Abstract
Following DNA damage, p53 translocates to the cytoplasm and mitochondria, where it triggers transcription-independent apoptosis by binding to Bcl-2 family proteins. However, little is known about how this exonuclear function of p53 is regulated. Here, we identify and characterize a p53-interacting protein called Hades, an E3 ligase that interacts with p53 in the mitochondria. Hades reduces p53 stability via a mechanism that requires its RING-finger domain with ubiquitin ligase activity. Hades polyubiquitinates p53 in vitro independent of Mdm2 and targets a critical lysine residue in p53 (lysine 24) distinct from those targeted by Mdm2. Hades inhibits a p53-dependent mitochondrial cell death pathway by inhibiting p53 and Bcl-2 interactions. These findings show that Hades-mediated p53 ubiquitination is a novel mechanism for negatively regulating the exonuclear function of p53.
Keywords: p53, ubiquitination, E3 ligase
The tumour suppressor p53 acts as a central switch for inducing apoptosis and cell cycle arrest in response to a variety of cellular signals, including DNA damage and hypoxia. The functions of p53 in the nucleus are known to have a crucial role in cellular homeostasis and organism survival.1, 2, 3, 4 In addition to these, p53 is involved in transcription-independent mechanisms outside the nucleus, collectively known as the exonuclear functions of p53.5 Several lines of evidence support such a role of p53, including p53-dependent cell death in the absence of protein synthesis and gene transcription,6 as well as potent induction of apoptosis by a transcriptionally defective p53 mutant.7, 8 In addition, activation of cytoplasmic p53 in cell-free system induces release of mitochondrial cytochrome C.9 In response to DNA damage, p53 translocates to the cytoplasm and mitochondria,10 and binds cytoplasmic Bcl-xl and Bad11 to inhibit the interaction between pro-apoptotic and anti-apoptotic Bcl-2 proteins, leading to oligomerization of pro-apoptotic Bcl-2 proteins.12, 13, 14
The mechanisms underlying regulation of cytoplasmic p53 and its exonuclear function have been only partially elucidated. Ubiquitination of p53 is thought to regulate its localization as well as its exonuclear function.15, 16 Upon ubiquitination, p53 cannot bind Bax.17 However, deubiquitination of p53 by the ubiquitin-specific protease HAUSP allows p53 to bind to BH3-domain proteins and increases mitochondrial permeability and cell death.17 Thus, p53 ubiquitination inhibits the interaction between p53 and Bcl-2 protein. Together, these observations suggest that ubiquitination has a central role in the exonuclear function and nucleo-cytoplasmic shuttling of p53, as well as in proteasomal degradation. Despite this, the mechanisms that regulate the exonuclear role of p53 remain incompletely understood. Here, we describe a novel ubiquitin-dependent mechanism for regulating the exonuclear role of p53.
Results
Hades is a p53-binding protein
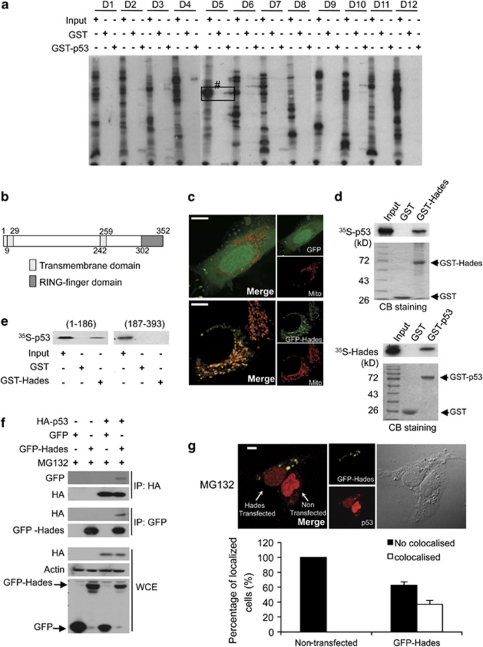
To identify proteins that bind p53, we prepared a human full-length cDNA library derived from HeLa and liver Chang cells. The in vitro transcribed and translated protein pool from the cDNA library was screened for p53-binding proteins using modified SMART technology (Supplementary Figure 1a). After identifying the potential p53-interacting proteins in the protein pool (e.g. pool no. D5 in Figure 1a), the corresponding cDNA pool was progressively subdivided and re-examined in the same manner until a single positive cDNA clone was isolated. Among the isolates, one clone was identified for a major p53-binding partner. This protein, which we have named Hades, contains several predicted functional domains: a transmembrane (TM) domain or a signal peptide at the N-terminus, a second TM domain in the middle of the protein, and a RING-finger domain (i.e. the signature E3 ligase domain) at the C-terminus (Figure 1b). Confocal microscopy revealed that Hades was localized distinctly in the mitochondria (Figure 1c).
Figure 1.
Hades binds p53 in vitro and in vivo. (a) Screening for GST-p53 binding proteins among in vitro translated, 35S-labelled proteins derived from a cDNA library. Bound proteins were detected using autoradiography. The positive protein pool, no. D5, was progressively subdivided until a single positive cDNA clone (Hades) was isolated. Input was 10% of total protein in the binding reaction. (b) Hades contains one transmembrane (TM) domain at the N terminus, one TM domain in the central region, and one RING-finger domain at the C terminus. (c) Confocal microscopic images showing Hades localization in the mitochondria of U2OS cells, which were cotransfected with mitotracker and control GFP (upper panel) or GFP-Hades (lower panel) plasmid. The scale bar represents 10 μm. (d) In vitro pull-down assay shows that Hades interacts with p53. In vitro translated, 35S-labelled p53 interacted with GST-Hades (left panel). In vitro translated, 35S-labelled Hades interacted with GST-p53 (right panel). (e) The N-terminal region of p53 interacts with Hades. An in vitro translated, 35S-labelled N-terminal (aa 1–186) or C-terminal (aa 187–393) fragment of p53 was incubated with GST-Hades, and bound proteins were detected by autoradiography. (f) Hades interacts with p53 in vivo. p53 null H1299 cells were cotransfected with HA-p53 and GFP-Hades expression plasmids. At 24 h after transfection, anti-GFP or anti-HA immunoprecipitates were subjected to immunoblotting using anti-HA or anti-GFP antibody. (g) Hades is colocalized with p53. U2OS cells were transfected with GFP-Hades expression plasmids and, 24 h later, incubated in the presence (lower panel) of MG132 (10 μM) for 4 h. Cells were then fixed, successively probed with p53 antibody and Texas Red-conjugated secondary antibody, and visualized by confocal microscopy. The scale bar represents 10 μm. Statistical results are the average of three independent experiments with 600 cells scored (bottom panel). Error bars represent the means (S.E.M.) of at least three independent experiments
Next, we verified that Hades is a bona fide p53-interacting protein in vitro and in vivo. Pull-down assays revealed that GST-p53 interacted with in vitro translated Hades and GST-Hades interacted with in vitro translated p53 (Figure 1d). To identify the binding site between p53 and Hades, we performed in vitro binding assays using deletion mutants of p53. GST-N-p53 (aa 1–186), in which the C-terminal half of p53 was deleted, associated with immobilized GST-Hades, whereas GST-C-p53 (aa 187–393), in which the N-terminal half of p53 was absent, did not (Figure 1e, lane 6). Binding assays using cell lysates and immobilized Hades demonstrated that endogenous p53 was bound to GST-Hades (Supplementary Figure 1b). The in vivo interaction between Hades and p53 was also detected by coimmunoprecipitation. That is, p53 coimmunoprecipitated with ectopically expressed GFP-Hades in H1299 cells (Figure 1f). Moreover, interaction between endogenous p53 and Hades was still detected in MG132-treated normal breast MCF10A and normal human lung fibroblast (NHLF) cells (Supplementary Figure 1c). We examined the subcellular distribution of Hades and p53 using confocal microscopy. Endogenous p53 was mainly observed in the nucleus of MG132-treated, Hades-non-transfected U2OS cells (data not shown). However, p53 was colocalized with Hades outside of the nucleus, as shown in a merged image (Figure 1g). Quantitation of these results demonstrated that more than 30% of cells were analyzed by the colocalization of p53 and Hades (Figure 1g bottom). Also, colocalization of ectopically expressed p53 and Hades was determined in U2OS cells (Supplementary Figure 1d). Collectively, our data reveal that Hades interacts with p53 both in vitro and in vivo.
Hades reduces the level of p53 through its RING-finger domain with ubiquitin ligase activity
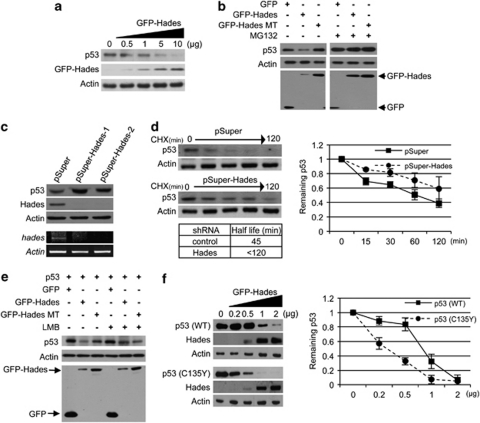
To investigate the possible regulation of p53 by Hades, we determined the effect of Hades on endogenous p53 level in U2OS cells. As shown in Figure 2a, ectopically expressed Hades reduced the p53 level in a dose-dependent manner. This decrease in p53 was also observed in immortalized breast epithelial cells (MCF10A) and normal human lung fibroblast (NHLF) cells (Supplementary Figure 2a).
Figure 2.
Hades reduces the cytoplasmic p53 level. (a) Immunoblot analysis showed that ectopically expressed Hades reduces the level of endogenous p53. U2OS cells were transfected with increasing amounts of GFP-Hades plasmid. At 24 h after transfection, the level of p53 protein was examined by immunoblot analysis. (b) Hades decreases the level of ectopically expressed p53, depending upon its ubiquitin ligase activity. H1299 cells were cotransfected with 1 μg of each indicated plasmid for 24 h and treated with 10 μM MG132 for 4 h. (c) Generation of stable Hades-knockdown MCF7 cells using the shRNA, pSuper-Hades. After selection for 14 days in media containing G418 (500 μg/ml), hades mRNA levels were measured by RT-PCR and the levels of endogenous Hades and p53 proteins were measured by immunoblotting. (d) Ablation of Hades increases p53 stability. Stable Hades-knockdown MCF7 cells were incubated with cycloheximide (CHX, 150 μg/ml) for the indicated times. Endogenous levels of p53 were measured by immunoblotting, while residual cellular levels of p53 were quantitated by densitometry. The half-life of p53 was measured based on the decay of normalized (to Actin control) p53 levels to 50% of their original level. Values are an average of two independent experiments. (e) Hades-mediated degradation of p53 is recovered in the presence of leptomycin B (LMB). H1299 cells were cotransfected with 1 μg of each indicated plasmid. After 24 h, cells were treated with LMB (20 nM), for 4 h (lanes 3–6), and the level of p53 and GFP-Hades was measured by immunoblotting using anti-p53 and anti-GFP antibodies. An anti-Actin antibody was used as a loading control. (f) p53 stability in H1299 cells cotransfected with 0.5 μg wild-type or mutant (C135Y) p53 plasmid and the indicated amounts of GFP-Hades plasmid. At 24 h after transfection, ectopically expressed p53 and GFP-Hades were measured by immunoblotting. In addition, stability was determined as in (d). Values are the average of two independent experiments
We next investigated whether the E3 ligase activity of Hades is involved in mediating this reduction in p53. Thus, an E3 ligase-defective Hades mutant was generated by site-directed mutagenesis at specific residues (C302S/C305S) in the RING-finger domain that are critical for E3 ligase activity (Supplementary Figure 2b).18 Unlike wild-type Hades, the mutant Hades on RING-finger domain failed to reduce the cellular p53 level and Hades function (Supplementary Figure 2c). To gain a more detailed insight into the function of Hades in p53 downregulation, we investigated whether the proteasomal degradation pathway participates in the regulation of p53 stability by Hades. Indeed, we found that exposure of H1299 cells to MG132 restored the p53 levels in the presence of Hades (Figure 2b). Using two different Hades shRNAs, we confirmed that p53 was stabilized by silencing of Hades (Figure 2c). A kinetic analysis of the p53 stabilization induced by Hades silencing in cells treated with cycloheximide (CHX) revealed that the half-life of p53 protein was greater in Hades shRNA-treated cells than in control shRNA-treated cells (Figure 2d). As p53 and Hades colocalized outside of the nucleus (Figure 1g), we further examined whether cytoplasmic localization of p53 is important for the reduction of p53 level by Hades. Treatment with the Crm1 inhibitor leptomycin B (LMB), which blocks nuclear export, restored the level of p53 (Figure 2e). Furthermore, we used a p53 expression plasmid encoding a mutant form of p53 (p53 C153Y) that is localized mainly to the cytoplasm but not to the nucleus.19 Ectopically expressed Hades reduced p53 more effectively in p53 C153Y-expressing H1299 cells than in wild-type p53-expressing H1299 cells (Figure 2f). Together, these data indicate that Hades downregulates cytoplasmic p53 through a ubiquitin-dependent degradation pathway.
Hades promotes polyubiquitination of p53
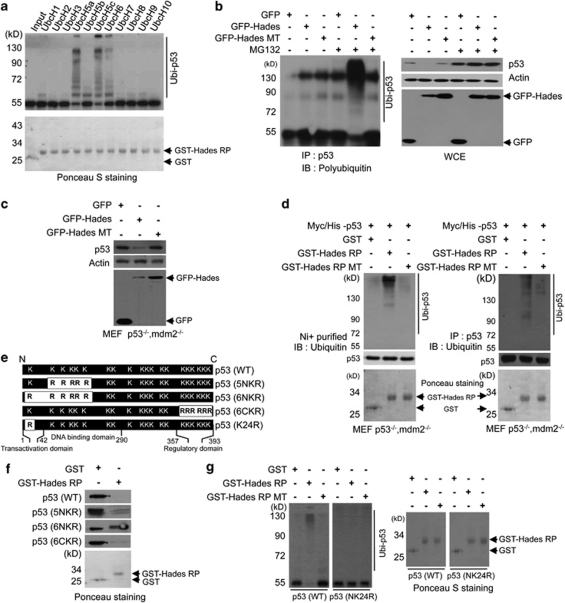
Next, we investigated whether Hades is directly responsible for p53 ubiquitination. Autoubiquitination assay showed that both immobilized full-length (lane 2, GST-Hades) and a RING-finger domain (aa 271–351) (lane 4, GST-Hades Ring protein (RP)) exhibited autoubiquitination, whereas the RING mutant peptide (lane 6 C302S/C305S; GST-Hades RP MT) did not (Supplementary Figure 2b). We then conducted in vitro ubiquitination assays using GST-Hades RP, which was found to interact with p53 protein (Supplementary Figure 3a). We attempted to identify the E2 enzyme that is responsible for the ubiquitination of p53 by Hades, as several E2-conjugating enzymes participate in ubiquitin-mediated degradation.20 In vitro translated, 35S-labelled p53 was incubated with GST or GST-Hades RP in the presence of ATP, purified His-tagged ubiquitin, E1, and one of several E2 enzymes (Figure 3a). Our data demonstrated that the 35S-labelled p53 was polyubiquitinated in the presence of UbcH5a or UbcH5c (lanes 5 and 7). As UbcH5c is usually responsible for general ubiquitin-mediated degradation as well as Mdm2-mediated p53 ubiquitination (Supplementary Figure 3b), we used UbcH5c in further in vitro ubiquitination assays. To confirm a direct role for Hades in p53 ubiquitination, we performed in vitro ubiquitination assay of Escherichia coli-derived recombinant p53 (Supplementary Figure 3c). This experiment verified the involvement of Hades in p53 ubiquitination in vitro in the presence of E1, E2 (UbcH5c) and ubiquitin. Moreover, in vivo ubiquitination assays performed with GFP-Hades and GFP-Hades MT in H1299 cells (Figure 3b) showed that polyubiquitinated p53 was detected only in MG132-treated Hades-transfected cells.
Figure 3.
Hades promotes polyubiquitination of p53. (a) Hades ubiquitinates p53 in vitro. We incubated in vitro translated, 35S-labelled p53 with GST-tagged Hades RING finger peptide (aa 271–351) (GST-Hades RP), ATP, His-ubiquitin, E1, and E2 enzymes. Reaction products were analyzed by autoradiography. GST and GST-Hades RP were measured as controls. (b) Hades ubiquitinates p53 in vivo. p53 null H1299 cells were cotransfected with plasmids for p53 (2 μg) and GFP-Hades or GFP-Hades MT (4 μg). At 24 h after transfection, cells were incubated with or without MG132 (10 μM) for 4 h. Anti-p53 immunoprecipitates were analyzed using anti-polyubiquitin antibody. GFP-Hades and p53 protein levels in whole-cell extracts were measured. (c) Mdm2 is dispensable for Hades-mediated p53 degradation. MEF p53−/− mdm2−/− cells were transfected with 1 μg of indicated plasmid. After 24 h, GFP and p53 levels were analyzed. (d) Hades ubiquitinates p53 in MEF p53−/−mdm2−/− cells. MEF p53−/−mdm2−/− cells were transfected with plasmid for His-p53. At 24 h after transfection, cells were harvested and the lysates were incubated for 2 h with GST, GST-Hades RP, or GST-Hades RP MT. p53 was pulled down with Ni+-conjugated beads (left) or immunoprecipitated using anti-p53 antibody (right). Precipitated proteins were analyzed using anti-ubiquitin antibody. Ponceau S staining of GST, GST-Hades RP, and GST-Hades RP MT were measured as loading controls. (e) Schematic of p53 lysine mutants. 5NKR was mutated at five N-terminal lysine residues (aa 101, 120, 132, 139, and 164), 6NKR at six N-terminal lysine residues (aa 24, 101, 120, 132, 139, and 164), 6CKR at six C-terminal lysine residues (aa 370, 372, 373, 381, 382, and 386), and K24R at one N-terminal lysine residue (aa 24). (f) Hades ubiquitinates p53 at the N-terminal residue, lysine 24. In vitro ubiquitination was assessed using in vitro translated p53 and GST-Hades RP. Naïve proteins in reaction product were analyzed by immunoblotting using anti-p53 antibody. GST and GST-Hades RP were measured as controls. (g) Hades cannot ubiquitinate the p53 NK24R mutant. In vitro ubiquitination was assessed with in vitro translated, 35S-labelled WT or N24KR mutant p53 and GST, GST-Hades RP, or GST-Hades RP MT. Reaction products were separated and analyzed by autoradiography. GST, GST-Hades RP, and RP mutant were measured as controls
We tested whether the mechanism of p53 ubiquitination by Hades differed from that of ubiquitination by Mdm2. Plasmids expressing p53 and GFP-Hades or GFP-Hades MT were transfected in MEF cells lacking both p53 and Mdm2 (MEF p53−/−mdm2−/− cells). We found reduced p53 level in the presence of GFP-Hades in MEF p53−/−mdm2−/− cells (Figure 3c, lane 2). Moreover, in vitro ubiquitination assays demonstrated polyubiquitinated p53 in the presence of GST-Hades RP but not GST-Hades RT MT in MEF p53−/−mdm2−/− cells (Figure 3d). Altogether, these data reveal that Mdm2 is dispensable for ubiquitination of p53 by Hades. Furthermore, synergistic effects on p53 degradation were shown after transfection with Mdm2 and Hades (Supplementary Figure 3d).
Previous reports indicated that six specific lysine residues in the C-terminus of p53 become ubiquitinated by Mdm2.21 Therefore, we proceeded to identify which lysine residue is critical for Hades-mediated degradation of p53 (Figure 3e). Interestingly, the data suggest that none of these lysine residues are necessary for this process as Hades was still able to ubiquitinate a form of p53 in which all six lysines were mutated (6CKR) (Figure 3f). Instead, a comparison of p53 mutants with substitutions at five or six of the N-terminal lysines (5NKR or 6NKR) showed that lysine-24 is a critical residue for stabilization of p53 by Hades (Figure 3f). To examine whether the p53 degradation depends on Hades-induced p53 ubiquitination, we performed in vitro ubiquitination assays with wild-type p53 and mutant p53 in which lysine 24 residue is substituted with arginine (mutant p53 K24R). As expected, mutant p53 K24R was efficiently ubiquitinated by Mdm2 but not by Hades (Figure 3g and Supplementary Figure 3e). These results support the hypothesis that Mdm2 is dispensable for Hades-mediated polyubiquitination of p53 and that the lysine 24 residue in p53 is critical for ubiquitin-dependent degradation.
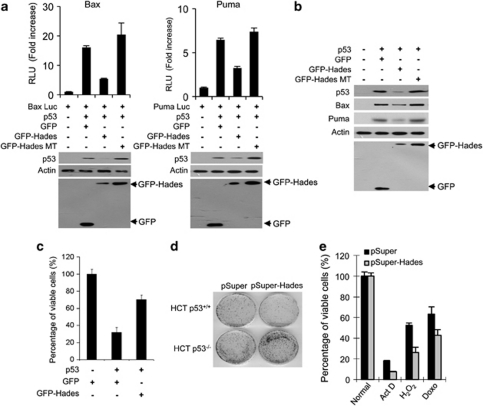
Hades negatively regulates p53-dependent growth suppression. As E3 ligase activities that are necessary for p53 ubiquitination can also regulate p53 function,22, 23 we explored the effect of Hades on p53-dependent growth suppression. First, we investigated whether Hades affects p53-mediated transcriptional activation using luciferase reporter assay (Figure 4a). H1299 cells were cotransfected with plasmids expressing p53, wild-type Hades or mutant Hades, and p53-luciferase reporter driven by p53 response element in the bax (Bax-luc) or puma promoters (Puma-luc). As shown in Figure 4a, p53-mediated transactivation was suppressed by Hades and was dependent on the ubiquitin ligase activity of this enzyme. Moreover, the expressions of endogenous p53 downstream genes Bax and Puma were downregulated by wild-type, but not mutant, Hades (Figure 4b). Our data also indicate that Hades-mediated inhibition of p53 transactivation is caused by the destruction of cytoplasmic p53 as transactivation of p53 was not inhibited by Hades in the presence of LMB (Supplementary Figure 4a). Moreover, Hades-mediated degradation of p53 was independent of its transactivation ability (Supplementary Figure 4b).
Figure 4.
Hades restores p53-dependent growth suppression. (a) Hades suppresses p53-dependent transcriptional activity. H1299 cells were cotransfected with reporter plasmid (Bax-luc or Puma-luc) and plasmids encoding p53, GFP, GFP-Hades, and GFP-Hades MT as indicated. At 24 h after transfection, the relative luciferase activities were measured. Also, the level of p53 proteins was measured using anti-p53 antibody. (b) Hades suppresses endogenous p53 downstream genes. H1299 cells were cotransfected with 1 μg plasmids encoding p53, together with expression plasmid of GFP, GFP-Hades, or GFP-Hades MT. At 24 h after transfection, the protein levels of Puma and Bax were analyzed by immunoblotting. (c) Hades inhibits p53-mediated growth suppression. MTS cell proliferation assays were performed in U2OS cells at 72 h after cotransfection with 1 μg plasmids encoding p53, GFP, GFP-Hades, and GFP-Hades MT as indicated. (d) Effect of Hades expression on the clonogenic growth of HCT116 p53+/+ and HCT116 p53−/− cells, as assessed by colony formation assays. Cells were cotransfected with 1 μg pSuper control plasmid or pSuper-Hades plasmid, further incubated in media containing G418 (500 μg/ml) for 14 days, and stained with crystal violet (0.05%). (e) Hades knockdown reduces cell viability under various stress conditions. Stable Hades-knockdown MCF7 cells (gray bar) and control MCF7 cells (black bar) were exposed to H2O2 (10 μM), actinomycin D (1 μM), or doxorubicin (500 nM) for 24 h, and cell viability was measured using the MTS assay. Error bars represent the means (S.E.M.) of at least three independent experiments
As p53 accumulation leads to an inhibition of cell growth,24 we investigated whether Hades regulates p53-dependent growth suppression using the MTS assay. Ectopically expressed Hades inhibited p53-mediated growth suppression in U2OS cells (Figure 4c) and other various cell lines (Supplementary Figure 4c). We confirmed that Hades inhibits p53-mediated growth suppression using colony-formation assays in H1299 cells (Supplementary Figure 4d). Next, using the RNA interference system, we investigated whether Hades inhibits p53-dependent growth suppression. Wild-type p53-expressing HCT116 p53+/+ and p53 null HCT116 p53−/− cells were transfected with plasmids expressing Hades-targeted shRNA, and colony formation was examined. The data demonstrate that Hades knockdown restored the p53-mediated growth suppression in HCT116 p53+/+ cells but not in HCT116 p53−/− cells (Figure 4d). We found that stable Hades knockdown MCF7 cells were more sensitive to p53-mediated growth suppression induced by several stresses than in control cells (Figure 4e), thus supporting that p53 acts as a central switch to induce growth suppression in response to a variety of cellular signals.1, 2, 3 Collectively, these data suggest that Hades negatively regulates p53-mediated growth suppression and functions as a negative regulator of p53, protecting cells from apoptosis under various stress conditions.
Hades regulates the exonuclear function of p53
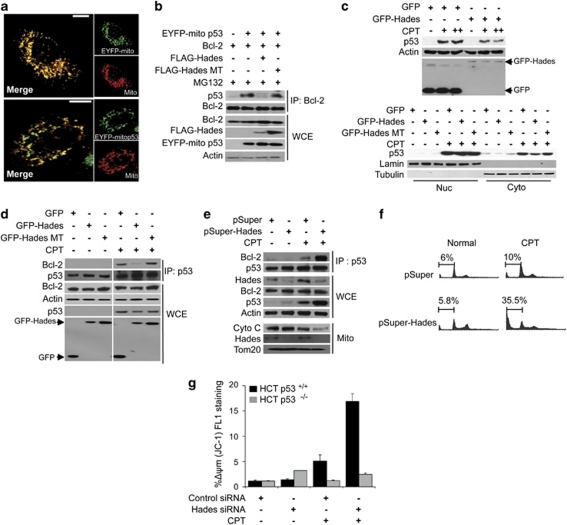
Recent reports have indicated that p53 is distributed throughout the nucleus and cytoplasm under normal conditions.25, 26 Our results showed that Hades is colocalized with p53 in the exonuclear region (Figure 1g). This, taken with the fact that cytoplasmic and mitochondrial p53 triggers stress apoptosis by interacting with the bcl-2 family,5 suggests that Hades interacts with p53 under certain conditions to control p53 function. Therefore, we investigated whether Hades regulates mitochondrial p53 by examining U2OS cells transfected with pEYFP-mito-p53, which encodes p53 coupled to the mitochondria-targeting signal (Figure 5a). Interestingly, ectopically expressed Hades reduced the interaction between pEYFP-mito-p53 and Bcl-2, although the level of p53 is abundant enough as control cells in the presence of MG132 (Figure 5b, lane 3). Moreover, expression of the RING-finger domain-deleted mutant Hades did not affect the interaction between mitochondrial p53 and Bcl-2 (Figure 5b, lane 4). We also tested the effect of Hades on cell proliferation and found that colony formation was reduced by ectopically expressed Hades in pEYFP-mito-p53-transfected H1299 cells (Supplementary Figure 5a). Moreover, clonogenic growth inhibition by ectopically expressed mitochondria localizing mutant p53 (pEYFP-mito-p53 (K24R)) was not attenuated by Hades in p53−/−, Mdm2−/− double-knockout MEF cells (Supplementary Figure 5b). These data suggest that Hades-mediated negative regulation of mitochondrial p53 by ubiquitination results in inhibition of p53 function.
Figure 5.
Hades regulates the exonuclear function of p53. (a) Confocal microscopic images of pEYFP-mito p53 localization in U2OS mitochondria at 24 h after transfection with mitotracker and pEYFP-mito p53. (b) Hades inhibits interaction between p53 and Bcl-2. HEK293 cells were cotransfected with expression plasmids for EYFP-mito-p53, Bcl-2, FLAG-Hades, and FLAG-Hades MT as indicated. The cell lysate was immunoprecipitated with anti-Bcl-2 antibody, then immunoblotted using anti-p53 antibody. (c) Hades attenuates camptothecin (CPT)-induced increases in p53. MCF7 cells were transfected with expression plasmid for GFP-Hades or GFP-Hades RING MT. At 24 h after transfection, cells were treated with CPT (1 or 5 μM (upper), 5 μM (lower)) for 6 h. The p53 protein level was analyzed by immunoblotting in whole-cell extracts (upper) and in nuclear and cytoplasmic fractions (lower). Anti-Lamin and anti-Tubulin antibodies were used as fractionation and loading controls, respectively. (d) Hades inhibits interaction between p53 and Bcl-2 in CPT-treated cells. MCF7 cells were transfected with expression plasmids for GFP-Hades or GFP-Hades MT for 24 h then further incubated with 1 μM CPT for 24 h. Anti-p53 immunoprecipitates were immunoblotted using anti-Bcl-2 and anti-p53 antibodies. (e) Stable Hades knockdown stimulates the interaction between Bcl-2 and p53 following 1 μM CPT treatment for 24 h in MCF7 cells. Immunoprecipitate complexes using anti-p53 antibody were subjected to immunoblotting as in (d) (upper). Whole-cell extracts were probed for p53, Hades, and Bcl-2 (center), whereas mitochondrial fractions were probed for cytochrome c (lower). (f) Hades ablation increases apoptosis among CPT-exposed cells. Stable Hades-knockdown MCF7 cells were exposed to 1 μM CPT for 24 h, stained with propidium iodide, and analyzed by FACS to measure the sub-G0 fraction. (g) siRNA-induced knockdown of Hades promotes CPT-induced mitochondrial damage in a p53-dependent manner. HCT116 p53+/+ and HCT116 p53−/− cells were transfected with 40 nM Hades siRNA or control siRNA, incubated for 24 h, exposed to 1 μM CPT for 24 h, stained with JC-1 for 30 min, and analyzed by flow cytometry. Error bars represent the means (S.E.M.) of at least three independent experiments
As p53 is rapidly translocated to the mitochondria upon camptothecin (CPT) treatment to carry out its exonuclear function,11 we investigated whether Hades prevents the exonuclear function of p53 after CPT treatment. Total intracellular level of p53 was accumulated following CPT treatment in MCF7 cells, and this effect was attenuated by Hades expression (Figure 5c, upper panel). Analysis of the subcellular fractions revealed that, under normal conditions and in the presence of CPT, Hades also reduced the cytoplasmic level of p53 (Figure 5c, lower panel). Moreover, the interaction between p53 and Bcl-2 upon CPT treatment was inhibited by Hades through its ubiquitin ligase activity (Figure 5d). Hades also inhibited the CPT-induced apoptosis (Supplementary Figure 5c). These results were confirmed by examining MCF7 cells transfected with Hades-specific shRNA. Coimmunoprecipitation followed by immunoblot analysis revealed that CPT treatment elicited a greater increase in the level of p53–Bcl-2 complexes in stable Hades knockdown MCF7 cells than in control cells (Figure 5e, lane 3 versus 4). Furthermore, CPT treatment induced a considerable increase in the p53 level and mitochondrial cytochrome c release in the absence of Hades (Figure 5e, lower panel). The negative role of Hades in p53-dependent apoptosis was further supported by results from FACS analysis. More specifically, Hades-knockdown MCF7 cells exhibited a higher percentage of apoptotic cells in the sub-G0 population after CPT treatment than in control cells (Figure 5f). Moreover, JC-1 staining in HCT116 p53+/+ and HCT116 p53−/− cells at 48 h after transfection with Hades or control siRNA showed that CPT induced a detectable loss of mitochondrial membrane potential in the presence of p53 but not in its absence (Figure 5g). Taken together, our data show that Hades inhibits the exonuclear tumor suppressor function of p53 through ubiquitination and subsequent proteasomal degradation of p53.
Discussion
Here, we have identified Hades (UniProtKB accession number Q969V5) as a p53-interacting protein that acts as an E3 ligase for exonuclear p53 and degrades it through the ubiquitin-dependent pathway. Although our study has focused on the functional communications between Hades and p53, other groups have identified Hades as MULAN/MAPL, a mitochondrial protein with a RING-finger domain (Figure 1c) that SUMOylates dynamin-related protein 1 to regulate mitochondrial dynamics.27, 28, 29 As the ubiquitin ligase activity of Hades is critical for its own stability, the ectopic expression of wild-type Hades was lower than that of Hades RING mutant (Figure 2b). The RING domain-deleted mutant form of MULAN was shown to have robust expression in mitochondria, also supporting our observations.27
Our study demonstrates that Hades has a novel function in modulating p53. Immunofluorescence analysis showed that Hades is colocalized with p53 outside of the nucleus (Figure 1g). Furthermore, we showed that ectopic expression of Hades causes degradation of cytoplasmic p53 (Figure 5c, lower panel). These observations are consistent with the hypothesis that ubiquitination of cytoplasmic p53 by Hades is responsible for reducing the p53 level in cytoplasmic and mitochondrial compartments.
Interestingly, we observed that mutant Hades lacking the signal peptide and TM domain (GFP-ΔSP-TM Hades) still induces the p53 degradation in 293T cells (Supplementary Figure 6a). Also, we showed that the RING-finger domain (RP) of Hades can bind to wild-type p53 as well as ubiquitinate it in vitro (Figures 3d and g, and Supplementary Figures 3b and c). These observations suggest that the subcellular localization of Hades is not critical for the degradation of p53.
Although several E3 ligases, including Mdm2, have been reported to target p53,22, 23, 30 we believe that Hades and Mdm2 use distinct mechanisms for the following reasons. First, Mdm2 reduces the nuclear p53 level,31 whereas Hades reduces the cytoplasmic p53 level (Figure 5c, lower panel) and ubiquitinates this tumor suppressor in an Mdm2-independent manner (Figure 3e). Also, Hades inhibits p53 function in Mdm2 null cells (Supplementary Figures 4c and b). Second, ubiquitination by Hades, but not Mdm2, is dependent on lysine 24 residue in p53 (Figure 3g and Supplementary Figure 3e). Third, Hades does not participate in a negative feedback loop. More specifically, neither H2O2 treatment nor ectopic expression of p53 affected the transcriptional expression of Hades (Supplementary Figures 6b and c). It is important to mention that several candidate E3 ligases that target p53, including ARF-bp1 and p300,32, 33 have been reported to be E3 ligases under normal conditions. Therefore, we cannot rule out the possibility that the other E3 ligase pathways may also contribute to Hades-mediated p53 modulation.
Therefore, our data point to a role for Hades in regulating the exonuclear function of p53. Hades inhibited the tumour suppressor function of mitochondrial p53 and interfered with the interaction between p53 and Bcl-2 (Figure 5b). Our data indicate that Hades attenuates CPT-induced accumulation of p53 (Figure 5c) and the interaction between p53 and Bcl-2 (Figure 5d). A previous report has shown that the interaction of p53 with Bcl-2 protein and the subsequent loss of mitochondrial membrane potential are hallmarks of the transcription-independent cell death activated by p53.14 We found that Hades knockdown resulted in mitochondrial cytochrome c release upon CPT exposure (Figure 5e) as well as loss of mitochondrial membrane potential in HCT116 p53+/+ cells, but not in HCT116 p53−/− cells (Figure 5g). Consistent with these findings, CPT treatment led to a greater increase in apoptotic cells in stable Hades knockdown MCF7 cells than in control cells (Figure 5f). Although the defined biochemical pathway underlying p53-induced, transcription-independent cell death remains elusive, our data indicate that Hades can prevent CPT-induced cell death by inducing ubiquitin-dependent proteasome-mediated degradation of p53. This result supports the idea that Hades negatively regulates the exonuclear function of p53.
In conclusion, our study suggests that Hades is an E3 ligase that acts on the tumour suppressor p53. This is the first evidence of Hades-mediated regulation of cytoplasmic and exonuclear p53 function. Hades has a unique role in the exonuclear p53 pathway, acting as a novel regulator of p53-dependent mitochondrial cell death. Thus, the physiological significance of Hades warrants further investigation.
Materials and Methods
Plasmids and construction of a human full-length cDNA library
The Hades gene was isolated from HeLa cell cDNA by RT-PCR and subcloned into vectors. The primers and vectors are as follows: pCITE4 (Novagen, USA; forward: 5′-GGAATTCCATGGAGAGCGGAGGC-3′, reverse: 5′-GGAATTCCTTAGCTGTTGTACAGGGGTATC-3′), pGEX6p3 (Amersham Bioscience, Piscataway, NJ, USA; forward: 5′-GGAATTCCATGGAGAGCGGAGGGCGGC-3′, reverse: 5′-GGAATTCCTTAGCTGTTGTACAGGGGTATC-3′), pEGFP C1 (Clontech, Mountain View, CA, USA; forward: 5′-GGAATTCCATGGAGAGCGGAGGGCGGC-3′, reverse: CGCGGATCCGCGTTAGCTGTTGTACAG-3′), and pcDNA3.1 FLAG vectors (Invitrogen, Carlsbad, CA, USA; forward: 5′-CGGGATCCCGACCATGGAGAGCGGAGG-3′, reverse: 5′-CCGCTCGAGCGTTACTTATCGTCGTCATCCTTGTAATCGCTGTTGTACAGGGG-3′). To generate the RING inactive mutant (C302S/C305S), we performed site-directed mutagenesis using pGEX-Hades RP or pEGFP-Hades and the following primers: C305S, forward: 5′-TGAAGCTGCTCAGACACACTACAGAGGCGCTCTTCAGACTCTCCCTG-3′ and reverse: 5′-CAGGGAGAGTCTGAAGAGCGCCTCTGTAGTGTGTCTGAGCAGCTTCA-3′ or C305S, forward: 5′-CTAAAGAGCGCCTCTGTAGTGTCTCTGAGCAGCTTCAAGTCCTGC-3′ and reverse: 5′-GCAGGACTTGAAGCTGCTCAGAGACAGTACAGAGGCGCTCTTCAG-3′. To generate the signal sequence and TM-deleted form of Hades (GFP-ΔSP-TM Hades), we subcloned from GFP-Hades using two different set of primers. (Hades ΔSP, forward: 5′-GGAATTCCTACTCCGTGTACCGGCAG-3′, reverse: 5′-CGGGATCCCGTTAGCTGTTGTACAG-3′), (Hades ΔTM, forward: 5′-CGCGGATCCCGGAAGCAGTATCTGCAG-3′, reverse: 5′-CGGGATCCGCTCGACTCCTGCCTC-3′). To generate the shRNA expression plasmids targeting p53, we subcloned siRNA oligos into pSuper-GFP-neo vector (Hades-1, forward: 5′-GATCCCCCGUGUGUGUAGAGGACAAATTCAAGAGAUUUGUCCUCUACACACACGTTTTTA-3′, reverse: GGGGCACACACAUCUCCUGUUUAAGTTCTCTAAACAGGAGAUGUGUGUGCAAAAATTCGA-3′ and Hades-2, forward: 5′-GATCCCCGGGAUUUUUAUCUCGAGGCTTCAAGAGAGCCUCGAGAUAAAAAUCCCTTTTTA-3′, reverse: 5′-GGGCCCUAAAAAUAGAGCUCCGAAGTTCTCTCGGAGCUCUAUUUUUAGGGAAAAATTCGA-3′). pcDNA-p53 Myc/His, pEYFP mito-p53, and pEGX-p53 were amplified by PCR and subcloned into pcDNA HA-p53 (pcDNA-p53 Myc/His, forward: 5′-GGAATTCCACCATGGAGGAGCCGCAGT-3′, reverse: 5′-GGAATTCCGCGTCTGAGTCAGGCCCTT-3′ and pEYFP mito-p53, forward: 5′-CGCGGATCCGATGGAGGAGCCGCAG-3′, reverse: 5′-CGCGGATCCCGGTCTGAGTCAGGCCC-3′). pET28b-p53 and p53K24R were amplified by PCR using HA-p53 or HA K24R p53 as templates and subcloned into pET28b vector (pET28b-p53 and p53K24R mutants, forward: 5′-CGCGGATCCGATGGAGGAGCCGCAG-3′, reverse: 5′-CGCGGATCCCGGTCTGAGTCAGGCCC-3′). p53 K24R mutant was generated by point mutagenesis PCR using pcDNA p53-FLAG (forward: 5′-GACCTATGGAGACTACTTCCTG-3′, reverse: 5′-CAGGAAGTAGTCTCCATAGGTC-3′). pcDNA HA-p53 plasmids containing p53 fragments (1–185 and 185–393) were kindly provided by Dr. Gerald M Cohen (Leicester University, UK), p53 mutant (C135Y) by Dr. Carl G Maki (University of Chicago, USA), p53 lysine mutants (N5KR, N6KR, and C6KR) by Dr. Randy YC Poon (Hong Kong University of Science and Technology, Hong Kong), pGL3-Bax reporter plasmid by Dr. Anastasis Stephanou (University College London, UK), pPV-PUMA FLAG2-Luc by Dr. Bert Vogelstein (Johns Hopkins Medical Institutions, USA), and pCI–Bcl-2 by Dr. Hiroyuki Osada (Discovery Research Institute, RIKEN, Japan).
Cell culture and transfection
MCF7, H1299, A549, U2OS, and HeLa cells were purchased from the Korean Cell Line Bank (Korea). MEF p53−/− and MEF p53−/−mdm2−/− cells were kind gifts from Dr. Wei Gu (Columbia University, USA). MCF10A cells were kind gift from Dr. Sujae Lee (Hanyang Unversity, Korea). Stable pSuper-Hades MCF7 cells were maintained in DMEM supplemented with 10% fetal bovine serum and 500 μg/ml G418 (neomycin). siRNA for Hades and mock siRNA were obtained from Ambion (Austin, TX, USA) and had the following sequences: Hades siRNAs, 5′-GGGAUUUUUAUCUCGAGGC-3′ and 5′-CGUGUGUGUAGAGGACAAA-3′ mock siRNA, 5′-AUGAACGUGAAUUGCUCAAG-3′.
Antibodies and reagents
Mouse monoclonal antibodies against p53, tubulin, Bcl-2, HA, Puma, Bax, and ubiquitin; goat polyclonal antibodies against lamin; anti-goat IgG-HRP; normal mouse IgG; anti-mouse IgG-Texas Red; and protein A agarose and protein G agarose were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit polyclonal antibody against Hades was generated using the peptide sequences N-SGERPKGIQETEEM-C and N-SRAKPEDRESLKSAC-C. Anti-β-actin antibody and anti-FLAG antibody were from Sigma (St. Louis, MO, USA); anti-GFP antibody, anti-mouse IgG-HRP and anti-rabbit IgG-HRP from Cell Signaling Technology (Beverly, MA, USA); cytochrome c and Tom 20 antibodies from BD Pharmingen (San Diego, CA, USA); GFP antibody for immunoprecipitation was from Anaspec (Fremont, CA, USA) and polyubiquitin chain antibody from BioMol (Plymouth Meeting, PA, USA). E1, E2 family, and His-ubiquitin were from Boston Biochem (Cambridge, MA, USA); CPT, Actinomycin D, crystal violet, propidium iodine, and DAPI from Sigma; CHX from Biopure (Burlington, Ontario, Canada); MG132 from Calbiochem (La Jolla, CA, USA); the JC-1 staining kit from Cayman Chemical (Ann Arbor, MI, USA); Annexin V from BD Pharmingen; and LMB from Alexis Biochemical (San Diego, CA, USA).
Screening of putative p53-interacting proteins
Pools of human cDNAs from HeLa and liver Chang cells (see Supplementary Information) were transcribed and translated in vitro using the TnT-Coupled Reticulocyte Lysate System (Promega, Madison, WI, USA) in the presence of 35S-methionine (Perkin Elmer, Waltham, MA, USA). When a protein pool was confirmed positive in the in vitro binding assay, the corresponding cDNA pool was subdivided and re-examined in the same manner until a single positive cDNA clone was isolated. Positive clones were sequenced (Bionics, Seoul, Korea) and compared with known sequences by searching the NCBI gene databases.
Ubiquitination assays
For in vitro p53 ubiquitination assays, 1 μl of in vitro translated, 35S-labelled protein was incubated for 2 h at 30°C in 20 μl ubiquitination buffer (25 mM Tris-HCl, pH 7.5; 1 mM DTT; 2 mM ATP; 0.06% NP40; 5 mM MgCl2; and 15 μM ZnCl2) containing 1 μg of the indicated E3 ligase, 150 ng purified E1, 150 ng E2 enzymes, and 10 μg ubiquitin. The reaction products were analyzed by SDS-PAGE, followed by autoradiography or immunoblotting. For endogenous p53 ubiquitination assays, MCF7 cells were harvested, resuspended in ubiquitination buffer, briefly sonicated and centrifuged. Equal amounts of the supernatants were incubated with GST, GST-Hades, or GST-Hades MT for 2 h at 30 °C, and anti-p53 or anti-ubiquitin immunoprecipitates were analyzed by immunoblotting. For p53 ubiquitination assays in MEF p53−/−mdm2−/− cells, lysates were obtained after cells had been transfected with pcDNA myc/His-p53 expression plasmid for 48 h, and reactions were conducted as described above. Reaction products were subjected to immunoprecipitation with anti-p53 antibody or pulled down with Ni+-conjugated beads and then analyzed by immunoblotting. For in vivo p53 ubiquitination assays, HCT116 p53−/− or H1299 cells were transfected with GFP, GFP-Hades, or GFP-Hades RING MT for 48 h and treated with 10 μM MG132 for 4 h. Cells were lysed in SDS-containing lysis buffer and were boiled for 10 min, then diluted 10 times with NP40 lysis buffer for immunoprecipitation with anti-p53 antibody. Anti-p53 immunoprecipitates were then analyzed by immunoblotting using anti-polyubiquitin antibody.
Purification of recombinant proteins
Expression of GST-fused recombinant p53, Hades (WT, FL, RP, and RP MT), and Mdm2 proteins was induced by treating BL21 cells with 0.1 mM IPTG for 2 h at 37°C or overnight at 25°C. Cells were sonicated in bacterial lysis buffer (30 mM Tris-HCl, pH 8; 100 mM NaCl; 0.1 mM EDTA; 1 mM DTT; and 1% NP40), and proteins were purified using glutathione-Sepharose (Amersham Bioscience). Expression of His-tagging p53 and p53 (K24R) mutant was induced by treating BL21 cells with 0.1 mM IPTG for 4 h at 37°C. His-tagging p53 and p53 (mutant) were purified using Ni-NTA beads.
In vitro binding assay
35S-labelled proteins obtained by in vitro transcription/translation were incubated with 5 μg glutathione Sepharose 4B-bound GST, GST-p53, or GST-Hades recombinant proteins for 3 h at 37°C in binding buffer (30 mM Tris-HCl, pH 8.0; 0.1 mM EDTA; 0.1 mM NaCl; 1 mM DTT; 1% NP-40; 0.5 mM PMSF), washed five times, and boiled in SDS-sample buffer (60 mM Tris-HCl, pH 6.8; 25% glycerol; 2% SDS; 14 mM 2-mercaptoethanol; 0.1% bromophenol blue). The bound proteins were separated by SDS-PAGE and visualized by autoradiography.
Immunofluorescence assay
Transfected cells grown on glass coverslips were fixed with 4% paraformaldehyde for 5 min at room temperature, washed twice with PBS, and then incubated in permeabilization buffer (0.5% Triton X-100 in PBS) for 3 min. Cells were blocked with 5% BSA and 2% goat serum in PBS for 30 min, incubated with anti-p53 antibody (1:200 in PBS) overnight at 4 °C, washed three times with PBS, and incubated with fluorescein Texas red-labelled secondary antibodies (1:500 in PBS) for 1 h. Coverslips were inverted, mounted on slides with Vectashield (Vector Laboratories, Burlingame, CA, USA), and fixed with nail polish. Fluorescence was monitored using a confocal laser-scanning microscope (FV-1000 spectral, Olympus, Tokyo, Japan).
Luciferase assay
Aliquots of 5 × 104 cells were seeded in 24-well plates and transfected, for 24 h, with the following: 500 ng of pGL plasmid containing the promoter p53-binding site for Bax or Puma, 100 ng of p53 plasmid, and 200 ng of Hades or Hades MT plasmid. Cells were harvested and cell extracts were prepared using 50 μl Passive lysis buffer (Promega). Luciferase activities were measured using a Biotek synergy HT microplate reader. The relative luciferase activities were normalized to β-galactosidase activities, which were determined using the Luminescent β-galactosidase detection kit II (Mountain View, CA, USA).
Immunoprecipitation and immunoblotting
Cells were lysed with SDS-containing buffer (20 mM Tris-HCl, pH 7.4; 1% SDS; 2 mM EDTA) for 5 min on ice. Lysates were diluted 10-fold with resuspension buffer (20 mM Tris-HCl, pH 7.4; 0.5% NP40; 150 mM NaCl; 2 mM EDTA) and centrifuged at 1200 r.p.m. for 30 min. The supernatants were incubated with antibody overnight at 4 °C, incubated with protein A- or protein G-conjugated agarose for 2 h, washed six times with resuspension buffer, supplemented with SDS-sample buffer, and boiled for 10 min. Immunoblotting was performed as previous described.34
Cell viability assays
For the MTS assay, cells were incubated with CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay reagent (Promega) for 1 h, and the OD was measured at 490 nm. Apoptosis, DNA content, and mitochondrial membrane potential were determined by flow cytometry using annexin V-PE, propidium iodine, and JC-1 staining as previously described.35
Subcellular fractionation
Cells were harvested, rinsed with PBS, pelleted, and resuspended in CLB buffer (10 mM HEPES, 5 mM NaHCO3, 10 mM NaCl, 1 mM CaCl2, 1 mM KH2PO4, 0.5 mM MgCl2, and 5 mM EDTA). Cells were allowed to swell on ice for 5 min, supplemented with NP40 at a final concentration of 0.5%, and incubated for an additional 2 min on ice. The lysate was centrifuged at 1300 r.p.m. for 5 min, and the resulting supernatant was further centrifuged at 12 000 r.p.m. for 30 min. The supernatant from this tube was collected as the cytoplasmic fraction. The crude nuclear pellet was resuspended in 1 ml of CLB buffer and washed three times. Nuclear and cytoplasmic fractions were supplemented with SDS sample buffer. Mitochondrial fractionation was performed as previously described.35 The mitochondrial fraction (pellet) was washed three times, supplemented with SDS sample buffer, boiled for 10 min at 100 °C, and analyzed by immunoblotting.
Colony formation assay
Either 5 × 103 or 1 × 104 HCT 116 p53+/+ or HCT 116 p53−/−, H1299 or p53−/− mdm2−/− MEF cells were cultured in a 60-mm dish and transfected with 500 ng of the indicated plasmid for 24 h. Cells were placed under G418 selection (500 μg/ml) for 14 days.
PCR analysis
RNA was isolated from stable Hades knockdown and control cells using Trizol (Invitrogen), according to the manufacturer's instructions. RNA (2 μg) was reverse transcribed and cDNA equivalent to 100 ng of RNA was used for quantitative RT-PCR. Primers for Hades no. 1 were forward: 5′-CACAAGATGGTGTGGAATCG-3′, reverse: 5′-TCAGCATCTCCTCGGTCTCT-3′, and for β-actin, forward: 5′-GGATTCCTATGTGGGCGACGA-3′, reverse: 5′-CGCTCGGTGAGGATCTTCATG-3′ were used. All PCR reactions were performed according to the manufacturer's instructions for the Taq-polymerase system from TaKaRa (Japan). The amplification products were subjected to agarose gel electrophoresis for visualization.
Statistical analysis
Statistical analysis was performed using the two-tailed Student's t-test. P<0.05 was considered significant.
Acknowledgments
We thank Dr. Wei Gu (Columbia University, USA) for kindly providing MEF p53−/− cells and MEF p53−/− mdm2−/− cells, Dr. Gerald M. Cohen (Leicester University, UK) for pcDNA HA-p53 plasmids, Dr. Carl G Maki (University of Chicago, USA) for p53 mutant (C135Y) plasmid, Dr. Randy YC Poon (Hong Kong University of Science and Technology, Hong Kong) for p53 lysine mutant (N5KR, N6KR, C6KR) plasmids, Dr. Anastasis Stephanou (University College, London, UK) for the pGL3-Bax reporter plasmid, Dr. Bert Vogelstein (Johns Hopkins Medical Institutions, USA) for the pPV-PUMA FLAG2-Luc plasmid, and Dr. Hiroyuki Osada (Discovery Research Institute, RIKEN, Japan) for the pcI-Bcl-2 plasmid. We are grateful to Drs. Hong-Duck Um and Yun-Sil Lee (Korea Institute of Radiological and Medical Sciences, Seoul, Korea) as well as all other members of our research group for their support and advice regarding this study. This work was supported by grants from the Ministry of Education, Science, and Technology (grants 2010-0020372, 2010-0015732, and 2010-0017593 to SA and 2010-0018768 to JHL); the Ministry of Knowledge Economy grant R-2006-1-043 to SA and Y-WJ; and the National R&D Program for Cancer Control, Ministry for Health, Welfare and Family Affairs grant 0720072 to SA.
Glossary
- Mdm2
murine double minute 2
- HEK293
human embryonic kidney 293
- Bcl-2
B-cell lymphoma 2
- RING-finger domain
really interesting new gene-finger domain
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Oren
Supplementary Material
References
- Dulic V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, et al. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Raycroft L, Wu HY, Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990;249:1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AJ, Kokontis JM, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- Schuler M, Bossy-Wetzel E, Goldstein JC, Fitzgerald P, Green DR. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J Biol Chem. 2000;275:7337–7342. doi: 10.1074/jbc.275.10.7337. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- Jiang P, Du W, Heese K, Wu M. The Bad guy cooperates with good cop p53: Bad is transcriptionally up-regulated by p53 and forms a Bad/p53 complex at the mitochondria to induce apoptosis. Mol Cell Biol. 2006;26:9071–9082. doi: 10.1128/MCB.01025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C, et al. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem. 2006;281:8600–8606. doi: 10.1074/jbc.M507611200. [DOI] [PubMed] [Google Scholar]
- Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko ND, Moll UM. The role of ubiquitination in the direct mitochondrial death program of p53. Cell Cycle. 2007;6:1718–1723. doi: 10.4161/cc.6.14.4503. [DOI] [PubMed] [Google Scholar]
- Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- Nie L, Sasaki M, Maki CG. Regulation of p53 nuclear export through sequential changes in conformation and ubiquitination. J Biol Chem. 2007;282:14616–14625. doi: 10.1074/jbc.M610515200. [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- Shaulsky G, Ben-Ze'ev A, Rotter V. Subcellular distribution of the p53 protein during the cell cycle of Balb/c 3T3 cells. Oncogene. 1990;5:1707–1711. [PubMed] [Google Scholar]
- Martinez J, Georgoff I, Martinez J, Levine AJ. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev. 1991;5:151–159. doi: 10.1101/gad.5.2.151. [DOI] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi TR, Zaika A, Moll UM. Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J. 2002;16:420–422. doi: 10.1096/fj.01-0617fje. [DOI] [PubMed] [Google Scholar]
- Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci USA. 2009;106:16275–16280. doi: 10.1073/pnas.0904305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SH, An S, Lee HC, Jin HO, Seo SK, Yoo DH, et al. A truncated form of p23 down-regulates telomerase activity via disruption of Hsp90 function. J Biol Chem. 2009;284:30871–30880. doi: 10.1074/jbc.M109.052720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HO, Seo SK, Woo SH, Lee HC, Kim ES, Yoo DH, et al. A combination of sulindac and arsenic trioxide synergistically induces apoptosis in human lung cancer H1299 cells via c-Jun NH2-terminal kinase-dependent Bcl-xL phosphorylation. Lung Cancer. 2008;61:317–327. doi: 10.1016/j.lungcan.2008.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.