Abstract
The ubiquitin-editing enzyme A20 (tumor necrosis factor-α-induced protein 3) serves as a critical brake on nuclear factor κB (NF-κB) signaling. In humans, polymorphisms in or near the A20 gene are associated with several inflammatory disorders, including psoriasis. We show here that epidermis-specific A20-knockout mice (A20EKO) develop keratinocyte hyperproliferation, but no signs of skin inflammation, such as immune cell infiltration. However, A20EKO mice clearly developed ectodermal organ abnormalities, including disheveled hair, longer nails and sebocyte hyperplasia. This phenotype resembles that of mice overexpressing ectodysplasin-A1 (EDA-A1) or the ectodysplasin receptor (EDAR), suggesting that A20 negatively controls EDAR signaling. We found that A20 inhibited EDAR-induced NF-κB signaling independent from its de-ubiquitinating activity. In addition, A20 expression was induced by EDA-A1 in embryonic skin explants, in which its expression was confined to the hair placodes, known to be the site of EDAR expression. In summary, our data indicate that EDAR-induced NF-κB levels are controlled by A20, which functions as a negative feedback regulator, to assure proper skin homeostasis and epidermal appendage development.
Keywords: skin, A20, knockout, NF-κB, EDA
The epidermis consists mainly of keratinocytes organized in different layers. The basal layer keratinocytes are mitotically active and provide new cells that gradually undergo differentiation toward the skin surface where they cornify and are shed. This well-orchestrated biological process is tightly controlled, and distortions in the sequence of events can result in different skin disorders, including inflammation and cancer. An exact control of the transcription factor nuclear factor κB (NF-κB) is essential to maintain skin homeostasis, witnessed by the fact that NF-κB overactivity as well as the ablation of essential NF-κB activators lead to skin inflammation.1, 2, 3 NF-κB belongs to a family of structurally related and evolutionarily conserved proteins (p100 or NF-κB2, p105 or NF-κB1, p65 or RelA, RelB, c-Rel), which exist as homo- or heterodimers. In resting cells, NF-κB is kept inactive by binding to inhibitor of κB (IκB) proteins, of which IκBα is the most studied. Numerous stimuli lead to phosphorylation and polyubiquitination of IκB proteins, which targets them for proteasomal degradation. This results in the release of NF-κB, which accumulates in the nucleus and transcriptionally activates target genes.
A central event in NF-κB signaling is the activation of the IκB kinase (IKK) complex, which consists of two kinases, IKK1 and IKK2 (also known as IKKα and IKKβ), and a regulatory platform protein, NF-κB essential modulator (NEMO, also known as IKKγ). Gene targeting experiments showed that IKK2 and NEMO, but not IKK1, are required for NF-κB activation by tumor necrosis factor (TNF) and by Toll-like receptors.3 Some receptors, such as those for lymphotoxin β and CD40, can activate an alternative NF-κB pathway involving IKK1 and leading to processing of p100 NF-κB to p52, which forms a transcriptionally active dimer with RelB.
As shown by skin-specific ablation of different NF-κB activators, cell autonomous activity of NF-κB seems to prevent keratinocyte apoptosis in both homeostatic and TNF-dependent inflammatory conditions.2 As expected, overactivation of NF-κB in keratinocytes results in skin inflammation. Besides its role in controlling inflammation and preventing apoptosis, NF-κB is also involved in the development of epidermal derivatives, such as hair, nails and sweat glands,4, 5 in which signaling is initiated by the binding of ectodysplasin-A1 (EDA-A1) to the epidermal-specific TNF-R family member ectodysplasin receptor (EDAR).6, 7 EDAR acts through ectodysplasin receptor-associated adapter protein (EDARADD), TRAF6 and TAB2/TAK1 to activate the IKK complex.8 Human mutations in the Eda1, Edar or Edaradd genes result in hypohidrotic ectodermal dysplasia (HED) characterized by defects in development of hair, teeth and sweat glands.8, 9 Similarly, mice carrying mutations in these genes, or lacking them or Traf6, acquire HED.10, 11, 12, 13, 14, 15, 16 Transgenic mice overexpressing EDA-A1 or EDAR develop morphogenetic alterations in a variety of ectodermal organs, including hair and teeth, and sweat, mammary and sebaceous glands,17, 18, 19, 20, 21, 22 which indicates that overactivation of the EDA/EDAR pathway is detrimental for the development of epidermal appendages.
As NF-κB is crucial in development and inflammation, tight regulation of its activation is essential. This tight control is achieved by the combined action of positive and negative regulators. During recent years, ubiquitination and de-ubiquitination (DUB) of several NF-κB signaling components have been identified as crucial steps in the control of NF-κB signaling pathways.23 One critical brake on NF-κB activation is the ubiquitin-editing enzyme A20, which has been characterized as an inhibitor of both NF-κB activation and apoptosis. A20 is required for terminating NF-κB signaling in response to TNF and microbial products, such as lipopolysaccharide and muramyl dipeptide. The in vivo importance of the A20 anti-inflammatory activities is illustrated by severe multi-organ inflammation and perinatal death seen in A20−/− mice.24 Recent genetic studies demonstrate an association between the human A20/tumor necrosis factor-α-induced protein 3 (TNFAIP3) gene and autoimmune pathology and psoriasis, which suggests that A20 might have an important role in human autoimmune and inflammatory diseases.25 These findings emphasize the importance of A20 in the control of normal tissue homeostasis and identify it as a potential target for the treatment of multiple inflammatory pathologies, including psoriasis.
We used K14-driven Cre/LoxP-mediated gene targeting in mice to investigate the in vivo function of A20 in epidermal development and homeostasis. We show that A20 is essential for controlling keratinocyte proliferation and for proper development of ectodermal appendages. Our data indicate that keratinocyte-specific deletion of A20 results in excessive EDA-A1-induced NF-κB signaling, thereby phenocopying EDA-A1 transgenic mice.
Results
Generation of epidermis-specific A20-deficient mice
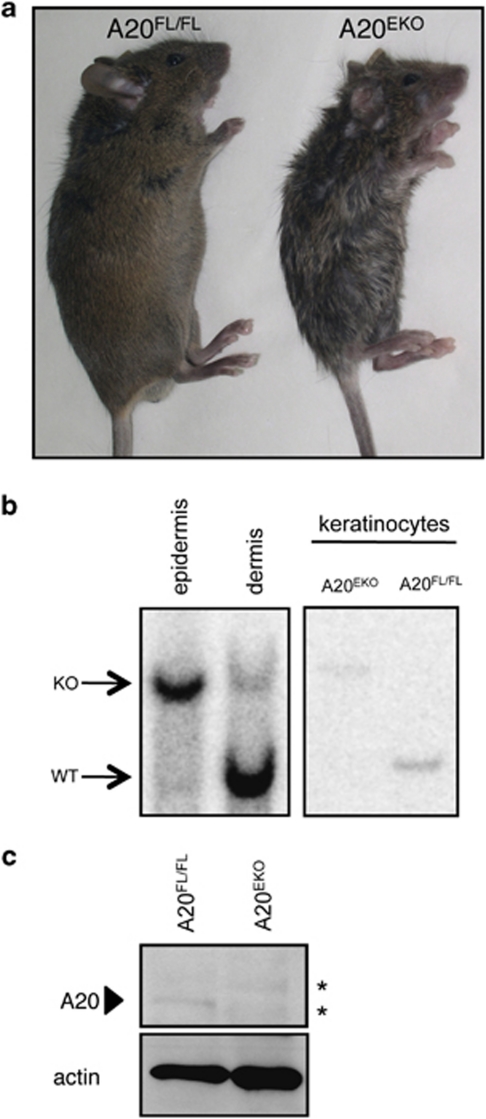
To identify the tissue-specific functions of A20, we generated mice carrying a conditional A20 allele in which exons IV and V of the mouse A20 gene are flanked by LoxP sites.26 To study the function of A20 in skin and epidermal appendages, we crossed the A20FL/FL mice with a transgenic mouse line that expresses Cre under the control of the human keratin 14 gene promoter (K14-Cre).27 Epidermal keratinocyte-specific A20-knockout mice (A20EKO) were born with normal Mendelian segregation (data not shown). To verify keratinocyte-specific A20 deletion in A20EKO mice (Figure 1a), we confirmed by Southern blot analysis that there was efficient Cre-mediated recombination in epidermis and in primary keratinocytes (Figure 1b). Consequently, we could not detect endogenous A20 in epidermal extracts from A20EKO mice (Figure 1c).
Figure 1.
Generation of A20EKO mice. (a) Photograph of A20EKO mouse and A20FL/FL control mouse. (b) Southern blot analysis of DNA from epidermis and dermis from A20EKO mice, and DNA from isolated keratinocytes of A20FL/FL (WT) and A20EKO mice. (c) Protein extracts from epidermal tissue, derived from keratinocyte (A20EKO)-specific knockouts, were analyzed by western blotting with an A20-specific antibody. The A20 protein band is indicated with an arrowhead, asterisk (*) indicates aspecific bands
A20EKO mice show keratinocyte hyperplasia
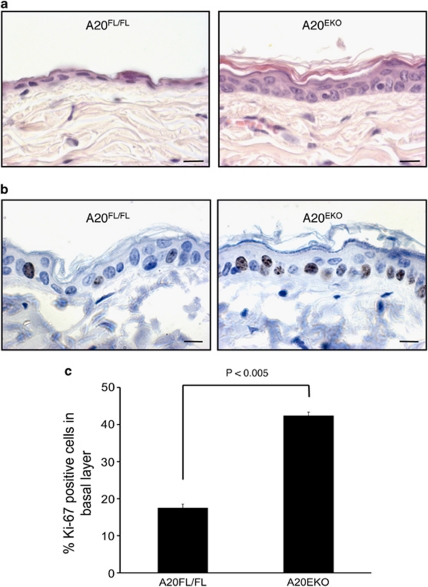
Macroscopic signs of skin inflammation, such as redness, scaling or excessive scratching due to itch, were not observed in A20EKO mice (Figure 1a). Histological analysis of back skin sections showed a thickened epidermis because of the increased proliferation (2.5-fold) of the basal cells (Figure 2), but similar very low levels of apoptosis (data not shown). The expression patterns of the differentiation markers caspase-14, filaggrin and keratin 10 were normal, and infiltrating immune cells were not observed (data not shown). These data indicate that absence of A20 does not lead to spontaneous skin inflammation, or that other DUB enzymes, such as cylindromatosis (CYLD),28 act redundantly to prevent spontaneous inflammation.
Figure 2.
A20EKO display mild keratinocyte hyperproliferation. (a) Sections of back skin of A20FL/FL and A20EKO mice were stained with hematoxylin and eosin, and analyzed by light microscopy (scale bar: 100 μm). (b) Ki-67-positive staining (dark brown nuclei) of back skin sections from A20FL/FL and A20EKO mice (scale bar: 100 μm). (c) Quantification of the percentage of Ki-67-positive basal layer keratinocytes in sections of back skin of A20FL/FL and A20EKO mice
A20EKO mice display abnormalities in ectodermal appendages
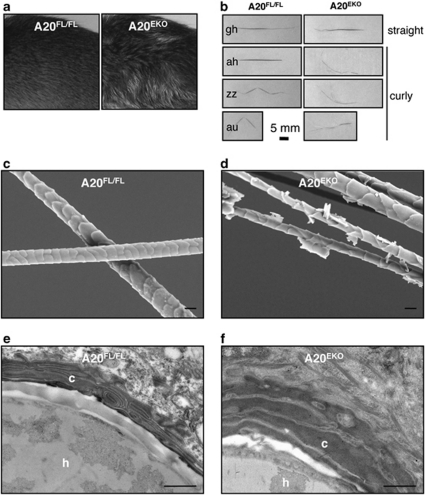
The coat of A20EKO mice was disorganized (Figures 1a and 3a). This disheveled hair phenotype was never observed in A20FL/FL control littermates. Mice were monitored up to the age of 12 months and did not exhibit signs of hair loss or alopecia. A20FL/FL mice were also crossed to a K5-Cre transgenic line,29 with which a similar phenotype was observed (data not shown). All further experiments were carried out on A20FL/FL/K14-Cre mice.27
Figure 3.
A20EKO mice have thinner hair with defective cuticle. (a) Picture of the coat hairs of A20FL/FL and A20EKO mice. (b) Hair from the back of A20FL/FL and A20EKO mice was epilated. Microscopic examination shows the presence of guard hair, awl hair, zigzag hair and auchene hair in A20FL/FL control mice. A20EKO mice have only straight or curly hair. ah, awl hair; au, auchene hair; gh, guard hair; zz, zigzag hair. (c and d) SEM picture of hair from A20FL/FL and A20EKO mice (scale bar: 10 μm). (e and f) TEM of sections through back skin of A20FL/FL and A20EKO mice showing detail of a section through a hair follicle (scale bar: 500 nm). The A20EKO mice have a hyperplasic and disorganized cuticular layer. h, hair shaft; c, cuticular layer
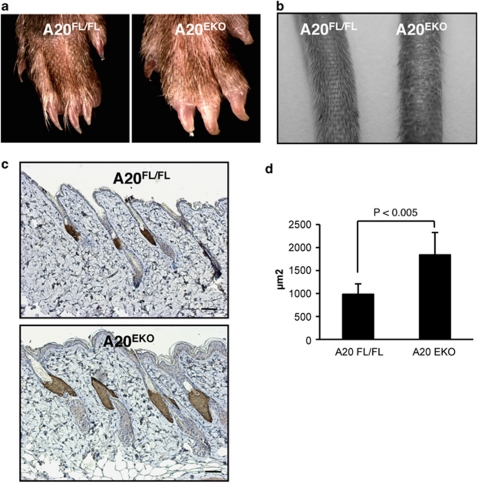
Four hair types can be distinguished in the mouse coat. Awl and guard hairs are straight, whereas auchene and zigzag hairs have one or multiple bends, respectively. Although straight hairs were found in A20EKO mice, other hair types had a curly instead of a bended morphology (Figure 3b). On epilating A20EKO mice, we noticed that the hair was more fragile and thinner than the hair of A20FL/FL mice. The increased fragility of A20EKO hair was further confirmed by testing its resistance to sonication (data not shown). Scanning electron microscopy (SEM) analysis confirmed extensive cuticle damage in hair fibers of A20EKO mice (Figures 3c and d). This hair damage can be explained by the amorphous nature of the hair cuticle, which, as observed by transmission electron microscopy, lacked a clear structure (Figures 3e and f). The medulla of the A20EKO hair is similar to that of A20FL/FL mice (data not shown). The A20EKO hair phenotype is strikingly similar to that of mice with keratinocyte-specific transgenes for EDA-A1 or EDAR.17, 18, 19, 30, 31 The EDA-A1-induced NF-κB pathway is required for the development of epidermal appendages, including hair, and its hyperactivity results in disheveled hair, longer nail growth, sebocyte hyperplasia, and the induction of additional molar-like teeth and nipples in mice.17, 18, 19, 20, 21, 22 A20EKO mice also display longer nails (Figure 4a), larger scale-like folds and more curly hair on the tails (Figure 4b), and sebocyte hyperplasia (Figures 4c and d). Extra teeth or nipples were never observed in A20EKO mice.
Figure 4.
Additional hallmarks of the A20EKO phenotype that phenocopy the EDA transgenes. (a) Picture of paws viewed with a binocular microscope shows that A20EKO mice have longer nails than control littermates. (b) Macroscopic analysis of the tails shows more curly tail hairs and larger scale-like folds on A20EKO mice. (c) Sebocytes in skin sections from A20EKO and control mice were stained with an anti-adipophillin antibody. (Scale bar: 500 μm.) (d) The surface areas of the sections through the sebaceous gland were determined based on this staining using Volocity image analysis software, and the mean surface areas in A20EKO and control mice were compared
A20 acts as a regulator of the EDA pathway to NF-κB activation
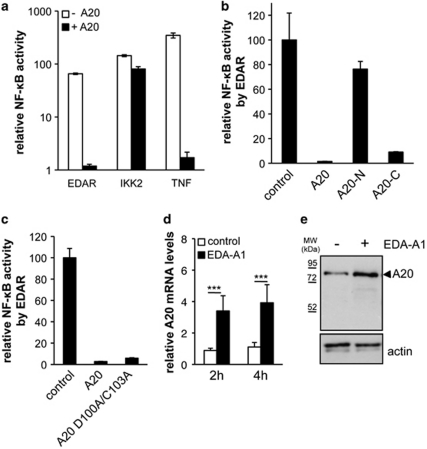
As the loss of A20 in the skin results in partial phenocopy of epidermis-specific EDA-A1 overexpressing mice we hypothesized that A20 might be a negative regulator of the EDA pathway. Indeed, by using an NF-κB-dependent luciferase (Luc) reporter assay, we demonstrated that A20 blocks EDAR-induced activation of NF-κB in HEK293T cells to a similar extend as TNF-induced NF-κB activation (Figure 5a). Making use of deletion mutants, we could demonstrate that EDAR-induced NF-κB activation was inhibited by the C-terminal domain of A20, containing the Zn-fingers, but not by the N-terminal DUB domain (Figure 5b and Supplementary Figure 1), suggesting that the DUB activity is not required for inhibition of NF-κB activation. To verify this, we also tested a double point mutant (A20 D100A/C103A), which has no DUB activity32 (Figure 5c and Supplementary Figure 1). A20 D100A/C103A was as effective as wild-type A20 in inhibiting EDAR-induced NF-κB, confirming that the A20 DUB activity is not required for this effect.
Figure 5.
EDA-A1 induces A20 as a negative regulator of EDAR-induced NF-κB activation. (a) Measurement of NF-κB luciferase activity in lysates of HEK293T cells transfected with combinations of an NF-κB-dependent luciferase reporter plasmid and plasmids encoding A20, IKK2 or EDAR. Cells were stimulated with 10 000 U/ml TNF for 4 h. (b) Inhibition of EDAR-induced NF-κB luciferase activity, by different A20 domains. A20-N: DUB domain of A20, A20-C: C-terminal Zn-finger domains. (c) Inhibition of EDAR induced NF-κB luciferase activity by an A20 DUB mutant (A20 D100A/C103A). (d) A20 expression levels were determined by quantitative PCR analysis of E14 EDA−/− skin explants treated with Fc-EDA-A1 for 2 or 4 h. Error bars indicate S.D. ***P<0001. (e) Skin explants were put in hanging drops and left untreated or treated with 250 ng/ml Fc-EDA-A1 for 6 h. Protein extracts were analyzed for A20 expression by western blot
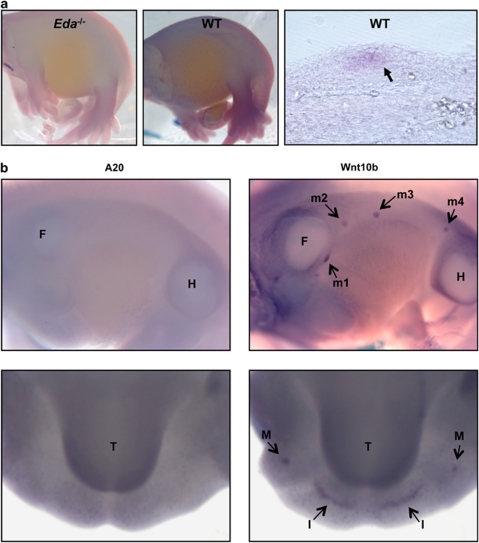
In addition, both A20 mRNA and protein levels were increased after EDA-A1 treatment of embryonic skin explants of Eda-deficient mice (Tabby; hereafter Eda−/−, Figures 5d and e). This indicates that A20 is an NF-κB response gene in the EDA pathway acting in negative feedback regulation of EDAR-induced NF-κB signaling. Interestingly, the expression pattern of A20 in the skin of developing embryos (E14) appears to be limited to the hair placode (Figure 6a), that is, at the sites of EDAR expression and NF-κB activity.4, 33 This indicates that under homeostatic conditions, A20 functions mainly in the hair follicle to prevent overactivation of EDAR. We could not detect A20 in mammary buds or tooth placodes of E12 embryos (Figure 6b). However, it was previously reported that A20 is expressed at later stages of tooth development, as well as in secondary hair follicles and vibrissae.34 In conclusion, these data indicate that the phenotype of the A20EKO mice is due, at least in part, to overactivity of the EDA pathway in the absence of A20.
Figure 6.
EDA-A1 stimulation induces A20 expression in developing hair placodes. (a) Expression of A20 was visualized by whole-mount in situ hybridization on EDA-deficient and WT E14 embryos, dark bluish color indicates a positive in situ signal. The right panel depicts a vibratome section through a hair placode (arrow). (b) Whole-mount in situ hybridization was performed on E12 embryos using an anti-sense probe for A20 (left panels) or Wnt10b (right panels) as a positive control for expression in the tooth placode (upper panels) and mammary bud (lower panels). F, forelimb; H, hindlimb; I, incisor placode; m, mammary bud; M, molar placode; T, tongue
Discussion
Similar to IκBα, A20 is an important negative regulator of NF-κB signaling and inflammation.25 However, their modes of action are totally different. IκBα functions by binding NF-κB directly and sequestering it away from the nucleus, whereas A20 is a ubiquitin-editing enzyme that inactivates multiple targets in the NF-κB pathway.25 IκBα is always present in unstimulated cells to restrain uncontrolled NF-κB activation in the absence of a specific inflammatory trigger. In contrast, in most unstimulated cells, expression of A20 is weak, but it is strongly induced once NF-κB signaling is switched on. As IκBα expression is further induced on NF-κB triggering, both IκBα and A20 are involved in a negative feedback loop of NF-κB activation. The hyperinflammation in IκBα or full A20 null mice, and their early death, point to critical functions of IκBα and A20 in NF-κB regulation.24, 35, 36, 37
Excessive NF-κB signaling in the skin, such as in keratinocyte-specific IκBα-knockout mice and K5-IKK2-transgenic mice, induces skin inflammation.37, 38 Unexpectedly, though A20EKO mice developed epidermal hyperproliferation, they did not develop spontaneous inflammation. The presence of a functional IκBα might be sufficient to control minor inflammatory responses against environmental insults. Alternatively, redundant DUB enzymes that control NF-κB activation, such as CYLD, might be active in the skin. Although CYLD is prominently expressed in the epidermis, CYLD−/− mice do not exhibit skin abnormalities.28 It is possible that ablation of both A20 and CYLD is required for spontaneous severe skin inflammation.
We show that epidermis-specific A20 deletion leads to multiple abnormalities in ectodermal organs, similar to overactivation of the EDA pathway.18, 19, 20, 21, 22, 39 The resemblance of A20EKO mice to mice overexpressing EDA-A1 or EDAR and the inhibition of EDAR signaling by A20 in vitro strongly suggest that hyperactivation of the EDA pathway is the primary cause of the A20EKO phenotype. This conclusion is further supported by our finding that EDA-A1 induces A20 expression and that A20 mRNA co-localizes with Edar and NF-κB signaling activity in developing hair follicles. A20 is known to inhibit NF-κB signaling induced by different signaling pathways, and was shown to depend on the DUB activity of its OTU domain and the ubiquitin-ligating activity of the C-terminal Zn-fingers.40 We could demonstrate that the DUB activity of A20 is not involved in inhibition of EDAR signaling.
Similarities between A20EKO mice and EDA-A1-transgenic mice include nail and hair defects, and sebaceous gland hyperplasia. EDA-A1 transgenics also develop extra teeth and nipples,18 but these abnormalities were never observed in A20EKO mice. It is possible that EDA-A1 signaling is not regulated by A20 at these sites, which correlates with the lack of detectable A20 expression in tooth placodes and mammary buds. It is also possible that formation of supernumerary tooth and mammary buds requires more potent EDAR activity, which can be achieved by excessive ligand production but not by removal of a negative feedback mechanism that attenuates signaling. Another explanation is that K14-Cre-mediated deletion of A20 may have taken place too late to allow induction of ectopic organ primordia, because K14-Cre expression is strongest at E15, whereas induction of tooth and mammary placodes starts at E11 to E12.41
In conclusion, we identified A20 as an EDA-A1-induced protein acting as an inhibitor of EDAR-dependent NF-κB signaling, independent from its DUB activity. As a consequence, genetic deletion of A20 in keratinocytes affects normal epidermal appendage development, similar as observed in transgenic mice overexpressing EDA-A1 or EDAR.
Materials and Methods
A20EKO and Eda−/− mice
Mice with a conditional A20 allele, in which exons IV and V of A20 are flanked with two LoxP sites, were generated as described.26 All experiments were performed on mice backcrossed into the C57BL/6 background for at least five generations. Mice were housed in individually ventilated cages at the VIB Department for Molecular Biomedical Research in a specific pathogen-free animal facility. EDA-deficient mice were purchased from the Jackson Laboratories (Bar Harbor, ME, USA; Tabby, stock #JR0314). All experiments on mice were conducted according to institutional, national and European animal regulations, and were approved by the local Ethics Committee.
Southern blotting
Genomic DNA (10 μg) was digested with BamHI to differentiate between the 6.5-kb and 13.5-kb fragments of A20FL and A20EKO alleles, respectively. DNA was separated in agarose gels and transferred to nitrocellulose. DNA was hybridized with a32 P-labeled probe.
Hair analysis
Hairs from A20FL/FL and A20EKO mice were obtained from the back and examined with a stereomicroscope.
NF-κB Luc reporter assay
Human A20 truncation mutants A20-N (AA 1–376) and A20-C (AA 377–790), and A20 D100A/C103A double mutant, with amino-terminal E-tag were cloned in the pCAGGS expression plasmid. HEK293T cells were transfected with the indicated expression vectors combined with 100 ng of reporter plasmids for NF-κB Luc and pACT-β galactosidase (Gal). After 24 h, the cells were collected, washed in PBS and lysed in Luc lysis buffer (25 mM Tris phosphate (pH 7.8), 2 mM DTT, 2 mM CDTA, 10% glycerol and 1% Triton-X-100). Substrate buffer was added (658 mM luciferin, 378 mM coenzyme A and 742 mM ATP) and Luc activity was assayed in a GloMax 96 Microplate Luminometer (Promega, Leiden, The Netherlands). β-Gal activity in cell extracts was assayed with chlorophenol red β--galactopyranoside substrate (Roche Applied Science, Basel, Switzerland) and the optical density was read at 595 nm in a Benchmark microplate Reader (Bio-Rad Laboratories, Nazareth, Belgium). Luc values were normalized for Gal values to correct for differences in transfection efficiency (plotted as Luc/Gal). The data represent the average±S.D. of triplicates.
Histological and immunohistochemical analysis
Tissues were fixed for 2 h in 4% paraformaldehyde and embedded in paraffin. Sections of 5 μm were processed for immunohistochemical analysis, and anti-Ki-67 (Dako EnVision System, Glostrup, Denmark) or anti-adipophillin was used as primary antibody (Fitzgerald Industries Int., Acton, MA, USA). The primary antibody was detected with the appropriate HRP (DAB) kit (Dako EnVision System). The number of Ki-67-positive and -negative cells in the basal layer was counted. The percentage of Ki-67-positive cells was determined and results are expressed as the mean±S.D. The surface area of the sebaceous gland was measured with Volocity software (Perkin Elmer, Zaventem, Belgium) based on adipophillin staining. The total number of sebaceous glands measured was 139 in four A20FL/FL mice and 319 in five A20EKO mice. Mean surface area±S.E.M. is shown. Statistical significance between experimental groups was assessed using an unpaired two-sample Student's t-test. P-values <0.05 were considered to indicate statistical significance.
Transmission electron microscopy
Mice were anesthetized and then killed by perfusion fixation through the vascular system with 4% paraformaldehyde. After washing, skin samples were post fixed in 1% OsO4 with K3Fe(CN)6 in 0.1 M NaCacodylate buffer (pH 7.2). Samples were dehydrated through a graded ethanol series, including en-bloc staining with 2% uranyl acetate at the 50% ethanol step, and then embedded in Spurr's resin. Ultrathin sections of ∼150 nm were cut using an ultramicrotome (Leica EM UC6, Leica Microsystems, Mannheim, Germany), and collected on formvar-coated copper grids. They were post-stained with uranyl acetate and lead citrate in a Leica EM AC20 autostainer and viewed with a JEOL 1010 transmission electron microscope (JEOL, Tokyo, Japan) at 80 kV and digital images were captured on reusable image plates.
Scanning electron microscopy
Samples of hair for SEM were mounted on specimen stubs with double-sided tape and coated with gold using a Polaron SC 7620 Sputter coater (Quorum Technologies, East Sussex, UK). Samples were viewed with a Hitachi TM-1000 scanning electron microscope (Hitachi High-Technologies Gmbh, Krefeld, Germany) and images were captured directly as digital files.
Skin explant culture and quantitative RT-PCR
Embryonic skin explants were cultured as previously described.42 In brief, E14 EDA−/− back skins were dissected, cut into halves along the midline and cultured for 2 (10 samples) or 4 h (9 samples) in a 30-μl hanging drop of DMEM, 10% FCS, glutamine and penicillin–streptomycin. One-half was supplemented with 250 ng/ml of recombinant Fc-Eda-A1 protein,43 and the control half with PBS, 0.1% BSA. For RNA isolation, skin explants were harvested in 350 μl of Rneasy (Qiagen, Venlo, The Netherlands) lysis buffer as specified by the manufacturer. Dnase-treated RNA (300 ng) was reverse transcribed with Superscript II (Invitrogen, Merelbeke, Belgium), and quantitative PCR was performed in a LightCycler480 (Roche). Transcript number was quantified by comparison with PCR product dilutions of the genes of interest using Lightcycler480 (Roche) software provided by the manufacturer. Expression of each gene was normalized against Ranbp1. The primer sequences were as follows: A20 forward: 5′-AGGCTATGACAGCCAGCACT-3′ A20 reverse 5′-AAACCTACCCCGGTCTCTGT-3′. Statistical significance between experimental groups was assessed using a paired Student's t-test.
In situ hybridization
Embryos were fixed overnight in 4% paraformaldehyde and dehydrated in a methanol series. Whole-mount in situ hybridization was carried out by the InSituPro robots (Intavis AG, Germany) as described earlier.44 The following digoxigenin-labeled probes were used: a 656-bp probe specific for A20 (nt 3223-3879 of NM_009397). A sense probe, used as a negative control, showed no positive signal in any of the hybridizations (data not shown). Some samples were embedded in 0.5% gelatin, 30% albumin, 20% sucrose and 2% glutaraldehyde in PBS and sectioned at 30 μm using a vibratome.
Western blotting
Mouse epidermis was separated from dermis after 15 min of incubation in 3.8% ammonium thiocyanate. Epidermal extracts were prepared using lysis buffer containing 5 mM Hepes, 0.1% NP-40, 10% glycerol, 150 mM NaCl and 5 mM EDTA. Epithelia of the mouse embryos were separated from the mesenchyme as described previously.45 Isolated epithelia were incubated for 6 h in accordance with the hanging drop protocol then harvested in 2% SDS in PBS using syringe and needle. Protein concentration was determined using the BCA Protein Assay kit (Pierce, Rockford, IL, USA) according to the manufacturer instructions. Protein (20 μg) was separated in 10% SDS-PAGE, transferred onto a Hybond-C-extra membrane (Amersham, Uppsala, Sweden), and probed with an anti-A20 antibody (Santa Cruz, Santa Cruz, CA, USA; sc-166692; 1:150) followed by HRP-conjugated anti-mouse secondary antibody (Jackson Laboratories, 1:6000). Blots were developed using enhanced chemiluminescence (SuperSignal West Pico, Thermo Scientific, Asse, Belgium).
Acknowledgments
We thank Dr. A Bredan for editing the manuscript. We are grateful to Dr. Jos Jonkers for sharing the K14-Cre transgenic mice, to Dr. Pascal Schneider for donating recombinant Fc-EDA-A1 and to Ingrid Fliniaux for technical help. We thank Ingo Haase for helpful discussions and suggestions. We are grateful to Dimitri Huyghebaert, Laetitia Bellen and Carine Van Laere for animal care. SL and GvL are postdoctoral researchers with the ‘Fonds voor Wetenschappelijk Onderzoek-Vlaanderen'. LV and KV are PhD fellows with the ‘Instituut voor Innovatie door Wetenschap en Technologie', CM is a PhD fellow with the FWO, and SLe was supported by the Marie Curie Early Stage Training program (FP6) and the Finnish Cultural Foundation. This research has been supported by the Flanders Institute for Biotechnology (VIB), Ghent University and several grants: FWO Odysseus Grant (to GvL), FP6 Integrated Project Epistem LSHB-CT-2005-019067, COST action SKINBAD BM0903, FP6 ApopTrain, MRTN-CT-035624; FP7 EC RTD Integrated Project, Apo-Sys, FP7-200767, ‘Interuniversity Attraction Poles program' (IAP6/18), ‘Belgian Foundation against Cancer', FWO (3G.0218.06, G.0089.10, G.0619.10), ‘Strategic Basis Research program' of the IWT, ‘Centrum voor Gezwelziekten', ‘Concerted Research Actions' and ‘Group-ID Multidisciplinary Research Platform' programs of Ghent University, and Academy of Finland and Sigrid Jusélius Foundation grants (to MLM). PV holds a Methusalem grant (BOF09/01M00709) from the Flemish Government.
Glossary
- CYLD
cylindromatosis
- EDA
ectodysplasin
- EDAR
ectodysplasin receptor
- EDARADD
ectodysplasin receptor-associated adapter protein
- EKO
epidermal knockout
- HED
hypohidrotic ectodermal dysplasia
- IκB
inhibitor of κB α
- IKK
IκB kinase
- NEMO
NF-κB essential modulator
- NF-κB
nuclear factor κB
- TNF
tumor necrosis factor
- TNFAIP3
TNFα-induced protein 3
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Seitz CS, Freiberg RA, Hinata K, Khavari PA. NF-kappaB determines localization and features of cell death in epidermis. J Clin Invest. 2000;105:253–260. doi: 10.1172/JCI7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippens S, Hoste E, Vandenabeele P, Agostinis P, Declercq W. Cell death in the skin. Apoptosis. 2009;14:549–569. doi: 10.1007/s10495-009-0324-z. [DOI] [PubMed] [Google Scholar]
- Pasparakis M, Luedde T, Schmidt-Supprian M. Dissection of the NF-kappaB signalling cascade in transgenic and knockout mice. Cell Death Differ. 2006;13:861–872. doi: 10.1038/sj.cdd.4401870. [DOI] [PubMed] [Google Scholar]
- Pispa J, Pummila M, Barker PA, Thesleff I, Mikkola ML. Edar and Troy signalling pathways act redundantly to regulate initiation of hair follicle development. Hum Mol Genet. 2008;17:3380–3391. doi: 10.1093/hmg/ddn232. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Aebischer T, Hulsken J, Birchmeier W, Klemm U, Scheidereit C. Requirement of NF-kappaB/Rel for the development of hair follicles and other epidermal appendices. Development. 2001;128:3843–3853. doi: 10.1242/dev.128.19.3843. [DOI] [PubMed] [Google Scholar]
- Mikkola ML. TNF superfamily in skin appendage development. Cytokine Growth Factor Rev. 2008;19:219–230. doi: 10.1016/j.cytogfr.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Yan M, Wang LC, Hymowitz SG, Schilbach S, Lee J, Goddard A, et al. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science. 2000;290:523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- Mikkola ML. Molecular aspects of hypohidrotic ectodermal dysplasia. Am J Med Genet A. 2009;149A:2031–2036. doi: 10.1002/ajmg.a.32855. [DOI] [PubMed] [Google Scholar]
- Smahi A, Courtois G, Rabia SH, Doffinger R, Bodemer C, Munnich A, et al. The NF-kappaB signalling pathway in human diseases: from incontinentia pigmenti to ectodermal dysplasias and immune-deficiency syndromes. Hum Mol Genet. 2002;11:2371–2375. doi: 10.1093/hmg/11.20.2371. [DOI] [PubMed] [Google Scholar]
- Headon DJ, Emmal SA, Ferguson BM, Tucker AS, Justice MJ, Sharpe PT, et al. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature. 2001;414:913–916. doi: 10.1038/414913a. [DOI] [PubMed] [Google Scholar]
- Mikkola ML, Thesleff I. Ectodysplasin signaling in development. Cytokine Growth Factor Rev. 2003;14:211–224. doi: 10.1016/s1359-6101(03)00020-0. [DOI] [PubMed] [Google Scholar]
- Monreal AW, Ferguson BM, Headon DJ, Street SL, Overbeek PA, Zonana J. Mutations in the human homologue of mouse dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia. Nat Genet. 1999;22:366–369. doi: 10.1038/11937. [DOI] [PubMed] [Google Scholar]
- Naito A, Yoshida H, Nishioka E, Satoh M, Azuma S, Yamamoto T, et al. TRAF6-deficient mice display hypohidrotic ectodermal dysplasia. Proc Natl Acad Sci USA. 2002;99:8766–8771. doi: 10.1073/pnas.132636999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Pispa J, Hartung AJ, Du Y, Ezer S, Jenks T, et al. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc Natl Acad Sci USA. 1997;94:13069–13074. doi: 10.1073/pnas.94.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I, Mikkola ML. Death receptor signaling giving life to ectodermal organs. Sci STKE. 2002;2002:pe22. doi: 10.1126/stke.2002.131.pe22. [DOI] [PubMed] [Google Scholar]
- Yan M, Zhang Z, Brady JR, Schilbach S, Fairbrother WJ, Dixit VM. Identification of a novel death domain-containing adaptor molecule for ectodysplasin-A receptor that is mutated in crinkled mice. Curr Biol. 2002;12:409–413. doi: 10.1016/s0960-9822(02)00687-5. [DOI] [PubMed] [Google Scholar]
- Cui CY, Durmowicz M, Ottolenghi C, Hashimoto T, Griggs B, Srivastava AK, et al. Inducible mEDA-A1 transgene mediates sebaceous gland hyperplasia and differential formation of two types of mouse hair follicles. Hum Mol Genet. 2003;12:2931–2940. doi: 10.1093/hmg/ddg325. [DOI] [PubMed] [Google Scholar]
- Mustonen T, Pispa J, Mikkola ML, Pummila M, Kangas AT, Pakkasjarvi L, et al. Stimulation of ectodermal organ development by ectodysplasin-A1. Dev Biol. 2003;259:123–136. doi: 10.1016/s0012-1606(03)00157-x. [DOI] [PubMed] [Google Scholar]
- Newton K, French DM, Yan M, Frantz GD, Dixit VM. Myodegeneration in EDA-A2 transgenic mice is prevented by XEDAR deficiency. Mol Cell Biol. 2004;24:1608–1613. doi: 10.1128/MCB.24.4.1608-1613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS, Headon DJ, Courtney JM, Overbeek P, Sharpe PT. The activation level of the TNF family receptor, Edar, determines cusp number and tooth number during tooth development. Dev Biol. 2004;268:185–194. doi: 10.1016/j.ydbio.2003.12.019. [DOI] [PubMed] [Google Scholar]
- Pispa J, Mustonen T, Mikkola ML, Kangas AT, Koppinen P, Lukinmaa PL, et al. Tooth patterning and enamel formation can be manipulated by misexpression of TNF receptor Edar. Dev Dyn. 2004;231:432–440. doi: 10.1002/dvdy.20138. [DOI] [PubMed] [Google Scholar]
- Chang SH, Jobling S, Brennan K, Headon DJ. Enhanced Edar signalling has pleiotropic effects on craniofacial and cutaneous glands. PLoS One. 4;10:e7591. doi: 10.1371/journal.pone.0007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereecke L, Beyaert R, van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 2009;30:383–391. doi: 10.1016/j.it.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Vereecke L, Sze M, Guire CM, Rogiers B, Chu Y, Schmidt-Supprian M, et al. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. J Exp Med. 2010;207:1513–1523. doi: 10.1084/jem.20092474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Ramirez A, Page A, Gandarillas A, Zanet J, Pibre S, Vidal M, et al. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 2004;39:52–57. doi: 10.1002/gene.20025. [DOI] [PubMed] [Google Scholar]
- Zhang M, Brancaccio A, Weiner L, Missero C, Brissette JL. Ectodysplasin regulates pattern formation in the mammalian hair coat. Genesis. 2003;37:30–37. doi: 10.1002/gene.10230. [DOI] [PubMed] [Google Scholar]
- Mou C, Thomason HA, Willan PM, Clowes C, Harris WE, Drew CF, et al. Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Hum Mutat. 2008;29:1405–1411. doi: 10.1002/humu.20795. [DOI] [PubMed] [Google Scholar]
- Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Tobin DJ, Lenhard D, Schneider P, Paus R, Scheidereit C. NF-kappaB transmits Eda A1/EdaR signalling to activate Shh and cyclin D1 expression, and controls post-initiation hair placode down growth. Development. 2006;133:1045–1057. doi: 10.1242/dev.02278. [DOI] [PubMed] [Google Scholar]
- Tewari M, Wolf FW, Seldin MF, O′Shea KS, Dixit VM, Turka LA. Lymphoid expression and regulation of A20, an inhibitor of programmed cell death. J Immunol. 1995;154:1699–1706. [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Baltimore D. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- Klement JF, Rice NR, Car BD, Abbondanzo SJ, Powers GD, Bhatt PH, et al. IkappaBalpha deficiency results in a sustained NF-kappaB response and severe widespread dermatitis in mice. Mol Cell Biol. 1996;16:2341–2349. doi: 10.1128/mcb.16.5.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebholz B, Haase I, Eckelt B, Paxian S, Flaig MJ, Ghoreschi K, et al. Crosstalk between keratinocytes and adaptive immune cells in an IkappaBalpha protein-mediated inflammatory disease of the skin. Immunity. 2007;27:296–307. doi: 10.1016/j.immuni.2007.05.024. [DOI] [PubMed] [Google Scholar]
- Page A, Navarro M, Garin M, Perez P, Casanova ML, Moreno R, et al. IKKbeta leads to an inflammatory skin disease resembling interface dermatitis. J Invest Dermatol. 2010;130:1598–1610. doi: 10.1038/jid.2010.28. [DOI] [PubMed] [Google Scholar]
- Cui CY, Schlessinger D. EDA signaling and skin appendage development. Cell Cycle. 2006;5:2477–2483. doi: 10.4161/cc.5.21.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Mustonen T, Ilmonen M, Pummila M, Kangas AT, Laurikkala J, Jaatinen R, et al. Ectodysplasin A1 promotes placodal cell fate during early morphogenesis of ectodermal appendages. Development. 2004;131:4907–4919. doi: 10.1242/dev.01377. [DOI] [PubMed] [Google Scholar]
- Fliniaux I, Mikkola ML, Lefebvre S, Thesleff I. Identification of dkk4 as a target of Eda-A1/Edar pathway reveals an unexpected role of ectodysplasin as inhibitor of Wnt signalling in ectodermal placodes. Dev Biol. 2008;320:60–71. doi: 10.1016/j.ydbio.2008.04.023. [DOI] [PubMed] [Google Scholar]
- Gaide O, Schneider P. Permanent correction of an inherited ectodermal dysplasia with recombinant EDA. Nat Med. 2003;9:614–618. doi: 10.1038/nm861. [DOI] [PubMed] [Google Scholar]
- Pummila M, Fliniaux I, Jaatinen R, James MJ, Laurikkala J, Schneider P, et al. Ectodysplasin has a dual role in ectodermal organogenesis: inhibition of Bmp activity and induction of Shh expression. Development. 2007;134:117–125. doi: 10.1242/dev.02708. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Pispa J, Jung HS, Nieminen P, Mikkola M, Wang X, et al. Regulation of hair follicle development by the TNF signal ectodysplasin and its receptor Edar. Development. 2002;129:2541–2553. doi: 10.1242/dev.129.10.2541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.