Abstract
Background
Pancreatic stellate cells are emerging as key players in pathophysiopathological processes underlying the development of pancreatic disease, including pancreatitis and cancer. The cells are scarce in the pancreas making their isolation time and resource use consuming.
Methods
Therefore, with the ultimate goal of facilitating mechanistic studies, here we report the isolation, characterization, and immortalization of stellate cell lines from rat and mouse origin.
Results
These cell lines display morphological and molecular markers as well as non-tumorigenic characteristics similar to the frequently used hepatic counterparts. In addition, we have tested their robustness as a model for transcriptional regulatory studies. We find that these cells respond well to TGFβ signaling by triggering a distinct cascade of gene expression, some genes overlap with the TGFβ response of LX2 cells. These cells express several key chromatin proteins and epigenetic regulators involved in the regulation of gene expression, including co-repressors such as Sin3A (short-term repression), HP1 (long-term repression), as well as CBP/p300 (activation). Furthermore, these cells are well suited for Gal4-based transcriptional activation and repression assays.
Conclusions
The cell model reported here may therefore help fuel investigations in the field of signaling, transcription, and perhaps other studies on similarly exciting cellular processes.
Key Words: Pancreatic stellate cells, Cancer, Desmoplasia, Transforming growth factor-β, Transcription factors, Epigenetics
Introduction
The discovery and study of pancreatic stellate cells (PSC) was originally inspired by descriptions on the existence of a similar cell in the liver which mediates fibrotic reconstitution of organ damage [1,2,3,4,5]. Under normal conditions in the liver, these cells act as pericytes, whereas in the pancreas they are located in close proximity to the acini. Besides this difference in location, these cells look and for the most part appear to behave functionally similar, though this topic is an area that warrants further investigation. Therefore, studies on stellate cells are emerging as an exciting and prolific research theme in normal cell biology and the investigation of disease mechanisms, particularly in the liver and pancreas.
In the pancreas, our area of interest, key cell biological roles for stellate cells include extracellular matrix (ECM) remodeling, vitamin A metabolism, and secretory processes [4,6,7,8,9]. During disease, for instance, stellate cells are part of a well-balanced, orchestrated response to injury [5,10,11]. Some symptoms in chronic pancreatitis may have their cellular basis, at least in part, in a paracrine or autocrine signaling cascade that involves stellate cells [5,12]. In pancreatic cancer, stellate cells have recently been identified as an ‘accomplice’ to epithelial tumor cells in the stimulation of their growth and aggressiveness [6,13]. In addition, in a genetic engineered model of pancreatic cancer, a reduction of stellate cells and their accompanying ECM limit the aggressiveness of this disease [14]. Collectively, these studies have significantly fuelled a large number of new investigations in stellate cells.
Currently, however, studies on transcriptional regulation, epigenetics, and chromatin dynamics in stellate cells, which are key to gaining a better understanding of how genes turn on a hierarchical cascade of gene expression that regulates the remodeling of the ECM composition, still remain an underrepresented area of research. Importantly, some of these studies, including our own on signaling and transcriptional regulation, are in need of well-characterized stellate cell models. For this purpose, here we report the development of a novel subset of stellate cell models for mechanistic studies. As a proof of principle, we demonstrate that these cells constitute excellent models for studies on chromatin-mediated transcriptional regulation. Lastly, the combined observations regarding the behavior of these cell lines, as reported here, strongly suggest that these models will be useful for other types of basic and translational studies in pancreatic physiology, pancreatitis, and cancer.
Materials and Methods
Tissue Culture and Reagents
The human hepatic stellate cell line LX2 was obtained as a generous gift from Dr. Steve Freeman (Mount Sinai, N.Y., USA). The human pancreatic cancer cell line, L3.6, originally isolated by Bruns et al. [15], was cultured in appropriate media. Stellate cells were cultured at 37°C and 5% CO2 in complete media, DMEM (Invitrogen) with 10% FBS and antibiotics. For defining doubling times, hepatic and immortalized mouse PSC (imPSC) (1 × 106) were plated and incubated at 37°C for up to 72 h. At various times, cells were trypsinized, trypan blue (Cellgro) stained and counted with a hemacytometer. Bright field images were obtained with a Canon EOS Rebel Xsi camera mounted on a Nikon Eclipse TS100 microscope at 20× magnification.
Stellate Cell Isolation
Primary stellate cells were isolated from rat and mouse pancreas as previously described [1,16]. Briefly, pancreata were excised from anesthetized animals, minced and digested in Grey's balanced salt solution containing 0.1% DNase, 0.05% collagenase and 0.02% pronase. Tissue was digested with agitation for 20–30 min at 37°C, centrifuged and then filtered to remove undigested tissue. Stellate cells were isolated by Accudenz (Accurate Chemical & Scientific Corporation) density gradient centrifugation with the stellate cells, containing intracellular lipid vesicles, located in a cloudy band above the interface. Stellate cells were cultured at 37°C with 5% CO2 in DMEM containing L-glutamine, 10% FBS, antibiotics and antimycotics.
Primary Cell Immortalization
To immortalize rat and mouse PSC, primary cells were incubated with ecotropic retrovirus containing SV40 large T antigen (EcoPack2-293; Clontech) for 24 h at culture conditions. Media and virus were replenished 3–5 times to ensure viral uptake and gene incorporation. Heterogeneous populations of immortalized stellate cells were serially diluted and plated as single cells per well to establish clones. Three independent clones were propagated for each rat and mouse PSC line. Immortalized stellate cells were frozen in cryoprotectant media containing 45% complete media, 50% FBS and 5% DMSO.
Stellate Cell Markers PCR
Total RNA was extracted from cells according to manufacturer's instructions using an RNeasy Kit (Qiagen) with on-column DNase digestion (Qiagen). RNA (2 μg) was converted to cDNA using an oligo (dT) primer and SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) as per manufacturer's protocol. Primers were designed to stellate-specific markers (online suppl. table 1, www.karger.com/doi/10.1159/320540): desmin (DES), glial fibrillary acidic protein (GFAP), smooth muscle actin alpha 2 (ACTA2), collagen type 1a1 (COL1A1), collagen type 2a1 (COL2A1), vimentin (VIM), laminin alpha 1 (LAMA1), nestin (NES). Additional genes tested were hypoxanthine guanine phosphoribosyl transferase (HPRT) and large T antigen (SV40gp6). PCR was performed using Platinum Taq DNA Polymerase (Invitrogen). Cycle conditions were as follows: 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min. Positive bands were visualized on 1.5% agarose with ethidium bromide.
Tumor Formation Assay
Athymic nu/nu mice (n = 2–3) were injected with L3.6 (pancreatic cancer cells), immortalized rat PSC (irPSC), or imPSC clones (5 × 106 cells) resuspended in 50% v/v growth factor reduced, phenol red free Matrigel (BD Biosciences). Tumor growth was monitored for 3 weeks, then animals were sacrificed and tumors excised and measured. Tumor volume (mm3) was estimated from the length (l), width (w), and height (h) of the tumor using the formula: V = (π/6) × (l) × (w) × (h).
Transfection and Viral Transduction
To test exogenous gene expression, stellate cells were transiently transfected with up to 10 μg pEGFP (Clontech). Cells were electroporated in 0.4-cm cuvettes at either 200 V with one 10-ms pulse or 180 V with four 5-ms pulses using an ECM 830 square-wave electroporator (BTX Harvard Apparatus, Holliston, Mass., USA). Cells were also transfected using Lipofectamine (Invitrogen) according to manufacturer's protocols. For viral transduction of stellate cells, 1 × 104 cells were plated on poly-L-lysine-coated coverslips for 12–24 h. Thereafter, cells were incubated for 48 h with retrovirus or adenovirus expressing GFP. Cells were infected with adenovirus, Ad5CMVeGFP [17] University of Iowa, Gene Transfer Vector Core), at a multiplicity of infection of 200:1. Coverslips were mounted and nuclei counterstained with DAPI mounting medium (Vector Laboratories Inc.). Stellate cells were imaged at 10× magnification on the Zeiss Axioskop 40 CFL with a ProgResC3 Jenoptix camera. Cells were counted (100–500 per condition) and evaluated for GFP expression.
Western Blot
Total protein extracts were prepared by lysing cells in Laemmli buffer and sonicating for 10–15 s. Cellular lysates were subjected to 4–20% SDS-PAGE (Lonza) after boiling in Laemmli buffer and separated proteins were transferred to Immobilon-P transfer membrane (Millipore). Membranes were incubated 1 h in blocking solution (Tris-buffered saline solution containing 5% non-fat dried milk and 0.1% Tween-20). After blocking, immunoblotting was performed with specified primary antibodies overnight at 4°C. Primary antibodies were used at the following dilutions: transcriptional regulator SIN3 homolog A (Sin3A) (Santa Cruz Biotech), 1:250; CREB binding protein (CBP) (Santa Cruz Biotech), 1:500; histone deacetylase 1 (HDAC1) (Santa Cruz Biotech), 1:500; chromobox homolog 5 (HP1α) (Abcam), 1:1,000; β-actin (Sigma), 1:1,000. Membranes were then washed three times, re-blocked and incubated with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotech). After three final washes, immune complexes were visualized by using an enhanced chemiluminescence kit (Denville) and resolved on X-ray film.
Luciferase Assay
To confirm activity of transcriptional machinery, stellate cells were transfected with constructs that either activate or repress transcription in a luciferase reporter assay as described previously [18]. Briefly, stellate cells were electroporated with 4 μg of the pGL3 reporter plasmid containing five tandem GAL4 DNA binding domains (DBD) and 8 μg of the pM/GAL4 effector plasmid fused to either herpes simplex virus protein VP16 or chromobox homolog 5 (HP1α) [19,20,21]. Co-transfection with the empty effector vector allowed for normalization to basal transcriptional activity. Following electroporation, cells were allowed to express plasmids for 48 h and then lysed. The Luciferase Reporter Assay was completed according to manufacturer's protocol (Promega) and read using a Turner 20/20 luminometer.
Expression Profiling Using Real-Time PCR Array and Pathway Reconstruction
Hepatic (LX2) and PSC clones (imPSCc1, imPSCc2, imPSCc3) (1 × 106) were plated and serum starved for 12 h prior to 6 h treatment with TGFβ (10 ng/ml) or vehicle (4 mM HCl + 0.1% BSA). Following incubation, cells were lysed and total RNA collected according to manufacturer's protocols (RNeasy, Qiagen). cDNA was synthesized from 1 μg of RNA using the RT2 First Strand kit (SABiosciences). Transcript levels were compared between conditions utilizing SABioscience's pathway-focused Extracellular Matrix and Adhesion PCR Arrays. Briefly, cDNA was mixed with SYBR green qPCR master mix and pipetted into 96-well plates containing gene primers. PCR cycles were completed and SYBR detected with the Bio-Rad CFX96 real-time PCR machine. Using the Bio-Rad CFX manager software, threshold values for the PCR Arrays were set in the lower third of the linear portion of amplification plots. Threshold cycles (Ct) were further analyzed with SABioscience's RT2 Profiler PCR Array Data Analysis Software. Data analysis calculated fold up- and downregulation (ΔΔCt) for each gene in TGFβ-treated cells as compared to vehicle treatment. Each of the three imPSC clones produced similar fold change values, therefore these were considered representative biological replicates (n = 3). The arbitrary cut-off for fold change values considered no change was between −1.5 and 1.5. Pathway reconstruction was completed with Pathway Studio 6.2 (Ariadne Genomics), focusing on direct physical and expression regulation interactions among genes identified in the arrays.
Results
Immortalized Stellate Cells Display Their Characteristic Morphological Features and Express Typical Gene Markers
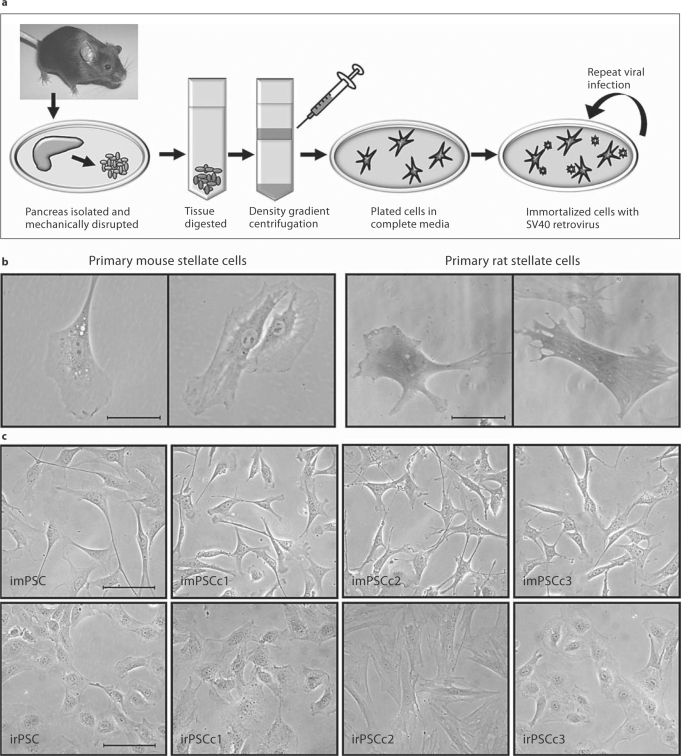
PSC were isolated from excised C57BL/6 mice and Sprague-Dawley rat pancreas by Accudenz gradient centrifugation (fig. 1a). Cells were imaged after 2–5 days in culture, where lipid vesicles were occasionally evident (fig. 1b). Activation of the stellate cells, through repeated culture on plastic [4,22,23], resulted in decreased size and number of lipid vesicles. In an effort to establish immortalized cultures, primary stellate cells were incubated with SV40gp6 containing ecotropic lentivirus and heterogeneous populations of cells were maintained for both mouse and rat pancreas, imPSC and irPSC, respectively (fig. 1c). These rapidly growing cell populations were then serially diluted to establish three cell lines derived from single cells for each mouse and rat PSC, hereafter designated imPSC cloned expansion (imPSCc): imPSCc1, imPSCc2, imPSCc3; and irPSC cloned expansion (irPSCc): irPSCc1, irPSCc2, and irPSCc3 (fig. 1c). Clonal stellate cells have been passaged in culture >50 times over the course of more than 15 months. Additionally, cells remained viable when cryogenically preserved in 5% DMSO and frozen in liquid nitrogen for up to 1 year.
Fig. 1.
Methodology and resultant PSC isolation and immortalization. a As shown, PSC were isolated from rat and mouse. Briefly, the pancreas was isolated, mechanically disrupted, enzymatically digested, and stellate cells isolated by Accudenz gradient centrifugation. b Microscopy images of primary rat and mouse stellate cells, with some cells containing vitamin-A-containing vacuoles. Scale bar represents 50 μm. c To establish immortalized cell lines and clones, primary stellate cells were infected with ecotropic virus containing the SV40 large T antigen. From a heterogeneous population of cells, three clonally expanded cell lines were established for each the mouse and rat pancreas. Phase images of heterogeneous and clonally expanded imPSC and irPSC clones 1, 2, and 3. Scale bar represents 100 μm.
While the PSC had a star-like shape, similar to the highly characterized hepatic stellate cell line (HSC), LX2 [24], the immortalized cells were further characterized for the presence or absence of various stellate cell marker genes. Each population of imPSC and irPSC, heterogeneous and clonally expanded, were characterized by RT-PCR amplification of common stellate cell marker genes (table 1), including desmin (DES), glial fibrillary acidic protein (GFAP), alpha smooth muscle actin (ACTA2), collagens 1a1 and 2a1 (COL1A1, COL2A1), vimentin (VIM), laminin (LAMA1), and nestin (NES). These marker genes were identified in previous publications as commonly expressed in primary and immortalized stellate cell populations, regardless of the originating organ [4,5,6,13,22,25,26]. All genes were confirmed present in the immortalized rat and mouse PSC through transcript analysis (table 1). Hypoxanthine guanine phosphoribosyl transferase (HPRT), a widely used housekeeping gene, was also readily transcribed in these stellate cells. Large T-antigen (SV40gp6) was transcribed by the imPSC and irPSC cells, confirming its integration and thus the immortalization of these PSC. Growth rates among the PSC were compared, with the majority of cell lines doubling in under 48 h (fig. 2a). Specifically, the doubling time in hours was 37 for imPSC, 45 for imPSCc1, 29 for imPSCc2, and 51 for imPSCc3; in comparison, LX2 hepatic stellate cells doubled in 68 h. Rat PSC grow at similar rates and double in approximately 48–72 h (data not shown). The imPSC and clones were also cultured in DMEM with serum concentrations as low as 5% with only a slight reduction in doubling time (data not shown). Therefore, these cells are able to replicate rapidly in vitro and used as a robust model for various studies.
Table 1.
Immortalized PSC expressed well-characterized stellate cell gene markers
| Human |
Immortal mouse PSC |
I mmortal rat PSC |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| HSC (LX2) | imPSC | c1 | c2 | c3 | irPSC | c1 | c2 | c3 | |
| DES | + | + | + | + | + | + | + | + | + |
| GFAP | – | + | + | + | + | + | + | + | + |
| ACTA2 | + | + | + | + | + | + | + | + | + |
| COL1A1 | + | + | + | + | + | + | + | + | + |
| COL2A1 | + | + | + | + | + | + | + | + | + |
| VIM | + | + | + | + | + | + | + | + | + |
| LAMA1 | + | + | + | + | + | + | + | + | + |
| NES | + | + | + | + | + | + | + | + | + |
| HPRT | + | + | + | + | + | + | + | + | + |
| SV40gp6 | – | + | + | + | + | + | + | + | + |
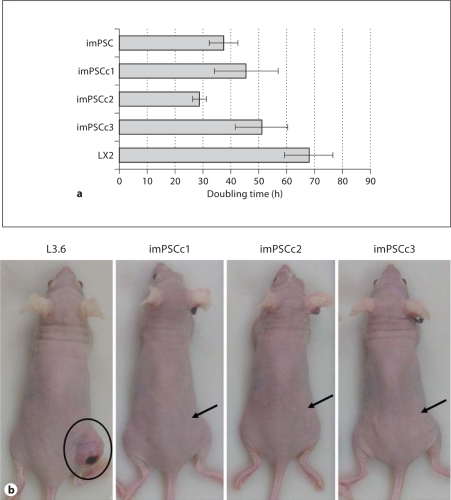
Fig. 2.
PSC lines have distinct in vitro and in vivo growth characteristics. a PSC rapidly divided and doubled every 24–48 h. Hepatic (LX2) and pancreatic (imPSC) stellate cells were plated and incubated for up to 72 h. At 24-hour intervals, cells were trypsinized, trypan blue stained and manually counted with a hemacytometer. Exponential best-fit curves from three independent experiments were averaged and doubling time calculated ± SEM. b Immortalized PSC were unable to establish subcutaneous tumors in nude mice. Athymic nu/nu mice were injected with control L3.6 and immortalized PSC lines and observed after 3 weeks. Tumor formation was evident in L3.6 mice (837.8 mm3) and essentially absent in all imPSC clones (imPSCc1, none; imPSCc2, 9.4 mm3; imPSCc3, none). Arrows indicate injection site on mouse flanks.
Immortalized PSC Do Not Form Tumors in Athymic Nude Mice
To understand the role of the tumor microenvironment in pancreatic cancer, stellate cells must not form tumors on their own when injected as xenografts in mice. Our research with cancer models required us to determine whether the immortalized PSC isolated here were tumorigenic. Athymic nu/nu mice were injected with control, tumor-forming, L3.6 pancreatic cancer cells (n = 2 mice) or imPSC clones (n = 3 mice per clone) resuspended in 50% v/v Matrigel and observed for 3 weeks. Tumor size was measured and images taken of the mice injected with control and imPSC clones (fig. 2b). Mice developed large tumor burden (volume: 837.7 and 282.2 mm3) when injected with L3.6 cells, while most imPSC clones failed to produce tumors. Injection of imPSCc2 cells resulted in small, unremarkable tumors (volume range: 4.7–20.9 mm3) that largely contained unabsorbed Matrigel, while the imPSCc1 and imPSCc3 cells developed no tumor. Similarly, no tumors developed when athymic nu/nu mice were injected with the immortalized rat PSC clones (data not shown). The ability of the imPSC clones to maintain growth in a variety of serum concentrations, have a rapid doubling time, and the lack of tumor formation in nude mice suggest that these cells can be effectively utilized to study the role of the tumor microenvironment and cancer progression. We are particularly interested in the ability of the stellate cells to form tumors when co-injected with a cancer epithelial cell as xenografts in mice.
Immortalized PSC Are Readily Amenable to Gene Transfer
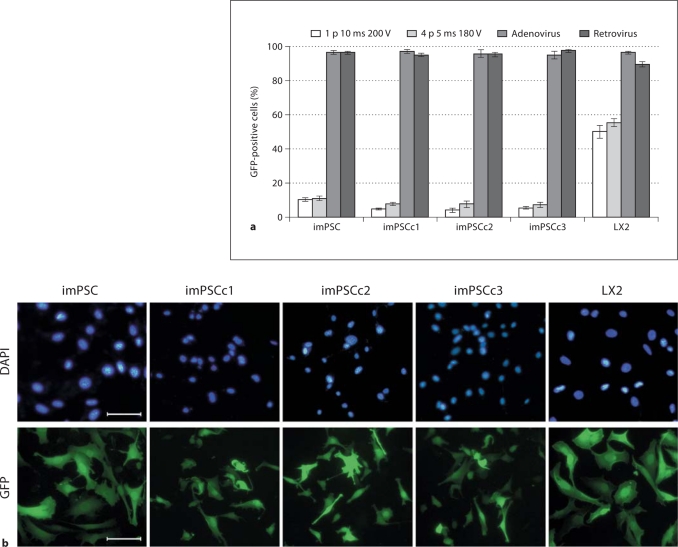
Transfection and transduction are often necessary for the testing and understanding of cellular models, thus we proceeded to calculate the efficiency of the immortalized PSC for expressing exogenously introduced GFP. We tested standard and more advanced methods of gene transfer, including lipid transfection, electroporation, and viral transduction. Gene transfer efficiencies were determined by imaging and counting 100–500 cells in a given condition and determining what percentage of those cells also fluoresced green. We found that stellate cells expressed GFP in all the experimental groups; however, the efficiency and viability of cells varied greatly among groups. For instance, lipid transfections were highly toxic for the heterogeneous imPSC population and the imPSCc3 clonal cells, and efficiency of GFP expression was generally low (<10%, data not shown). Electroporation was tolerated well, with the majority of cells remaining viable. However, the efficiency was again relatively low, with 5–10% of cells expressing GFP (fig. 3a). Finally, the transduction of the immortalized mouse cells with adenovirus and retrovirus was well tolerated and the most efficient method of gene delivery, as measured by GFP expression. Cells transduced with adenovirus or retrovirus containing GFP resulted in greater than 90% of cells expressing GFP after a 48 h infection (fig. 3a, b). Similarly, irPSC transduced with GFP containing adenovirus or retrovirus showed high efficiency of expression and minimal cell death (data not shown). The imPSC clones are able to express GFP and, while the lipid and electroporation transductions were not optimized, there was sufficient evidence that any of the tested methods may provide widespread, high levels of target gene expression.
Fig. 3.
Immortalized mPSC efficiently expressed exogenous GFP after adenoviral and retroviral transductions. Stellate cells were subjected to a variety of transfection and transduction protocols to determine the ability of these cells to express exogenous GFP. Cells expressed GFP 48 h after electroporation, at two different voltages and pulse rates, or incubation with adenovirus or retrovirus. a Cells were counted (100–500 per replicate) and percentage of total population expressing GFP calculated, n ≥ 3 independent replicates. b Representative images of an adenoviral infection of the stellate cells and clones. Scale bar represents 100 μm.
Key Transcriptional and Chromatin Remodeling Pathways Are Intact in Immortalized Stellate Cells
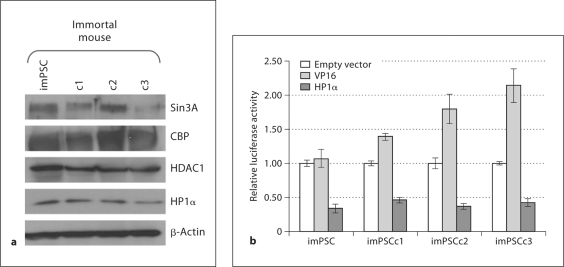
In order to demonstrate that these PSC can constitute a model for studies on chromatin-mediated transcriptional regulation, we set out to confirm the presence and functionality of various transcription-regulating proteins. Lysates from untreated stellate cells demonstrated that all four mouse cell lines expressed key chromatin proteins, including transcriptional regulator SIN3 homolog A (Sin3A), CREB binding protein (CBP), histone deacetylase 1 (HDAC1), and chromobox homolog 5 (HP1α) (fig. 4a), thus suggesting that key chromatin proteins, which act on both activation and repression, were present in these cells. To further confirm the functionality of transcriptional pathways, we used the Gal4-based transcriptional regulatory assay to test for transcriptional activation using VP16 and for repression, HP1α. At 48 h, the PSC were able to enhance (VP16, increased 1.5- to 2-fold) or repress (HP1α, decreased 2-fold) luciferase expression as compared to the empty vector alone (fig. 4b). These cells therefore may be utilized to study the effects of transcriptional regulation on stellate cell function.
Fig. 4.
Immortalized mPSC expressed functional transcriptional machinery and chromatin cofactors. a Lysates were isolated from untreated, immortalized stellate cells and subjected to protein analysis by Western blot. Antibodies to transcriptional machinery and chromatin cofactors (Sin3A, CBP, HDAC1, HP1α) confirm the presence of these proteins in the mouse PSCs; β-actin was completed as a loading control. b The GAL4 DBD alone or fused with VP16 or HP1α was co-transfected into imPSC along with the pGL3 reporter plasmid. Luciferase activity was calculated relative to the empty vector construct and level of repression or activation graphed. Six independent experiments were averaged and error bars represent ± SEM.
Cell Surface Signaling by TGFβ Induces Transcript Changes in Immortalized PSC
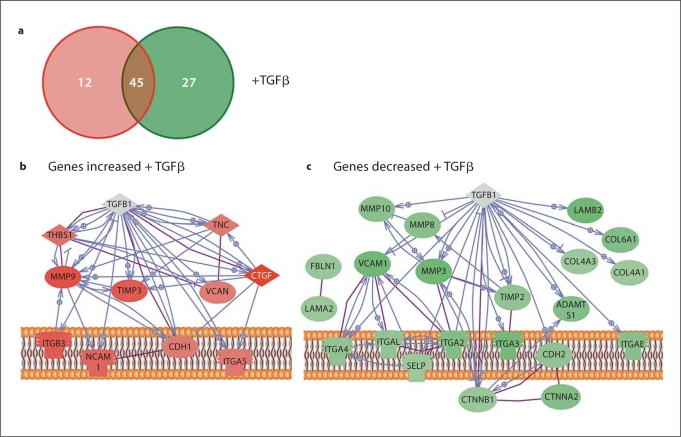
To consider the transcriptional response of the stellate cells to growth factor treatments, we analyzed the transcript levels of ECM and adhesion genes by quantitative RT-PCR (SABioscience). The stellate cells altered transcripts in response to TGFβ as compared to untreated controls. Since the transcriptional profiles of each of the three imPSC clones were relatively similar, the fold change values were calculated as an average of these three independent, biological replicates. Of the 84 genes tested on the array, 14% are upregulated and 32% downregulated following TGFβ treatment (fig. 5a). Genes were considered altered if the fold change, compared to untreated, was >1.5 or <−1.5 (table 2, upregulated; table 3, downregulated). These genes can provide insight into the mechanisms by which TGFβ control matrix deposition and fibrosis in stellate-rich environments. In order to better characterize the stellate cell genes involved in TGFβ signaling, we reconstructed pathways using software that retrieves information from a large variety of databases and can gather proteins into pathways according to physical interactions and or expression regulation (fig. 5b, upregulated; fig. 5c, downregulated). For TGFβ, the stromelysin matrix metalloproteinases (MMPs), MMP3 and MMP10, and a collagenase, MMP8 were downregulated and a gelatinase, MMP9, was upregulated [27]. Tissue inhibitors of MMP (TIMP) transcripts were altered in both directions, with TIMP2 downregulated, and TIMP3 upregulated. The relationships between TGFβ and ECM must be explored further at the protein level to determine if the balance was shifted from matrix deposition to degradation. Although extremely useful as an example, one limitation of this experiment was that we measured gene expression at only one time point after treatment (6 h). Therefore, we might have detected changes in some transcripts that are known targets of a particular pathway, but lost other similarly known target simply because they were expressed at a different time point. Nevertheless, this data serves as a proof of principle for more ambitious expression profiling.
Fig. 5.
Immortalized mPSC altered ECM genes in response to TGFβ. The imPSCs were conditioned with low serum, stimulated with the growth factor TGFβ for 6 h, and the transcript levels of ECM and adhesion genes measured by real-time PCR arrays (SABioscience). a Summary of the number of genes changing was represented by red (transcript increased) and green circles (transcript decreased), with no change indicated in the overlapping region. Pathway diagrams were constructed from the genes increased (b) or decreased (c) following TGFβ treatment of imPSC. In the diagram, direct physical interactions are indicated with purple lines and expression regulation in blue. Genes with no interactions, therefore not diagramed, included: ADAMTS8, EMILIN1, ECM1, SYT1, NCAM2, ADAMTS5, THBS3, and CNTN1.
Table 2.
ECM and adhesion genes with increased transcript following TGFß treatment
| Symbol | Unigene | TGFß |
|---|---|---|
| ctgf | Mm.393058 | 4.69 |
| Mmp9 | Mm.4406 | 2.87 |
| Timp3 | Mm.4871 | 2.57 |
| Itgb3 | Mm.87150 | 2.50 |
| Ncam1 | Mm.4974 | 2.18 |
| Thbs1 | Mm.4159 | 1.82 |
| Itga5 | Mm.16234 | 1.62 |
| Adamts8 | Mm.100582 | 1.59 |
| Tnc | Mm.454219 | 1.58 |
| Vcan | Mm.158700 | 1.53 |
| Emilin1 | Mm.286375 | 1.52 |
| Cdh1 | Mm.35605 | 1.51 |
Table 3.
ECM and adhesion genes with decreased transcript following TGFß treatment
| Symbol | Unigene | TGFβ |
|---|---|---|
| Adamts5 | Mm.112933 | −7.14 |
| Mmp3 | Mm.4993 | −5.22 |
| Vcam1 | Mm.76649 | −3.77 |
| Itga3 | Mm.57035 | −3.33 |
| Lamb2 | Mm.425599 | −3.06 |
| Adamts1 | Mm.1421 | −2.58 |
| Col6a1 | Mm.2509 | −2.57 |
| Itgb2 | Mm.1137 | −2.54 |
| Ctnna2 | Mm.34637 | −2.49 |
| Itgae | Mm.96 | −2.35 |
| Mmp8 | Mm.16415 | −2.34 |
| Syt1 | Mm.289702 | −2.29 |
| Mmp10 | Mm.14126 | −2.18 |
| Timp2 | Mm.206505 | −2.00 |
| Col3a1 | Mm.249555 | −1.95 |
| Cdh2 | Mm.257437 | −1.87 |
| Itga4 | Mm.31903 | −1.86 |
| Itgal | Mm.1618 | −1.84 |
| Thbs3 | Mm.2114 | −1.73 |
| Fbln1 | Mm.297992 | −1.64 |
| Lama2 | Mm.256087 | −1.63 |
| Ncam2 | Mm.433941 | −1.62 |
| Ecm1 | Mm.3433 | −1.59 |
| Ctnnb1 | Mm.291928 | −1.54 |
| Cntn1 | Mm.416876 | −1.53 |
| Selp | Mm.3337 | −1.51 |
| Col4a3 | Mm.389135 | −1.50 |
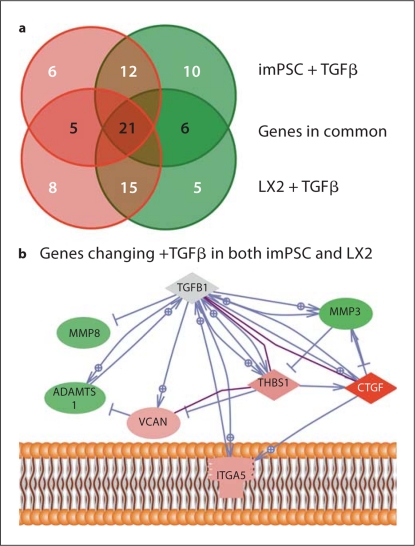
As a second comparison, we analyzed the transcript profile of our imPSC to that of the well-characterized stellate cell line, LX2 [24]. TGFβ and untreated LX2 cells were subject to quantitative RT-PCR, fold change calculated again as a comparison to the untreated cells, and expression profiles compared with those of the imPSC. Since the cell lines differ in source – pancreas vs. liver, and in species – mouse vs. human, some similarities and differences were observed. Of the 60 genes tested on both arrays, 53% shared common regulation (up, down, or no change) between the two cell types (fig. 6a). We considered the genes altered similarly in both imPSC and LX2 cell lines; specifically, 5 genes were increased and 6 genes were decreased to similar levels (table 4). These genes have a variety of established direct interactions and expression regulatory pathways (fig. 6b), thus stellate cells may use common pathways to establish and reorganize the ECM structure and composition in different species and organs.
Fig. 6.
Comparison of ECM genes altered in hepatic and PSC in response to TGFβ treatment. imPSC and LX2 cells were conditioned with low serum, stimulated with TGFβ for 6 h, and the transcript levels of ECM genes measured by real-time PCR arrays (SABioscience). Sixty genes were assayed in common on the human and mouse ECM arrays. a A summary of the number of genes changing was represented by red (transcript increased) and green circles (transcript decreased), with no change indicated in the overlapping region. Genes in common, changing in the same direction in either imPSC or LX2 cells, are indicated in black script within the overlapping circles. b A pathway diagram was constructed from the genes altered similarly following TGFβ treatment of imPSC and LX2 cells. In the diagram, direct physical interactions are indicated with purple lines and expression regulation in blue. Genes with no interactions, therefore not diagramed, included: ECM1, ADAMTS8, ITGB4, and ITGAL.
Table 4.
ECM genes with similarly altered transcript following TGFß treatment of hepatic and pancreatic stellate cells
| Symbol | Mouse | Human | imPSC | LX2 |
|---|---|---|---|---|
| Adamts8 | Mm.100582 | Hs.271605 | 1.59 | 1.97 |
| ctgf | Mm.393058 | Hs.591346 | 4.69 | 1.99 |
| Itga5 | Mm.16234 | Hs.505654 | 1.62 | 1.95 |
| Thbs1 | Mm.4159 | Hs.164226 | 1.82 | 2.61 |
| Vcan | Mm.158700 | Hs.643801 | 1.53 | 1.62 |
| Adamts1 | Mm.1421 | Hs.643357 | −2.58 | −2.26 |
| Ecm1 | Mm.3433 | Hs.81071 | −1.59 | −1.59 |
| Itgal | Mm.1618 | Hs.174103 | −1.84 | −1.71 |
| Itgb4 | Mm.213873 | Hs.632226 | −1.86 | −1.89 |
| Mmp3 | Mm.4993 | Hs.375129 | −5.22 | −2.32 |
| Mmp8 | Mm.16415 | Hs.161839 | −2.34 | −1.50 |
Discussion
The study of PSC has become central to the field of pancreatic cancer research. These cells, which are in close proximity to the acini, secrete vitamin A and other substances that influence the function of acinar cells. Conversely, under certain conditions, exocrine cells secrete a large variety of growth factors and cytokines that exert a function on stellate cells. Therefore, there is a clear interaction between both epithelial cell lines and mesenchymal cell lines with a functional outcome. For instance, during embryonic development, there is a cooperation of stellate cells and developing exocrine cells which influences the final formation of the organ [28,29]. At approximately day 15 in mice, there is an energetic exchange between these cells by which stellate cells secrete factors that influence acini formation and branching morphogenesis [28,29]. In developed tissue, these cells interact again whereby stellate cells secrete factors that modulate the malignancy of pancreatic cancer and perhaps some of the unknown premalignant features of chronic pancreatitis which can ultimately also develop into cancer. Therefore, discovering cellular mediators and characterizing an epithelial-mesenchymal interaction at the molecular levels is of paramount importance for understanding the pathophysiology of several pancreatic diseases.
During the last two decades, humoral mediators that stimulate stellate cell responses have been the focus of most investigations. Among these molecules we find collagens, MMPs, laminins, elastic fibers, fibronectin, and a plethora of growth factors. Unfortunately, however, little is known on how some of these growth factors and ECM proteins cooperate to impart to pancreatic cancer a more or less malignant phenotype. For example, mice overexpressing TGFα and TGFβ in the pancreas develop desmoplasia, however only the TGFα develop cancer [30,31,32,33]. How neoplastic cells interact with the stellate cells to provide the phenotype of this animal is unknown. There is the possibility that desmoplasia does not always stimulate pancreatic cell growth, but rather sometimes, it can act as a tumor suppressor. Therefore, these results suggest that currently unknown differences in the constitution of the fibrogenic response to various factors may dictate the biological outcome of the tumor. Notably, we expect an expansion of this intriguing and medically important area of pancreatic cancer research, where the use of new cells and new animal models will help to explore concepts that may ultimately impact clinical medicine.
Nuclear cell biology, chromatin dynamics, epigenetics, and transcription are the fundamental underlying mechanisms by which stellate cells respond to fibrogenic stimuli. Current models predict that nuclear responses to fibrogenic stimuli will ultimately dictate the composition of the ECM during development and diseases, most conspicuously the composition of the tumor microenvironment [34,35,36]. Unfortunately, there is a paucity of these studies, primarily due to the lack of development of appropriate tools. Thus, in the current manuscript we partially fill the gap in the existent knowledge, by not only developing a new panel of stellate cells, but having validated their significant usefulness for studies on transcription. We showed that these PSC models: (1) contain the morphology and behavior of bona-fide stellate cells, (2) transcribe genes that are commonly ascribed to primary stellate cells, (3) are transfectable and infectable, (4) do not develop tumors in nude mice, which make them ideal for co-culture and co-injection studies with pancreatic cancer cells, (5) can produce reliable transcriptional arrays after treatment with a growth factor, (6) contain chromatin proteins that mediate gene activation and repression, and (7) are useful to perform transcriptional reporter assays for both gene activation and silencing. Thus, the differentiation, presence of typical markers, and chromatin composition and dynamics of gene expression in these cells make them perform optimally in transcriptional regulatory studies, though they may also help other types of cellular and molecular studies not tested here.
Therefore, in summary, strong emerging evidence suggests that pancreatic cancer is a tissue disease where the behavior of cancer cells can be either negatively or positively modulated by the remodeling and composition of the desmoplastic reaction. The tumor microenvironment is rich in different pro- and antitumorigenic molecules which can be manipulated, and in the future, may lend itself to potential advantageous novel therapeutic interventions including those that target transcription, chromatin dynamics, and epigenetics.
Supplementary Material
The sequence of human, mouse, and rat primers used in the characterization of hepatic and pancreatic stellate cells.
References
- 1.Apte MV, Haber PS, Applegate TL, Norton ID, McCaughan GW, Korsten MA, Pirola RC, Wilson JS. Periacinar stellate-shaped cells in rat pancreas: identification, isolation, and culture. Gut. 1998;43:128–133. doi: 10.1136/gut.43.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachem MG, Schneider E, Gross H, Weidenbach H, Schmid RM, Menke A, Siech M, Beger H, Grunert A, Adler G. Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology. 1998;115:421–432. doi: 10.1016/s0016-5085(98)70209-4. [DOI] [PubMed] [Google Scholar]
- 3.Buchholz M, Kestler HA, Holzmann K, Ellenrieder V, Schneiderhan W, Siech M, Adler G, Bachem MG, Gress TM. Transcriptome analysis of human hepatic and pancreatic stellate cells: Organ-specific variations of a common transcriptional phenotype. J Mol Med. 2005;83:795–805. doi: 10.1007/s00109-005-0680-2. [DOI] [PubMed] [Google Scholar]
- 4.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omary MB, Lugea A, Lowe AW, Pandol SJ. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J Clin Invest. 2007;117:50–59. doi: 10.1172/JCI30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrow B, Rowley D, Dang T, Berger DH. Characterization of tumor-derived pancreatic stellate cells. J Surg Res. 2009;157:96–102. doi: 10.1016/j.jss.2009.03.064. [DOI] [PubMed] [Google Scholar]
- 7.McCarroll JA, Phillips PA, Santucci N, Pirola RC, Wilson JS, Apte MV. Vitamin A inhibits pancreatic stellate cell activation: implications for treatment of pancreatic fibrosis. Gut. 2006;55:79–89. doi: 10.1136/gut.2005.064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vonlaufen A, Joshi S, Qu C, Phillips PA, Xu Z, Parker NR, Toi CS, Pirola RC, Wilson JS, Goldstein D, Apte MV. Pancreatic stellate cells: partners in crime with pancreatic cancer cells. Cancer Res. 2008;68:2085–2093. doi: 10.1158/0008-5472.CAN-07-2477. [DOI] [PubMed] [Google Scholar]
- 9.Vonlaufen A, Phillips PA, Xu Z, Goldstein D, Pirola RC, Wilson JS, Apte MV. Pancreatic stellate cells and pancreatic cancer cells: an unholy alliance. Cancer Res. 2008;68:7707–7710. doi: 10.1158/0008-5472.CAN-08-1132. [DOI] [PubMed] [Google Scholar]
- 10.Jaster R. Molecular regulation of pancreatic stellate cell function. Mol Cancer. 2004;3:26. doi: 10.1186/1476-4598-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malhi H, Gores GJ. Cellular and molecular mechanisms of liver injury. Gastroenterology. 2008;134:1641–1654. doi: 10.1053/j.gastro.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reeves HL, Friedman SL. Activation of hepatic stellate cells – a key issue in liver fibrosis. Front Biosci. 2002;7:d808–d826. doi: 10.2741/reeves. [DOI] [PubMed] [Google Scholar]
- 13.Bachem MG, Zhou S, Buck K, Schneiderhan W, Siech M. Pancreatic stellate cells – role in pancreas cancer. Langenbecks Arch Surg. 2008;393:891–900. doi: 10.1007/s00423-008-0279-5. [DOI] [PubMed] [Google Scholar]
- 14.Spector I, Honig H, Kawada N, Nagler A, Genin O, Pines M. Inhibition of pancreatic stellate cell activation by halofuginone prevents pancreatic xenograft tumor development. Pancreas. 2010 doi: 10.1097/MPA.0b013e3181da8aa3. (in press). [DOI] [PubMed] [Google Scholar]
- 15.Bruns CJ, Harbison MT, Kuniyasu H, Eue I, Fidler IJ. In vivo selection and characterization of metastatic variants from human pancreatic adenocarcinoma by using orthotopic implantation in nude mice. Neoplasia. 1999;1:50–62. doi: 10.1038/sj.neo.7900005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramm GA. Isolation and culture of rat hepatic stellate cells. J Gastroenterol Hepatol. 1998;13:846–851. doi: 10.1111/j.1440-1746.1998.tb00747.x. [DOI] [PubMed] [Google Scholar]
- 17.Vasquez EC, Johnson RF, Beltz TG, Haskell RE, Davidson BL, Johnson AK. Replication-deficient adenovirus vector transfer of gfp reporter gene into supraoptic nucleus and subfornical organ neurons. Exp Neurol. 1998;154:353–365. doi: 10.1006/exnr.1998.6917. [DOI] [PubMed] [Google Scholar]
- 18.Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved α-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaczynski J, Zhang JS, Ellenrieder V, Conley A, Duenes T, Kester H, van der Burg B, Urrutia R. The Sp1-like protein BTEB3 inhibits transcription via the basic transcription element box by interacting with mSin3A and HDAC-1 co-repressors and competing with Sp1. J Biol Chem. 2001;276:36749–36756. doi: 10.1074/jbc.M105831200. [DOI] [PubMed] [Google Scholar]
- 20.Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol. 2006;8:407–415. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- 21.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 22.Kordes C, Sawitza I, Haussinger D. Hepatic and pancreatic stellate cells in focus. Biol Chem. 2009;390:1003–1012. doi: 10.1515/BC.2009.121. [DOI] [PubMed] [Google Scholar]
- 23.Masamune A, Shimosegawa T. Signal transduction in pancreatic stellate cells. J Gastroenterol. 2009;44:249–260. doi: 10.1007/s00535-009-0013-2. [DOI] [PubMed] [Google Scholar]
- 24.Xu L, Hui AY, Albanis E, Arthur MJ, O'Byrne SM, Blaner WS, Mukherjee P, Friedman SL, Eng FJ. Human hepatic stellate cell lines, lx-1 and lx-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Algul H, Treiber M, Lesina M, Schmid RM. Mechanisms of disease: chronic inflammation and cancer in the pancreas – a potential role for pancreatic stellate cells? Nat Clin Pract Gastroenterol Hepatol. 2007;4:454–462. doi: 10.1038/ncpgasthep0881. [DOI] [PubMed] [Google Scholar]
- 26.Farrow B, Albo D, Berger DH. The role of the tumor microenvironment in the progression of pancreatic cancer. J Surg Res. 2008;149:319–328. doi: 10.1016/j.jss.2007.12.757. [DOI] [PubMed] [Google Scholar]
- 27.Curran S, Murray GI. Matrix metalloproteinases in tumour invasion and metastasis. J Pathol. 1999;189:300–308. doi: 10.1002/(SICI)1096-9896(199911)189:3<300::AID-PATH456>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Cleaver O, MacDonald RJ. Developmental molecular biology of the pancreas. In: Neoptolemos JP, Urrutia R, Abbruzzese JL, Büchler MW, editors. Pancreatic Cancer. vol 1. New York: Springer Science + Business Media; 2010. pp. 71–118. [Google Scholar]
- 29.Rovasio RA. Development and structure of the pancreas. In: Neoptolemos JP, Urrutia R, Abbruzzese JL, Büchler MW, editors. Pancreatic Cancer. vol 1. New York: Springer Science + Business Media; 2010. pp. 27–38. [Google Scholar]
- 30.Greten FR, Wagner M, Weber CK, Zechner U, Adler G, Schmid RM. TGF-α transgenic mice. A model of pancreatic cancer development. Pancreatology. 2001;1:363–368. doi: 10.1159/000055835. [DOI] [PubMed] [Google Scholar]
- 31.Jhappan C, Stahle C, Harkins RN, Fausto N, Smith GH, Merlino GT. TGF-α overexpression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell. 1990;61:1137–1146. doi: 10.1016/0092-8674(90)90076-q. [DOI] [PubMed] [Google Scholar]
- 32.Lohr M, Schmidt C, Ringel J, Kluth M, Muller P, Nizze H, Jesnowski R. Transforming growth factor-β1 induces desmoplasia in an experimental model of human pancreatic carcinoma. Cancer Res. 2001;61:550–555. [PubMed] [Google Scholar]
- 33.Sanvito F, Nichols A, Herrera PL, Huarte J, Wohlwend A, Vassalli JD, Orci L. TGF-β1 overexpression in murine pancreas induces chronic pancreatitis and, together with TNF-α, triggers insulin-dependent diabetes. Biochem Biophys Res Commun. 1995;217:1279–1286. doi: 10.1006/bbrc.1995.2906. [DOI] [PubMed] [Google Scholar]
- 34.Abbott DE, Bailey CM, Postovit LM, Seftor EA, Margaryan N, Seftor RE, Hendrix MJ. The epigenetic influence of tumor and embryonic microenvironments: how different are they? Cancer Microenviron. 2008;1:13–21. doi: 10.1007/s12307-008-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingber DE. Cancer as a disease of epithelial-mesenchymal interactions and extracellular matrix regulation. Differentiation. 2002;70:547–560. doi: 10.1046/j.1432-0436.2002.700908.x. [DOI] [PubMed] [Google Scholar]
- 36.Lunt SJ, Chaudary N, Hill RP. The tumor microenvironment and metastatic disease. Clin Exp Metastasis. 2009;26:19–34. doi: 10.1007/s10585-008-9182-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequence of human, mouse, and rat primers used in the characterization of hepatic and pancreatic stellate cells.