Abstract
Background/Aims
Antibodies against infectious pathogens provide information on past or present exposure to infectious agents. While host genetic factors are known to affect the immune response, the influence of genetic factors on antibody levels to common infectious agents is largely unknown. Here we test whether antibody levels for 13 common infections are significantly heritable.
Methods
IgG antibodies to Chlamydophila pneumoniae, Helicobacter pylori, Toxoplasma gondii, adenovirus 36 (Ad36), hepatitis A virus, influenza A and B, cytomegalovirus, Epstein-Barr virus, herpes simplex virus (HSV)-1 and −2, human herpesvirus-6, and varicella zoster virus were determined for 1,227 Mexican Americans. Both quantitative and dichotomous (seropositive/seronegative) traits were analyzed. Influences of genetic and shared environmental factors were estimated using variance components pedigree analysis, and sharing of underlying genetic factors among traits was investigated using bivariate analyses.
Results
Serological phenotypes were significantly heritable for most pathogens (h2 = 0.17–0.39), except for Ad36 and HSV-2. Shared environment was significant for several pathogens (c2 = 0.10–0.32). The underlying genetic etiology appears to be largely different for most pathogens.
Conclusions
Our results demonstrate, for the first time for many of these pathogens, that individual genetic differences of the human host contribute substantially to antibody levels to many common infectious agents, providing impetus for the identification of underlying genetic variants, which may be of clinical importance.
Key Words: Pathogen, Infection, Antibody, Serology, Genetics, Heritability, Mexican Americans
Introduction
It is not always possible or convenient to determine the presence of a specific pathogen by direct means such as the presence or absence of DNA (or RNA) from a given pathogen within the host organism. One of the reasons is that many infections can become dormant over long periods of time, with the pathogen nonetheless persisting, often unnoticeably, within the body in specific, sometimes unknown and often inaccessible tissues. At this stage, microbial nucleic acids may be undetectable in blood samples. However, testing for the presence of antibodies directed against a pathogen is a commonly used and in many instances robust approach to establish exposure to an infectious agent. Antibodies can provide information regarding current infection as well as dormant and/or previous infections.
In many studies, the presence or absence of antibodies to a given pathogen is used as a proxy for exposure (i.e. seropositive individuals are assumed to be exposed and seronegative individuals are assumed to be unexposed). Levels of antibodies are often used to define exposures within populations and to compare exposures across different populations, with the assumption that the differences in antibody levels are reflective largely of differences in exposures. While this is adequate and appropriate for many types of studies, it is often not explicitly acknowledged that innate factors of the host organism may also influence the presence or absence, as well as the amount, of antibodies to a pathogen. Antibody levels may be partially genetically controlled, but little is known about the heritabilities (h2) of antibody levels in humans. Heritability studies are mainly limited to the genetic contribution to antibody response to vaccines [1,2,3]. Examples of heritability estimates of antibody response to naturally occurring infections are few and include studies of Helicobacter pylori (h2 = 0.57, [4]), malaria (h2 = 0.31–0.77, [5]), Trypanosoma cruzi seroprevalence (h2 = 0.56, [6]), and Epstein-Barr virus anti-viral capsid antigen IgG (h2 = 0.32–0.48, [7]).
The main focus of this paper is to test whether and to what degree genetic factors, measured in the aggregate as heritability, influence the serostatus and antibody level of individuals to 13 common infectious pathogens. We also provide estimates of the effect of shared environmental factors for these pathogens, as well as genetic correlations among pathogens. The pathogens were chosen in part based on suspected involvement in chronic inflammatory diseases including atherosclerosis. Infectious agents included two bacterial pathogens: Chlamydophila pneumoniae and Helicobacter pylori; one protozoan: Toxoplasma gondii; six herpesviruses: cytomegalovirus (CMV), Epstein-Barr virus (EBV), herpes simplex type I virus (HSV-1), herpes simplex type II virus (HSV-2), human herpesvirus 6 (HHV-6) and varicella zoster virus (VZV); and four other viruses: adenovirus 36 (Ad36), hepatitis A virus (HAV), influenza A virus, and influenza B virus.
Methods
Study Population
This study included 1,227 members of randomly ascertained, extended Mexican American families from around San Antonio, Tex., USA. They were recruited during the time period 1991–1995 for participation in the San Antonio Family Heart Study (SAFHS), which seeks to identify genetic risk factors for cardiovascular disease [8]. 482 participants were men and 745 were women, ranging in age from 15 to 94 years. The Institutional Review Board at the University of Texas Health Science Center at San Antonio approved the study and protocols, and the study participants signed statements of informed consent. Information of relevance to this paper collected on study participants included sex, age, and cohabitation.
Some findings were replicated in 648 participants from the San Antonio Family Diabetes/Gallbladder Study (SAFDGS), which seeks to investigate the genetic influences underlying type II diabetes mellitus, and more recently, gallbladder disease. This study is of similar design to the SAFHS, as it also focuses on Mexican American families from around San Antonio. However, the SAFDGS is enriched for diabetics as ascertainment occurred via a single diabetic proband in each pedigree [9,10].
Serology
At the time of recruitment (1991–1995), blood samples from participants were collected into EDTA vacutainers following an overnight fast. At that time, plasma samples were aliquoted and frozen as previously described [11], and stored at −80°C. These archived samples were later thawed just prior to use for IgG antibody level determinations, using commercially available ELISA kits for the following pathogens: C. pneumoniae (Bioclone Australia Pty Ltd., Marrickville, N.S.W., Australia); H. pylori and CMV (Inverness Medical Professional Diagnostics, Palatine, Ill., USA); T. gondii, EBV (specifically to EBV nuclear antigen 1 [EBNA-1]), VZV, influenza A, and influenza B (IBL America, Minneapolis, Minn., USA); HSV-1 and HSV-2 (Focus Diagnostics Inc., Philadelphia, Pa., USA); HAV (Bio-Rad Laboratories, Redmond, Wash., USA); and HHV-6 (Advanced Biotechnologies, Rockville, Md., USA). Ad36 antibodies were measured using a published serum neutralization test [12]. Dichotomous serostatus phenotypes (seropositive/seronegative) were determined according to the manufacturers’ instructions, based on the following absorbance values: seropositive if ≥1.1, seronegative if ≤0.9, and those in between classified as seroindeterminate (i.e. phenotypically unknown). For Ad36, samples were run in duplicate, with those having both neutralization levels ≥1:8 considered seropositive, otherwise they were classified as seronegative.
Statistical Methodology
For each pathogen, the quantitative antibody level and discrete serostatus (seronegative/seropositive) traits were analyzed. Heritability (h2), which we define here as aggregate additive autosomal genetic effects on the serological phenotypes, was estimated using a variance components approach implemented in the SOLAR software package [13]. Because variance components analyses are sensitive to non-normality, particularly high kurtosis, all quantitative antibody traits were transformed using an inverse normalization (rank-based normalization) prior to analysis, ensuring that the phenotype is distributed like a standard normal variable. For the analysis of discrete serostatus traits within the variance components framework, a threshold liability model was used (i.e. discrete traits were assumed to represent the readout of an unobservable underlying quantitative liability, with individuals above a threshold considered to be seropositive, and those below considered seronegative) [14,15]. Covariates used in all analyses included sex, age, age2, sex × age interaction, and sex × age2 interaction, as we have previously shown the substantial influence, especially of age, on antibody levels [16]. In discrete trait analysis, the covariates lead to individual-specific liability thresholds beyond which an individual is declared to be seropositive. The influence of shared environmental factors was modeled using a ‘household’ random effects component, based on information on co-habitation of family members at the time of sample collection [17]. Individuals residing in the same house were assumed to potentially share environmental exposures with one another, while absence of shared exposure was assumed for individuals in different residences. To determine whether there is genetic overlap between the pathogens in terms of host susceptibility to infection, bivariate analyses [18,19] were conducted for each pair of infectious agents, for both the quantitative and discrete traits. The resulting genetic correlation matrix for all pairs of pathogens was subjected to principal components analysis in R (using the function princomp).
Results
Heritability
In this study we examined antibody levels to 13 common pathogens among 1,227 randomly ascertained Mexican American participants of the SAFHS. Study participants represented 45 pedigrees with up to 95 phenotyped individuals, and up to 5 generations (table 1). The average kinship size was 3.2, ranging in size from 2 to 11 siblings. Participants resided in 658 different households, 289 of which had more than one phenotyped individual (with an average household size of 3.3 individuals). Infection seroprevalence ranged from 9% seropositive for T. gondii to 92% for VZV, as previously described (table 2).
Table 1.
Information on pedigree relationships and households for study participants
| Pedigree information | |
| Number of pedigrees | 45 |
| Maximum number of generations | 5 |
| Size of largest pedigree, n | 95 |
| Average sibship size (range) | 3.2 (2–11) |
| Familial relationships, observed pairs | |
| Parent-offspring | 1,034 |
| Monozygotic twins | 3 |
| Full siblings | 1,143 |
| Half siblings | 194 |
| Grandparent-grandchild | 357 |
| Avuncular | 2,411 |
| Half avuncular | 379 |
| First cousins | 2,608 |
| Household information | |
| Number of households | 658 |
| Average household size (range) | 1.9 (1–14) |
| Number of households >1 individual | 289 |
| Average household size for households >1 individual (range) | 3.3 (2–14) |
| Number of spousal pairs residing in same household | 115 |
Table values represent only study participants (n = 1,227), actual pedigree sizes, pairs of familial relationships and household sizes may actually be larger due to presence of unphenotyped relatives/co-inhabitants.
Table 2.
Seroprevalence estimates for pathogens examined in this study
| Pathogen | Seropositive, % | Seronegative, % | Seroindeterminate, % |
|---|---|---|---|
| C. pneumoniae | 85.7 | 8.8 | 5.5 |
| H. pylori | 57.1 | 38.7 | 4.2 |
| T. gondii | 9.2 | 88.0 | 2.8 |
| CMV | 55.4 | 38.2 | 6.4 |
| EBV | 46.4 | 30.1 | 23.5 |
| HSV-1 | 81.7 | 16.8 | 1.5 |
| HSV-2 | 21.7 | 77.1 | 1.2 |
| HHV-6 | 81.6 | 11.1 | 7.3 |
| VZV | 92.2 | 1.9 | 5.9 |
| Ad36 | 13.4 | 86.6 | 0.0 |
| HAV | 73.1 | 26.8 | 0.1 |
| Influenza A | 76.6 | 10.8 | 12.6 |
| Influenza B | 57.5 | 23.5 | 19.0 |
Using a variance components model allowing simultaneously for the influence of aggregate additive genetic effects (with expected sharing of genetic material estimated from pedigree relationships) and shared environmental influences (based on presence or absence of co-habitation of study participants), and after accounting for the influence of age and sex, the quantitative antibody levels to all pathogens were found to have a significant heritable component at significance level 0.05, with the exceptions of HSV-2 and Ad36 (table 3). The maximum heritability estimate was 39% for CMV. All significant heritabilities remain so after Bonferroni correction for the studied number of pathogens (data not shown).
Table 3.
Heritability estimates (with standard error, SE) for quantitative antibody level traits and discrete serostatus traits, including household effects
| Pathogen | Quantitative antibody level traits |
Discrete serostatus traits |
||||||
|---|---|---|---|---|---|---|---|---|
| h2 ± SE | p value (h2) | c2 ± SE | p value (c2) | h2 ± SE | p value (h2) | c2 ± SE | p value (c2) | |
| C. pneumoniae | 0.33 ± 0.06 | 2.7 × 10−11 | 0.06 ± 0.04 | 0.06 | 0.62 ± 0.18 | 2.1 × 10−5 | 0.10 ± 0.10 | 0.16 |
| H. pylori | 0.35 ± 0.06 | 6.0 × 10−15 | 0 | 0.50 | 0.45 ± 0.10 | 3.4 × 10−9 | 0 | 0.50 |
| T. gondii | 0.21 ± 0.05 | 1.3 × 10−6 | 0.11 ± 0.04 | 1.3 × 10−3 | 0.41 ± 0.94 | 1.3 × 10−3 | 0.01 ± 0.75 | 0.47 |
| CMV | 0.39 ± 0.06 | 1.3 × 10−14 | 0 | 0.50 | 0.60 ± 0.11 | 5.2 × 10−11 | 0 | 0.50 |
| EBV | 0.38 ± 0.06 | 2.7 × 10−12 | 0.05 ± 0.04 | 0.08 | 0.57 ± 0.11 | 2.1 × 10−8 | 0.09 ± 0.08 | 0.13 |
| HSV-1 | 0.17 ± 0.06 | 1.7 × 10−4 | 0.15 ± 0.05 | 3.6 × 10−4 | 0.22 ± 0.13 | 0.02 | 0.05 ± 0.09 | 0.28 |
| HSV-2 | 0.08 ± 0.05 | 0.05 | 0.21 ± 0.05 | 9.0 × 10−7 | 0 | 0.50 | 0.32 ± 0.09 | 1.4 × 10−4 |
| HHV-6 | 0.28 ± 0.06 | 8.2 × 10−10 | 0.10 ± 0.04 | 3.0 × 10−3 | 0.11 ± 0.13 | 0.19 | 0.15 ± 0.11 | 0.09 |
| VZV | 0.22 ± 0.06 | 4.0 × 10−7 | 0.20 ± 0.04 | 1.0 × 10−7 | 0.23 ± 0.45 | 0.29 | 0.03 ± 0.25 | 0.45 |
| Ad36 | 0.05 ± 0.04 | 0.06 | 0 | 0.50 | 0.14 ± 0.10 | 0.06 | 0 | 0.50 |
| HAV | 0.37 ± 0.06 | 2.7 × 10−12 | 0.01 ± 0.04 | 0.41 | 0.48 ± 0.11 | 3.0 × 10−8 | 0 | 0.50 |
| Influenza A | 0.26 ± 0.06 | 3.6 × 10−8 | 0.19 ± 0.04 | 2.2 × 10−6 | 0.19 ± 0.14 | 0.07 | 0.32 ± 0.11 | 1.1 × 10−3 |
| Influenza B | 0.20 ± 0.06 | 2.3 × 10−5 | 0.19 ± 0.04 | 2.1 × 10−6 | 0.27 ± 0.12 | 6.1 × 10−3 | 0.20 ± 0.09 | 9.0 × 10−3 |
h2 = Heritability; c2 = household effect.
Shared Environment
Shared environmental effects were significant for 7 of 13 quantitative antibody level traits (table 3), with a maximum estimate of 21% for HSV-2. For the majority of pathogens (C. pneumoniae, H. pylori, T. gondii, CMV, EBV, HHV-6 and HAV), the estimated influence of genes is much greater than that of shared environment. For other pathogens (HSV-1, VZV, influenza A virus, and influenza B virus), genes and shared environment account for a similar proportion of phenotypic variance. Only for HSV-2 do the shared environmental factors appear to be much more important than host genetic factors (21 and 8%, respectively). We did not detect significant genetic or shared environmental effects for Ad36.
Dichotomized Traits
We performed similar analyses on dichotomized antibody measurements (seropositive or seronegative), under the rationale that whether or not an immune response is triggered may be viewed as a dichotomous event. The binary characteristic of whether or not antibodies were produced could subsequently be obscured by other factors (i.e. assay noise, duration of time since infection, intensity of humoral immune response, etc.), resulting in a quantitative antibody level assay score. These analyses on the dichotomous traits showed a similar pattern of heritabilities and shared environmental influences, but typically with a reduced significance. Significant heritabilities ranged up to 62% for C. pneumoniae, and HHV-6, VZV, and the influenza A virus were no longer significantly heritable. The reduction in significance is likely due to the fact that dichotomous traits tend to provide less information than quantitative traits (especially if one of the two possible outcomes is rare, such as for VZV) and because the available sample sizes were slightly reduced, since some individuals’ serostatus fell into the indeterminate category and was coded as phenotypically unknown in the analyses (e.g. 30% indeterminates for EBV).
Bivariate Analysis
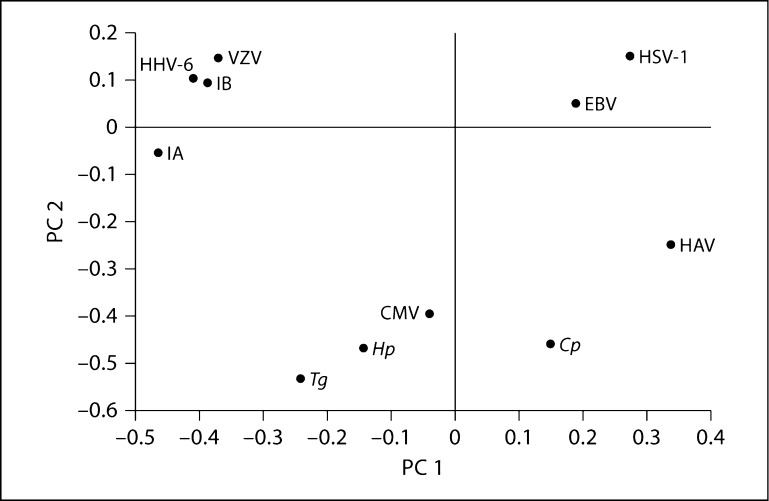
To investigate the degree to which genetic factors influencing the serological phenotypes might overlap between different pathogens, we performed bivariate variance components heritability analysis on pairs of pathogens. Genetic correlations in serological measures between pairs of pathogens are presented in table 4, which includes only those pathogens for which antibody levels/status were found to be significantly heritable in the previous univariate analyses, as only those are expected to provide useful information in pairwise genetic analyses. For the quantitative traits (table 4A), correlations in the genetic etiology underlying antibody levels to pairs of infectious agents ranged widely, from −0.42 to 0.54. It is apparent that the host genetic factors influencing serological measures of infection are significantly positively correlated (at p ≤ 0.05) for some pairs of pathogens (such as the two types of influenza viruses investigated), while being uncorrelated for the majority. Interestingly, we observed significant negative correlations for some pairs of pathogens (e.g. for VZV and influenza B), indicating shared genetic components with opposite effects on antibody response to different pathogens. In an attempt to obtain an overall interpretation, we performed principal components analysis on the genetic correlation matrix of all 11 heritable pathogens. A scatter plot of the first two principal components is shown in figure 1. No overall pattern suggesting an obvious explanation for the observed genetic correlations is apparent. The estimates of genetic correlations among the dichotomized serostatus phenotypes range from −0.32 to 0.74 and are shown in table 4B. The estimated correlations are often similar and typically in the same direction as for the quantitative measurements, but significance was generally lower, for the same reasons outlined for the univariate heritability analysis above.
Table 4.
Genetic correlations between quantitative traits (antibody levels) and between discrete traits (serostatus)
| A Quantitative traits (antibody levels) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cp | Hp | Tg | CMV | EBV | HSV-1 | HHV-6 | VZV | HAV | IA | IB | |
| Cp | 0.20 | 0.33 | 0.12 | 0.07 | −0.08 | −0.22 | −0.32 | 0.31 | −0.03 | −0.08 | |
| Hp | 0.10 | 0.43 | 0.30 | −0.01 | −0.14 | 0.12 | 0.15 | 0.26 | 0.11 | 0.08 | |
| Tg | 0.01 | 2.0 × 10−3 | 0.43 | −0.29 | −0.15 | −0.02 | 0.02 | −0.04 | 0.40 | 0.15 | |
| CMV | 0.32 | 0.02 | 1.7 × 10−3 | 0.18 | −0.05 | −0.08 | 0.03 | −0.07 | −0.06 | −0.05 | |
| EBV | 0.55 | 0.91 | 0.04 | 0.13 | 0.23 | −0.31 | 0.17 | 0.19 | −0.15 | 0.09 | |
| HSV-1 | 0.55 | 0.34 | 0.34 | 0.73 | 0.08 | −0.35 | −0.12 | 0.02 | −0.24 | −0.10 | |
| HHV-6 | 0.08 | 0.34 | 0.86 | 0.51 | 8.5 × 10−3 | 0.01 | 0.29 | −0.20 | 0.41 | 0.22 | |
| VZV | 0.01 | 0.23 | 0.87 | 0.80 | 0.16 | 0.38 | 0.02 | −0.17 | 0.27 | 0.54 | |
| HAV | 0.01 | 0.03 | 0.78 | 0.53 | 0.11 | 0.90 | 0.10 | 0.16 | −0.42 | −0.34 | |
| IA | 0.84 | 0.37 | 3.9 × 10−3 | 0.65 | 0.23 | 0.08 | 8.6 × 10−4 | 0.03 | 9.3 × 10−4 | 0.30 | |
| IB | 0.51 | 0.52 | 0.29 | 0.67 | 0.47 | 0.49 | 0.09 | 1.3 × 10−5 | 0.01 | 0.02 | |
| B Discrete traits (serostatus) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cp | Hp | Tg | CMV | EBV | HSV-1 | HAV | IB | |
| Cp | 0.40 | 0.20 | 0.24 | 0.352 | 0.45 | −0.45 | 0.14 | |
| Hp | 0.02 | 0.36 | 0.27 | 0.11 | −0.11 | 0.74 | −0.15 | |
| Tg | 0.37 | 0.12 | 0.11 | −0.32 | −0.18 | 0.34 | 0.01 | |
| CMV | 0.16 | 0.15 | 0.62 | 0.18 | 0.44 | 0.18 | −0.15 | |
| EBV | 0.04 | 0.48 | 0.13 | 0.25 | 0.44 | 0.09 | −0.18 | |
| HSV-1 | 0.10 | 0.65 | 0.56 | 0.08 | 0.06 | 0.04 | 0.32 | |
| HAV | 1.00 | 3.1 × 10−5 | 0.04 | 0.30 | 0.60 | 0.85 | −0.19 | |
| IB | 0.47 | 0.43 | 0.98 | 0.37 | 0.30 | 0.23 | 0.32 | |
Cp = C. pneumoniae; Hp = H pylori; Tg= T. gondii; IA = Influenza A; IB = Influenza B. Upper right (above and to the right of the diagonal) = correlations; lower left (below and to the left of the diagonal) = p values. Bold = Point-wise significant at p ≤ 0.05. Note that pathogens included here are only those for which the quantitative antibody levels (A) and the dichotomous serostatus phenotypes (B) were significantly heritable in the univariate analysis.
Fig. 1.
Principal components analysis of a pairwise genetic correlation matrix of significantly heritable antibody levels. The first two principal components are plotted. No obvious relationship among the host genetic factors to antibodies to these pathogens is apparent.
Replication
To assess the robustness of our heritability estimates, we measured antibody levels (using identical serological assays), and estimated heritabilities, for eight of the investigated pathogens in a separate Mexican American cohort consisting of participants in the SAFDGS [9,10]. Pathogens compared between the two studies included C. pneumoniae, H. pylori, T. gondii, CMV, EBV, HSV-1, HSV-2, and HAV. Heritability estimates of the quantitative antibody levels were similar between the two samples (e.g. 38% in the SAFHS vs. 34% in the SAFDGS for C. pneumoniae), but some discrete serostatus traits differed to a greater extent (table 5).
Table 5.
Comparison of heritability estimates (with standard error) between the SAFHS (this study) and SAFDGS for quantitative antibody level traits and discrete serostatus traits
| Pathogen | Quantitative antibody level traits |
Discrete serostatus traits |
||
|---|---|---|---|---|
| h2 (SAFHS) ± SE | h2 (SAFDGS) ± SE | h2 (SAFHS) ± SE | h2 (SAFDGS) ± SE | |
| C. pneumoniae | 0.38 ± 0.05 | 0.34 ± 0.07 | 0.72 ± 0.11 | 0.50 ± 0.13 |
| H. pylori | 0.35 ± 0.06 | 0.38 ± 0.08 | 0.45 ± 0.10 | 0.46 ± 0.001 |
| T. gondii | 0.27 ± 0.05 | 0.25 ± 0.08 | 0.41 ± 0.15 | 0.32 ± 0.19 |
| CMV | 0.39 ± 0.06 | 0.25 ± 0.07 | 0.59 ± 0.11 | 0.24 ± 0.14 |
| EBV | 0.41 ± 0.06 | 0.40 ± 0.09 | 0.64 ± 0.11 | 0.54 ± 0.18 |
| HSV-1 | 0.26 ± 0.05 | 0.22 ± 0.07 | 0.26 ± 0.11 | 0.55 ± 0.14 |
| HSV-2 | 0.18 ± 0.05 | 0.10 ± 0.07 | 0.07 ± 0.07 | 0.04 ± 0.11 |
| HAV | 0.38 ± 0.06 | 0.24 ± 0.07 | 0.48 ± 0.11 | 0.58 ± 0.14 |
Note that estimates for both studies are calculated without allowing for a shared household effect, as no such information is available for the SAFDGS. Therefore, SAFHS estimates provided here differ slightly from those in table 3.
Discussion
Previous research has demonstrated that there are host-related differences in susceptibility to infectious diseases between populations and among individuals [20]. Host factors that may be involved include age, sex, nutrition, stress, presence of other infections, hygiene, socioeconomic status, and ethnic background [21,22,23]. Also, in addition to genetic properties of the pathogen, host genetics may play a role in various aspects of infection risk in terms of host behavior (influencing exposure risk and intensity), host resistance to pathogen attachment and invasion, and subsequent regulation of the host's immune and non-immune response to infectious agents. Here we estimate the contribution of host genetic and shared environmental factors to infection as determined by the levels of antibodies to common infectious agents.
The measurement of antibodies is widely used in epidemiological studies to determine exposures to infectious agents and to compare exposures among different populations. In addition, these measures are often used to link exposures to chronic diseases, for example, CMV to risk of cardiovascular disease [24]. Antibody measurements are often interpreted as being indicative of pathogen exposure alone, without explicitly acknowledging that innate factors of the host, such as genetic factors, may influence antibody levels substantially. Our study clearly demonstrates that antibody levels to common infectious pathogens are significantly heritable. According to these results, genetic factors account for up to 40% of the individual variation in quantitative infectious disease phenotypes. Of the two pathogens in this study for which heritability was previously estimated, EBV has a similar heritability measure (h2 = 0.38) to an earlier study (h2 = 0.32–0.48), despite differences in antibody measurements between the two studies (antibodies against EBV nuclear antigen 1 [EBNA-1] vs. viral capsin antigen [VCA] in the previous study) [7], and the lower seroprevalence rate in this study (46 or 77% including the indeterminate samples). This seroprevalence rate is somewhat lower than for other adult populations [25], which may be due in part to differences in the serological measurements (e.g. differences in assay cutoff values, and some VCA-positive individuals may not generate an EBNA-1 response [26]). However, a similar EBV seropositivity rate was recently characterized for another US adult population, using the same EBNA-1 ELISA assay [R. Yolken, data not shown]. On the other hand, the heritability estimate for H. pylori in this study (h2 = 0.35) is lower than that reported for a previous study (h2 = 0.57), and while the other study reported significant shared environmental effects, this study does not [4].
The phenotypes examined here are largely the result of exposure to naturally occurring antibodies, in other words antibodies that were produced in response to infectious agents encountered in the environment, rather than through routine vaccination. A possible exception includes the influenza viruses, given that vaccine was available against these pathogens at the time the study samples were collected (1991–1995). However, it is not clear what effect this may have had, if any, on the influenza A and B antibody level measurements used in our study in part because annual influenza vaccines were less common when these samples were collected almost two decades ago, in particular among Hispanics who historically have significantly lower influenza vaccination rates than the general US public [27,28,29]. In any case, overall our study provides clear evidence that naturally acquired infectious disease antibody level traits are significantly heritable, and may therefore be viewed as partly genetic traits. The fact that antibody levels can vary from person to person as a function of genetics, rather than exposure alone, should be borne in mind when interpreting antibody test results in a clinical setting. Our bivariate analyses indicate that some genetic factors appear to be shared between some closely related pathogens, such as different influenza virus strains. However, we did not observe obvious evidence for genetic factors that influence antibody levels to all pathogens or even classes of pathogens (such as herpesviruses). For most pathogen pairs, our observations are consistent with host genetic factors influencing antibody levels being pathogen specific.
While our study demonstrates that genetic factors have a strong influence on antibody levels for many pathogens, shared environment (modeled as co-habitation) was also a significant contributing factor to the serological phenotypes for some pathogens. This may be due to direct transmission of infection between relatives, for example, through coughing or kissing, due to shared exposure to infectious agents such as via drinking water or pets in the household, or due to shared behavioral practices, such as hand washing, food preparation, etc., among family members.
The decision to focus on antibody levels, which are not a direct assessment of the presence or absence of infectious agents, complicates the interpretation of our results, due to the nature of serological data. The presence of antibodies indicates a past or present infection, but this does not necessarily correlate with protection [30]. In our study, it is not clear whether, for example, a high antibody level represents success in warding off infection, or whether the immune system was less efficient at dealing with the invading pathogen, resulting in a prolonged infection. It is also possible that elevated levels of antibody are related to more recent, repeated, or stronger dose exposures to the infectious agent [31,32]. Therefore, one has to be careful with equating heritability evidence for serological phenotypes with an altered susceptibility or resistance to infection and course of infectious disease.
Nonetheless, our demonstration of the heritability of antibody levels against infectious pathogens is consistent with the known role of a number of human genes in the immune response. Genes known to contribute to the immune response include numerous elements within the major histocompatibility complex as well as genes encoding proteins critical to the immune response such as cytokines – including chemokines, interferon and other mediators – and proteins that activate these factors, e.g. toll-like receptors and mannose-binding proteins [33,34,35,36]. Of the pathogens examined here, several have been previously associated with specific genes related to infection susceptibility. Susceptibility to C. pneumoniae has been linked to genes encoding toll-like receptors, cytokines, and mannose-binding proteins [37,36]. Polymorphisms in a number of genes are associated with H. pylori infection, including those encoding cytokines, metalloproteinases, glutathionine transferase, cyclooxygenase-2, and other metabolically active molecules [38,39,40]. Infection with T. gondii has been associated with genes encoding interferon gamma, toll-like receptors, and the purinergic receptor P2X7[41,42,43]. CMV susceptibility is attributed in part to polymorphisms in several genes including those encoding cytokines, toll-like receptors, and other immune response molecules [34,44,45]. Alterations in the HLA region and polymorphisms in genes encoding cytokines are linked to EBV susceptibility [46,47]. Susceptibility to infection with HSV-2 has been associated with a number of genes, including interferon gamma, mannose-binding proteins, transcription factor T-bet, and apolipoprotein E [33,48,49,50]. HAV infection is shown to be influenced by polymorphisms in TIM1, a gene that encodes the receptor for this virus [35].
Our demonstration, for the first time for many of these pathogens, that antibody levels to specific infectious agents are partially under genetic control provides further impetus for genetic searches of these variants. Identifying the underlying genetic factors influencing these infectious disease phenotypes may help to illuminate how the immune system functions, reveal factors influencing infection susceptibility or resistance, and suggest strategies for the prevention and treatment of infection.
Acknowledgements
We thank the participants of the SAFHS and the SAFDGS. In addition, we acknowledge Mr. Barry Grubbs, Ms. Bogdana Krivogorsky, and Dr. Olga Dubuisson for their assistance in performing the antibody assays. This work was supported by the National Institutes of Health (grants HL080149 to H.H.H.G.; HL045522 and MH059490 to J.B.; and DK53889 to R.D.) and the Stanley Medical Research Institute (to R.Y.). Analyses were conducted in facilities constructed with support from a Research Facilities Improvement Program Grant (RR017515) from the National Center for Research Resources, National Institutes of Health, and a gift from the SBC Foundation.
References
- 1.Klein NP, Fireman B, Enright A, Ray P, Black S, Dekker CL. A role for genetics in the immune response to the varicella vaccine. Pediatr Infect Dis J. 2007;26:300–305. doi: 10.1097/01.inf.0000257454.74513.07. [DOI] [PubMed] [Google Scholar]
- 2.Kimman TG, Vandebriel RJ, Hoebee B. Genetic variation in the response to vaccination. Community Genet. 2007;10:201–217. doi: 10.1159/000106559. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson RM, Ovsyannikova, Poland GA. Genetic basis for variation of vaccine response: our studies with rubella vaccine. Paediatr Child Health. 2009;19:S156–S159. doi: 10.1016/j.paed.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malaty HM, Engstrand L, Pedersen NL, Grahm DY. Helicobacter pylori infection: genetic and environmental influences. A study of twins. Ann Intern Med. 1994;120:982–986. doi: 10.7326/0003-4819-120-12-199406150-00002. [DOI] [PubMed] [Google Scholar]
- 5.Duah NO, Weiss HA, Jepson A, Tetteh KKA, Whittle HC, Conway DJ. Heritability of antibody isotype and subclass responses to Plasmodium falciparum antigens. PLoS ONE. 2009;4:e7381. doi: 10.1371/journal.pone.0007381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams-Blangero S, Vandeberg JL, Blangero J, Teixeira AR. Genetic epidemiology of seropositivity for Trypanosoma cruzi infection in rural Goias, Brazil. Am J Trop Med Hyg. 1997;57:538–543. doi: 10.4269/ajtmh.1997.57.538. [DOI] [PubMed] [Google Scholar]
- 7.Besson C, Amiel C, Le-Pendeven C, Plancoulaine S, Bonnardel C, Ranque B, Abbed K, Brice P, Fermé C, Carde P, Hermine O, Raphael M, Bresson J-L, Nicolas J-C, Gessain A, deThe G, Abel L. Strong correlations of anti-viral capsid antigen antibody levels in first-degree relatives from families with Epstein-Barr virus-related lymphomas. J Infect Dis. 2009;199:1121–1127. doi: 10.1086/597424. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 9.Hunt KJ, Lehman DM, Arya R, Fowler S, Leach RJ, Goring HH, Almasy L, Blangero J, Dyer TD, Duggirala R, Stern MP. Genome-wide linkage analyses of type 2 diabetes in Mexican Americans: the San Antonio Family Diabetes/Gallbladder Study. Diabetes. 2005;54:2655–2662. doi: 10.2337/diabetes.54.9.2655. [DOI] [PubMed] [Google Scholar]
- 10.Puppala S, Dodd GD, Fowler S, Arya R, Schneider J, Farook VS, Granato R, Dyer TD, Almasy L, Jenkinson CP, Diehl AK, Stern MP, Blangero J, Duggirala R. A genomewide search finds major susceptibility loci for gallbladder disease on chromosome 1 in Mexican Americans. Am J Hum Genet. 2006;78:377–392. doi: 10.1086/500274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng ML, Woodford SC, Hilburn JL, VandeBerg JL. A novel system for storage of sera frozen in small aliquots. J Biochem Biophys Methods. 1986;13:47–51. doi: 10.1016/0165-022x(86)90007-2. [DOI] [PubMed] [Google Scholar]
- 12.Dhurandhar NV, Israel BA, Kolesar JM, Mayhew GF, Cook ME, Atkinson RL. Increased adiposity in animals due to a human virus. Int J Obes Relat Metab Disord. 2000;24:989–996. doi: 10.1038/sj.ijo.0801319. [DOI] [PubMed] [Google Scholar]
- 13.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duggirala R, Williams JT, Williams-Blangero S, Blangero J. A variance component approach to dichotomous trait linkage analysis using a threshold model. Genet Epidemiol. 1997;6:987–992. doi: 10.1002/(SICI)1098-2272(1997)14:6<987::AID-GEPI71>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 15.Williams JT, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet. 1999;65:1134–1147. doi: 10.1086/302570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubicz R, Leach CT, Kraig E, Dhurandhar NV, Grubbs B, Blangero J, Yolken R, Goring HHH: Seroprevalence of 13 common pathogens in a rapidly growing U.S. minority population: Mexican Americans from San Antonio, TX. BMC Res Notes, in press. [DOI] [PMC free article] [PubMed]
- 17.Spence A, Westlake J, Lange K. Estimation of the variance components for dermal ridge count. Ann Hum Genet. 1977;41:111–115. doi: 10.1111/j.1469-1809.1977.tb01968.x. [DOI] [PubMed] [Google Scholar]
- 18.Lange K, Boehnke Extensions to pedigree analysis. IV. Covariance components models for multivariate traits. Am J Med Genet. 1983;14:513–524. doi: 10.1002/ajmg.1320140315. [DOI] [PubMed] [Google Scholar]
- 19.Boehnke M, Moll PP, Lange K, Weidman WH, Kottke BA. Univariate and bivariate analyses of cholesterol and triglyceride levels in pedigrees. Am J Med Genet. 1986;23:775–792. doi: 10.1002/ajmg.1320230306. [DOI] [PubMed] [Google Scholar]
- 20.Kimman TG. Genetics of Infectious Disease Susceptibility. Boston: Kluwer Academic Publishers; 2001. [Google Scholar]
- 21.Armstrong GL, Schillinger J, Markowitz L, Nahmias AJ, Johnson RE, McQuillan GM, St Louis ME. Incidence of herpes simplex virus type 2 infection in the United States. Am J Epidemiol. 2001;153:912–920. doi: 10.1093/aje/153.9.912. [DOI] [PubMed] [Google Scholar]
- 22.Dowd JB, Zajacova A, Aiello A. Early origins of health disparities: burden of infection, health, and socioeconomic status in U.S. children. Soc Sci Med. 2009;68:699–707. doi: 10.1016/j.socscimed.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McQuillan GM, Kruszon-Moran D, Kottiri BJ, Curtin LR, Lucas JW, Kington RS. Racial and ethnic differences in the seroprevalence of 6 infectious diseases in the United States: data from NHANES III, 1988–1994. Am J Public Health. 2004;94:1952–1958. doi: 10.2105/ajph.94.11.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts ET, Hann MN, Dowd JB, Aiello AE. Cytomegalovirus antibody levels, inflammation, and mortality among elderly Latinos over 9 years of follow-up. Am J Epidemiol. 2010;172:363–371. doi: 10.1093/aje/kwq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porter DD, Wimberly I, Benyesh-Melnick M. Prevalence of antibodies to EB virus and other herpesviruses. JAMA. 1969;208:1675–1679. [PubMed] [Google Scholar]
- 26.De Paschale M, Agrappi C, Manco MT, Agrappi C, Manco MT, Mirri P, Vigano EF, Clerici P. Seroepidemiology of EBV and interpretation of the ‘isolated VCA IgG’ pattern. J Med Virol. 2009;81:325–331. doi: 10.1002/jmv.21373. [DOI] [PubMed] [Google Scholar]
- 27.Centers for Disease Control and Prevention Influenza and pneumococcal vaccination levels among persons aged ≥65 years – United States, 2001. MMWR Morb Mortal Wkly Rep. 2002;51:1019–1024. [PubMed] [Google Scholar]
- 28.Link MW, Ahluwalia IB, Euler GL, Bridges CB, Chu SY, Wortley PM. Racial and ethnic disparities in influenza vaccination coverage among adults during the 2004–2005 season. Am J Epidemiol. 2006;163:571–578. doi: 10.1093/aje/kwj086. [DOI] [PubMed] [Google Scholar]
- 29.Chu SY, Barker LE, Smith PJ. Racial/ethnic disparities in preschool immunizations: United States, 1996–2001. Am J Public Health. 2004;94:973–977. doi: 10.2105/ajph.94.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen RT, Markowitz LE, Albrecht P, Stewart JA, Mofenson LM, Preblud SR, Orenstein WA. Measles antibody: reevaluation of protective titers. J Infect Dis. 1990;162:1036–1042. doi: 10.1093/infdis/162.5.1036. [DOI] [PubMed] [Google Scholar]
- 31.Whittle HC, Aaby P, Samb B, Jensen H, Bennett J, Simondon F. Effect of subclinical infection on maintaining immunity against measles in vaccinated children in West Africa. Lancet. 1999;353:98–102. doi: 10.1016/S0140-6736(98)02364-2. [DOI] [PubMed] [Google Scholar]
- 32.Marchant A, Pihlgren M, Goetghebuer T, Weiss HA, Ota MO, Schlegel-Hauter SE, Whittle H, Lambert PH, Newport MJ, Siegrist CA. Predominant influence of environmental determinants on the persistence and avidity maturation of antibody responses to vaccines in infants. J Infect Dis. 2006;193:1598–1605. doi: 10.1086/503775. [DOI] [PubMed] [Google Scholar]
- 33.Zhang SY, Boisson-Dupuis S, Chapgier A, Yang K, Bustamante J, Picard C, Abel L, Jouanguy E, Casanova JL. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol Rev. 2008;226:29–40. doi: 10.1111/j.1600-065X.2008.00698.x. [DOI] [PubMed] [Google Scholar]
- 34.Carvalho A, Cunha C, Carotti A, Aloisi T, Guarrera O, Di Ianni M, Falzetti F, Bistoni F, Aversa F, Pitzurra L, Rodrigues F, Romani L. Polymorphisms in Toll-like receptor genes and susceptibility to infections in allogeneic stem cell transplantation. Exp Hematol. 2009;37:1022–1029. doi: 10.1016/j.exphem.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 35.McIntire JJ, Umetsu DT, DeKruyff RH. TIM-1, a novel allergy and asthma susceptibility gene. Springer Semin Immunopathol. 2004;25:335–348. doi: 10.1007/s00281-003-0141-3. [DOI] [PubMed] [Google Scholar]
- 36.Worthley DL, Johnson DF, Eisen DP, Dean MM, Heatley SL, Tung JP, Scott J, Padbury RT, Harley HA, Bardy PG, Angus PW, Mullighan CG. Donor mannose-binding lectin deficiency increases the likelihood of clinically significant infection after liver transplantation. Clin Infect Dis. 2009;48:410–417. doi: 10.1086/596313. [DOI] [PubMed] [Google Scholar]
- 37.Poikonen K, Lajunen T, Silvennoinen-Kassinen S, Leinonen M, Saikku P. Effects of CD14, TLR2, TLR4, LPB, and IL-6 gene polymorphisms on Chlamydia pneumoniae growth in human macrophages in vitro. Scand J Immunol. 2009;70:34–39. doi: 10.1111/j.1365-3083.2009.02267.x. [DOI] [PubMed] [Google Scholar]
- 38.Tatemichi M, Iwasaki M, Sasazuki S, Tsugane S. Association between polymorphisms in glutathione S-transferase Mu3 and IgG titer levels in serum against Helicobacter pylori. J Hum Genet. 2009;54:557–563. doi: 10.1038/jhg.2009.77. [DOI] [PubMed] [Google Scholar]
- 39.Hu HM, Kuo CH, Lee CH, Wu IC, Lee KW, Lee JM, Goan YG, Chou SH, Kao EL, Wu MT, Wu DC. Polymorphism in COX-2 modifies the inverse association between Helicobacter pylori seropositivity and esophageal squamous cell carcinoma risk in Taiwan: a case control study. BMC Gastroenterol. 2009;9:37. doi: 10.1186/1471-230X-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh YC, Cheng HC, Chang WL, Yang HB, Sheu BS. Matrix metalloproteinase-3 promoter polymorphisms but not dupA-H. pylori correlate to duodenal ulcers in H. pylori-infected females. BMC Microbiol. 2010;10:218. doi: 10.1186/1471-2180-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albuquerque MC, Aleixo AL, Benchimol EI, Leandro AC, das Neves LB, Vicente R, Bonecini-Almeida Mda G, Amendoeira MR. The IFN-gamma +874T/A gene polymorphism is associated with retinochoroiditis toxoplasmosis susceptibility. Mem Inst Oswaldo Cruz. 2009;104:451–455. doi: 10.1590/s0074-02762009000300009. [DOI] [PubMed] [Google Scholar]
- 42.Peixoto-Rangel AL, Miller EN, Castellucci L, Jamieson SE, Peixe RG, Elias Lde S, Correa-Oliveira R, Bahia-Oliveira LM, Blackwell JM. Candidate gene analysis of ocular toxoplasmosis in Brazil: evidence for a role for toll-like receptor 9 (TLR9) Mem Inst Oswaldo Cruz. 2009;104:1187–1190. doi: 10.1590/s0074-02762009000800019. [DOI] [PubMed] [Google Scholar]
- 43.Jamieson SE, Peixoto-Rangel AL, Hargrave AC, Roubaix LA, Mui EJ, Boulter NR, Miller EN, Fuller SJ, Wiley JS, Castellussi L, Boyer K, Peixe RG, Krisits MJ, Elias Lde S, Coyne JJ, Correa-Oliveira R, Sautter M, Smith NC, Lees MP, Swisher CN, Heydemann P, Noble AG, Bardo D, Burrowes D, McLone D, Roizen N, Withers S, Bahia-Oliveira LM, McLeod R, Blackwell JM. Evidence for associations between the purinergic receptor P2X(7) (P2RX7) and toxoplasmosis. Genes Immun. 2010;11:374–383. doi: 10.1038/gene.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann TW, Halimi JM, Buchler M, et al. Association between a polymorphism in the IL-12p40 gene and cytomegalovirus reactivation after kidney transplantation. Transplantation. 2008;85:1406–1411. doi: 10.1097/TP.0b013e31816c7dc7. [DOI] [PubMed] [Google Scholar]
- 45.Zheng XQ, Zhu F, Shi WW, Lin A, Yan WH. The HLA-G 14 bp insertion/deletion polymorphism is a putative susceptible factor for active human cytomegalovirus infection in children. Tissue Antigens. 2009;74:317–321. doi: 10.1111/j.1399-0039.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- 46.Hatta K, Morimoto A, Ishii E, Kimura H, Ueda I, Hibi S, Todo S, Sugimoto T, Imashuku S. Association of transforming growth factor-beta1 gene polymorphism in the development of Epstein-Barr virus-related hematologic diseases. Haematologica. 2007;92:1470–1474. doi: 10.3324/haematol.11147. [DOI] [PubMed] [Google Scholar]
- 47.McAulay KA, Higgins CD, Macsween KF, Lake A, Jarrett RF, Robertson FL, Williams H, Crawford DH. HLA class I polymorphisms are associated with development of infectious mononucleosis upon primary EBV infection. J Clin Invest. 2007;117:3042–3048. doi: 10.1172/JCI32377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seppanen M, Lokki ML, Lappalainen M, Hiltunen-Back E, Rovio AT, Kares S, Hurme M, Aittoniemi J. Mannose-binding lectin 2 gene polymorphism in recurrent herpes simplex virus 2 infection. Hum Immunol. 2009;70:218–221. doi: 10.1016/j.humimm.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 49.Svensson A, Bergin AM, Lowhagen GB, Tunback P, Bellner L, Padyukov L, Eriksson K. A 3′-untranslated region polymorphism in the TBX21 gene encoding T-bet is a risk factor for genital herpes simplex virus type 2 infection in humans. J Gen Virol. 2008;89:2262–2268. doi: 10.1099/vir.0.2008/001305-0. [DOI] [PubMed] [Google Scholar]
- 50.Jayasuriya AN, Itzhaki RF, Wozniak MA, Patel R, Smit EJ, Noone R, Gilleran G, Taylor S, White DJ. Apolipoprotein E-epsilon 4 and recurrent genital herpes in individuals co-infected with herpes simplex virus type 2 and HIV. Sex Transm Infect. 2008;84:516–517. doi: 10.1136/sti.2008.032367. [DOI] [PubMed] [Google Scholar]