of HDAC or HDAC activity maintained the Zac1 promoter in a hypo-acetylated state and thus suppressed Zac1 expression, which accounted for the relatively high phosphorylation levels at Ser468 and Ser536 residues of p65 protein and the relatively high NF-κB activity
Materials and Methods
Cell culture and transfection
Murine F9 and P19 EC cells, 3T3-L1 fibroblasts and human embryonic kidney (HEK) 293T cells were cultivated in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (Invitrogen, Carlsbad, CA, USA) and supplied with penicillin (100 U/ml) and streptomycin (100 μg/ml). For F9 cells, dishes were coated with 0.1% gelatin. Murine AML-12 hepatocytes were cultured as previously described by Zhang et al.39 Cells were incubated at 37°C in a humidified atmosphere of 5% CO2. The concentrations of the HDAC inhibitors TSA and NaBt used in the experiments were 20 ng/ml and 1.5 mM, respectively, unless otherwise indicated. The cells were transfected with the Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. For transient transfected cells, experiments were performed 24–48 h after transfection. Stable transfected cells were obtained by G418 (800 μg/ml) selection after transfection.
Plasmids, antibodies, and reagents
To generate the GST-fusion protein of Zac1317–530 using a bacteria-expression system, appropriate PCR fragments were cloned into the pGEX-4T-1 vector. To knock down Zac1 expression in F9 cells, the pPGKsuper vector was used for the expression of shRNA. The target sequence of Zac1 is 5′-AAAAATGTGGCAAGTCCTTCG-3′, and the scrambled sequence (5′-GACCGAGTTATACGGCTTAAA-3′) was used as control. NF-κB p65 deletion mutants were amplified by PCR and subsequently cloned into the mammalian expression vector pcDNA3.1A. All of the above constructs were validated by DNA sequencing. Polyclonal rabbit anti-Zac1 and anti-p65 antibodies, and monoclonal mouse anti-HA-tag, anti-flag-tag and anti-myc-tag antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Polyclonal rabbit anti-acetylated histones and anti-phosphorylated p65 and monoclonal mouse anti-actin antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). TSA and NaBt were purchased from Sigma-Aldrich (Stockholm, Sweden).
Flow cytometry analysis (FACS)
Cells were trypsinized and fixed with 70% ethanol for over 2 h at 4°C. The cells were then pelleted and washed with PBS with 20 mM EDTA. RNA was removed by incubating the samples with RNase-A (1 mg/ml) at 37°C for at least 1 h. Cells were then stained with 30 μg/ml propidium iodine (PI) and detected by flow cytometry (Becton Dickinson FACSCalibur, Franklin Lakes, NJ, USA). The cell apoptotic rate was represented by the percentage of sub-G1 DNA content.
Caspase-3 activity assay
Cells were treated as indicated, and they were then trypsinized and washed once with PBS. The cells were collected and lysed with buffer containing 50 mM HEPES (pH 7.4), 100 mM NaCl, 0.1% CHAPS, 1 mM DTT, and 0.1 mM EDTA for 10 min on ice, and they were centrifuged at 12 000 × g for 10 min at 4°C. The supernatants were collected and protein concentrations were determined by Bradford's method. Then, 30 μg of each protein extract was incubated with 400 μM caspase substrate in 100 μl of caspase activity assay buffer (50 mM HEPES (pH 7.4), 100 mM NaCl, 0.1% CHAPS, 10 mM DTT, 0.1 mM EDTA, 10% glycerol) for 2 h at 37°C. Optical density (OD) was measured at 405 nm.
ChIP assay
ChIP assay was performed as previously described by Tang et al.40 A portion of the precipitated DNA was used for each PCR. The primer sequences used for detecting the precipitated DNA are as follows (5′–3′):
- p1f: CATGGCACTTGGCCTTTATT
p1r: CAAGTATCCTGCTGGGAAGC
- p2f: TGTTTCATGAGCCACCAGAG
p2r: CGAAGGACTTGCCACATTTT
Cell lysate preparation and western blotting
Cells cultured in 35-mm dishes were lysed in 0.2 ml of ice-cold lysis buffer containing 50 mM HEPES (pH 7.4), 5 mM EDTA, 50 mM NaCl, 1% Triton X-100, 50 mM NaF, 1 mM Na3VO4, 10 mM Na4P2O7, 10 mg/ml aprotinin, 10 mg/ml leupeptin, and 1 mM PMSF. The lysates were centrifuged at 12 000 × g for 15 min. The supernatants were collected and protein concentrations were determined by Bradford's method. The proteins were separated by sodium dodecyl sulfate (SDS)-PAGE and were transferred to a nitrocellulose membrane (Hybond ECL). The membrane was blocked for 30 min with 5% skimmed milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) and subsequently incubated with a primary antibody (1 : 2000 dilution) overnight at 4°C. After washing with TBST for 30 min at room temperature, the membrane was then incubated with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) for 2 h, followed by 45 min of washing (with three to five changes of the wash buffer). Protein bands were finally visualized by enhanced chemiluminescence (ECL) using the Super Signal Reagents (Pierce, Rockford, IL, USA).
Reverse transcription-PCR
Reverse transcription-PCR (RT-PCR) analysis was performed as described previously by Zhang et al.39 Briefly, total RNA was extracted using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. After RNA integrity was checked by agarose gel electrophoresis and ethidium bromide staining, 3 μg of RNA was reverse-transcribed at 42°C for 30 min using ReverTra Ace-α (Toyobo, Osaka, Japan). The primer sets for amplification are listed below (5′–3′):

GST pull-down assay
GST, the GST-fusion protein of Zac1317–530, and 6 × his-tagged p65372–551 were expressed in Escherichia coli BL21 strain and purified by affinity chromatography using glutathione or Ni-NTA agarose (Amersham Pharmacia, Buckinghamshire, England) according to the manufacturer's instructions. Cell lysates or purified 6 × his-p65372–551 proteins in 1 ml of binding buffer (20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Nonidet P-40) were incubated at 4°C for 3 h with GST or the GST-fusion protein of Zac1317–530 already bound to the glutathione beads. The beads were then washed and eluted in 50 μl of 1 × SDS-PAGE loading buffer and detected by western blotting.
Luciferase reporter gene assay
Luciferase activity was measured as described previously by Tang et al.40 Briefly, cells were seeded on 24-well plates and cultured for 24 h before transfection. To normalize luciferase activity, the luciferase reporter construct was transfected with the pRL-TK plasmid (Promega, Madison, WI, USA) containing the Renilla luciferase gene driven by the herpes simplex virus thymidine kinase promoter. After transfection, media were replaced and incubated with various stimuli for the time periods indicated. Luciferase activities were measured using the Dual Reporter assay system (Promega) according to the manufacturer's instructions.
Preparation of subcellular fractionation
Cells were harvested, washed twice with 1 × PBS, and resuspended on ice in 180 μl of CLB buffer (10 mM HEPES, 10 mM NaCl, 1 mM KH2PO4, 5 mM EDTA, 1 mM CaCl2, 0.5 mM MgCl2) for 5 min. Thirty strokes with a glass homogenizer were applied to the mixture, followed by adding 20 μl of 2.5 M sucrose. After centrifugation for 10 min at 6300 × g, the supernatants were removed as the cytoplasmic fraction. The pellet was resuspended in 0.5 ml of TSE buffer (10 mM Tris, 300 mM sucrose, 1 mM EDTA, 0.1% (v/v) Igepal-CA-630 (pH 7.5)) and homogenized with 50 strokes of a glass homogenizer. This suspension was then centrifuged at 4000 × g for 5 min. The resulting supernatant was discarded and the pellet was washed with the TSE buffer until the supernatant was clear. The resulting pellet was resuspended in 80 μl of the TSE buffer as the nuclear fraction.
Immunoprecipitation assay
Cell pellets were lysed in ice-cold RIPA buffer (phosphate-buffered solution containing 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with 50 mM NaF, 1 mM Na3VO4, 10 mM Na4P2O7, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 1 mM PMSF. After the insoluble fraction was removed by centrifugation at 4°C for 15 min (12 000 r.p.m.), whole-cell lysates were pre-cleared using protein-G sepharose. Immunoprecipitation was performed by incubating the above lysates with protein-G sepharose, pre-absorbed with 2 μg of the indicated primary antibodies at 4°C for 2 h, using an equal amount of normal IgG as negative control. After extensive washing, the sepharose beads were boiled in 50 μl of 1 × SDS-PAGE loading buffer. The eluted proteins were then subjected to western blotting.
All results shown are representative of or the statistics (mean value±S.E.) of at least three independent experiments.
Acknowledgments
We are grateful to Dr Shih-Ming Huang for providing Zac1 cDNA. We thank the members of the laboratory for helpful discussions. This work was supported by The Natural Science Foundation of China (grant number 30730023).
Glossary
- Zac1
zinc-finger protein regulator of apoptosis and cell-cycle arrest
- EC
embryonic carcinoma
- HDAC
histone deacetylase
- TSA
trichostatin-A
- NaBt
sodium butyrate
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by JC Marine
Supplementary Material
References
- Brown JM, Attardi LD. The role of apoptosis in cancer development and treatment response. Nat Rev Cancer. 2005;5:231–237. doi: 10.1038/nrc1560. [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Ten Dijke P. The dynamic roles of TGF-beta in cancer. J Pathol. 2011;223:205–218. doi: 10.1002/path.2785. [DOI] [PubMed] [Google Scholar]
- Prasad S, Ravindran J, Aggarwal BB. NF-kappaB and cancer: how intimate is this relationship. Mol Cell Biochem. 2010;336:25–37. doi: 10.1007/s11010-009-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta J, Fan Y, Gupta N, Fan G, Gelinas C. Current insights into the regulation of programmed cell death by NF-kappaB. Oncogene. 2006;25:6800–6816. doi: 10.1038/sj.onc.1209938. [DOI] [PubMed] [Google Scholar]
- Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawan C, Herceg Z. Histone modifications and cancer. Adv Genet. 2010;70:57–85. doi: 10.1016/B978-0-12-380866-0.60003-4. [DOI] [PubMed] [Google Scholar]
- Liu T, Kuljaca S, Tee A, Marshall GM. Histone deacetylase inhibitors: multifunctional anticancer agents. Cancer Treat Rev. 2006;32:157–165. doi: 10.1016/j.ctrv.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Martinez-Iglesias O, Ruiz-Llorente L, Sanchez-Martinez R, Garcia L, Zambrano A, Aranda A. Histone deacetylase inhibitors: mechanism of action and therapeutic use in cancer. Clin Transl Oncol. 2008;10:395–398. doi: 10.1007/s12094-008-0221-x. [DOI] [PubMed] [Google Scholar]
- Yao J, Duan L, Fan M, Wu X. NF-kappaB signaling pathway is involved in growth inhibition, G2/M arrest and apoptosis induced by trichostatin A in human tongue carcinoma cells. Pharmacol Res. 2006;54:406–413. doi: 10.1016/j.phrs.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Batra S, Sahu RP, Kandala PK, Srivastava SK. Benzyl isothiocyanate-mediated inhibition of histone deacetylase leads to NF-kappaB turnoff in human pancreatic carcinoma cells. Mol Cancer Ther. 2010;9:1596–1608. doi: 10.1158/1535-7163.MCT-09-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, Kang JK, Seo DW, Park JH, Park JW, Lee JC, et al. Depletion of embryonic stem cell signature by histone deacetylase inhibitor in NCCIT cells: involvement of Nanog suppression. Cancer Res. 2009;69:5716–5725. doi: 10.1158/0008-5472.CAN-08-4953. [DOI] [PubMed] [Google Scholar]
- Spengler D, Villalba M, Hoffmann A, Pantaloni C, Houssami S, Bockaert J, et al. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 1997;16:2814–2825. doi: 10.1093/emboj/16.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Ciani E, Boeckardt J, Holsboer F, Journot L, Spengler D. Transcriptional activities of the zinc finger protein Zac are differentially controlled by DNA binding. Mol Cell Biol. 2003;23:988–1003. doi: 10.1128/MCB.23.3.988-1003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Stallcup MR. Mouse Zac1, a transcriptional coactivator and repressor for nuclear receptors. Mol Cell Biol. 2000;20:1855–1867. doi: 10.1128/mcb.20.5.1855-1867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Schonthal AH, Stallcup MR. Enhancement of p53-dependent gene activation by the transcriptional coactivator Zac1. Oncogene. 2001;20:2134–2143. doi: 10.1038/sj.onc.1204298. [DOI] [PubMed] [Google Scholar]
- Barre B, Perkins ND. The Skp2 promoter integrates signaling through the NF-kappaB, p53, and Akt/GSK3beta pathways to regulate autophagy and apoptosis. Mol Cell. 2010;38:524–538. doi: 10.1016/j.molcel.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Cortez CC, Jones PA. Chromatin, cancer and drug therapies. Mutat Res. 2008;647:44–51. doi: 10.1016/j.mrfmmm.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikihara T, Arima T, Kato K, Matsuda T, Kato H, Douchi T, et al. Epigenetic silencing of the imprinted gene ZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int J Cancer. 2005;115:690–700. doi: 10.1002/ijc.20971. [DOI] [PubMed] [Google Scholar]
- Wilson AJ, Chueh AC, Togel L, Corner GA, Ahmed N, Goel S, et al. Apoptotic sensitivity of colon cancer cells to histone deacetylase inhibitors is mediated by an Sp1/Sp3-activated transcriptional program involving immediate-early gene induction. Cancer Res. 2010;70:609–620. doi: 10.1158/0008-5472.CAN-09-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Cheng H, Sampaio AV, Nielsen TO, Underhill TM. EGR1 reactivation by histone deacetylase inhibitors promotes synovial sarcoma cell death through the PTEN tumor suppressor. Oncogene. 2010;29:4352–4361. doi: 10.1038/onc.2010.204. [DOI] [PubMed] [Google Scholar]
- Varrault A, Ciani E, Apiou F, Bilanges B, Hoffmann A, Pantaloni C, et al. hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer. Proc Natl Acad Sci USA. 1998;95:8835–8840. doi: 10.1073/pnas.95.15.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdollahi A, Pisarcik D, Roberts D, Weinstein J, Cairns P, Hamilton TC. LOT1 (PLAGL1/ZAC1), the candidate tumor suppressor gene at chromosome 6q24-25, is epigenetically regulated in cancer. J Biol Chem. 2003;278:6041–6049. doi: 10.1074/jbc.M210361200. [DOI] [PubMed] [Google Scholar]
- Bilanges B, Varrault A, Basyuk E, Rodriguez C, Mazumdar A, Pantaloni C, et al. Loss of expression of the candidate tumor suppressor gene ZAC in breast cancer cell lines and primary tumors. Oncogene. 1999;18:3979–3988. doi: 10.1038/sj.onc.1202933. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Woods NT, Piluso LG, Lee HH, Chen J, Bhalla KN, et al. p53 acetylation is crucial for its transcription-independent proapoptotic functions. J Biol Chem. 2009;284:11171–11183. doi: 10.1074/jbc.M809268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Adachi M, Kawamura R, Imai K. Bmf is a possible mediator in histone deacetylase inhibitors FK228 and CBHA-induced apoptosis. Cell Death Differ. 2006;13:129–140. doi: 10.1038/sj.cdd.4401686. [DOI] [PubMed] [Google Scholar]
- Brazelle W, Kreahling JM, Gemmer J, Ma Y, Cress WD, Haura E, et al. Histone deacetylase inhibitors downregulate checkpoint kinase 1 expression to induce cell death in non-small cell lung cancer cells. PLoS One. 2010;5:e14335. doi: 10.1371/journal.pone.0014335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Mattioli I, Geng H, Sebald A, Hodel M, Bucher C, Kracht M, et al. Inducible phosphorylation of NF-kappa B p65 at serine 468 by T cell costimulation is mediated by IKK epsilon. J Biol Chem. 2006;281:6175–6183. doi: 10.1074/jbc.M508045200. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Matsushima-Nishiwaki R, Yamaguchi S, Iida H, Dohi S, Kozawa O. Mechanisms of tumor necrosis factor-alpha-induced interleukin-6 synthesis in glioma cells. J Neuroinflammation. 2010;7:16. doi: 10.1186/1742-2094-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Wong ET, Cheok CF, Tergaonkar V, Lane DP. R-Roscovitine simultaneously targets both the p53 and NF-kappaB pathways and causes potentiation of apoptosis: implications in cancer therapy. Cell Death Differ. 2008;15:263–273. doi: 10.1038/sj.cdd.4402257. [DOI] [PubMed] [Google Scholar]
- Hafeez BB, Siddiqui IA, Asim M, Malik A, Afaq F, Adhami VM, et al. A dietary anthocyanidin delphinidin induces apoptosis of human prostate cancer PC3 cells in vitro and in vivo: involvement of nuclear factor-kappaB signaling. Cancer Res. 2008;68:8564–8572. doi: 10.1158/0008-5472.CAN-08-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XX, Jiang HR, Li HB, Zhang TN, Zhou Q, Liu N. Apoptosis of stomach cancer cell SGC-7901 and regulation of Akt signaling way induced by bovine lactoferrin. J Dairy Sci. 2010;93:2344–2350. doi: 10.3168/jds.2009-2926. [DOI] [PubMed] [Google Scholar]
- Buss H, Dorrie A, Schmitz ML, Frank R, Livingstone M, Resch K, et al. Phosphorylation of serine 468 by GSK-3beta negatively regulates basal p65 NF-kappaB activity. J Biol Chem. 2004;279:49571–49574. doi: 10.1074/jbc.C400442200. [DOI] [PubMed] [Google Scholar]
- Gong R, Rifai A, Ge Y, Chen S, Dworkin LD. Hepatocyte growth factor suppresses proinflammatory NFkappaB activation through GSK3beta inactivation in renal tubular epithelial cells. J Biol Chem. 2008;283:7401–7410. doi: 10.1074/jbc.M710396200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid LV, Mayo MW, Reuther JY, Baldwin AS., Jr Akt stimulates the transactivation potential of the RelA/p65 Subunit of NF-kappa B through utilization of the Ikappa B kinase and activation of the mitogen-activated protein kinase p38. J Biol Chem. 2001;276:18934–18940. doi: 10.1074/jbc.M101103200. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274:30353–30356. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- Zhang KH, Tian HY, Gao X, Lei WW, Hu Y, Wang DM, et al. Ferritin heavy chain-mediated iron homeostasis and subsequent increased reactive oxygen species production are essential for epithelial–mesenchymal transition. Cancer Res. 2009;69:5340–5348. doi: 10.1158/0008-5472.CAN-09-0112. [DOI] [PubMed] [Google Scholar]
- Tang Y, Shu G, Yuan X, Jing N, Song J. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res. 2011;21:316–326. doi: 10.1038/cr.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.