Abstract
Holoprosencephaly (HPE), which results from failed or incomplete midline forebrain division early in gestation, is the most common forebrain malformation. The etiology of HPE is complex and multifactorial. To date, at least 12 HPE-associated genes have been identified, including TGIF (transforming growth factor beta-induced factor), located on chromosome 18p11.3. TGIF encodes a transcriptional repressor of retinoid responses involved in TGF-β signaling regulation, including Nodal signaling. TGIF mutations are reported in approximately 1–2% of patients with non-syndromic, non-chromosomal HPE. We combined data from our comprehensive studies of HPE with a literature search for all individuals with HPE and evidence of mutations affecting TGIF in order to establish the genotypic and phenotypic range. We describe 2 groups of patients: 34 with intragenic mutations and 21 with deletions of TGIF. These individuals, which were ascertained from our research group, in collaboration with other centers, and through a literature search, include 38 probands and 17 mutation-positive relatives. The majority of intragenic mutations occur in the TGIF homeodomain. Patients with mutations affecting TGIFrecapitulate the entire phenotypic spectrum observed in non-chromosomal, non-syndromic HPE. We identified a statistically significant difference between the 2 groups with respect to inheritance, as TGIF deletions were more likely to be de novo in comparison to TGIF mutations (χ2(2) = 6.97, ppermutated = 0.0356). In addition, patients with TGIF deletions were also found to more commonly present with manifestations beyond the craniofacial and neuroanatomical features associated with HPE (p = 0.0030). These findings highlight differences in patients with intragenic mutations versus deletions affecting TGIF, and draw attention to the homeodomain region, which appears to be particularly relevant to HPE. These results may be useful for genetic counseling of affected patients.
Key Words: Holoprosencephaly, Monosomy 18p, 18p, 18p deletion, TGIF
Introduction
Holoprosencephaly (HPE) is the most common human forebrain malformation, occurring in approximately 1 in 250 conceptions and 1 in 8,000 live births [Matsunaga and Shiota, 1977; Leoncini et al., 2008]. HPE results from failure of midline forebrain separation between days 18 and 28 of human gestation. Up to 90% of the children with HPE die during the first year of life, and a strong correlation exists between survival (as well as other clinical outcomes) and the degree of brain malformation [Plawner et al., 2002; Stashinko et al., 2004; Hahn et al., 2006].
Traditionally, HPE has been classified according to the degree of separation of the forebrain, which includes structures that later become the cerebrum, thalami, and basal ganglia. The 3 classic types of HPE include alobar (virtually no forebrain division), semilobar (some degree of hemispheric cleavage), and lobar HPE (more complete separation) [Hahn and Barnes, 2010]. Among both living and deceased patients with a form of HPE that cannot be attributed to chromosomal or syndromic etiologies (termed ‘non-syndromic, non-chromosomal HPE’), the frequency of alobar, semilobar, and lobar HPE approximates 18, 37, and 27%, respectively [Lazaro et al., 2004]. Additional milder and perhaps less common types include middle interhemispheric variant (MIHV) and septopreoptic types [Barkovich and Quint, 1993; Hahn et al., 2010]. The most mildly affected patients have normal central nervous system findings on conventional neuroimaging, but have microform features of HPE, which can include microcephaly, hypotelorism, a single central maxillary incisor, and cleft lip and/or palate [Solomon et al., 2010].
The phenotype of HPE varies dramatically, even within affected members of the same family [Solomon et al., 2009a]. In many cases, the severity of symptoms correlates with the severity of brain malformation. All patients born with structural brain differences consistent with HPE have some degree of cognitive impairment, but additional manifestations may vary. Neurological findings may include hydrocephalus, varying (but typically quite severe) degrees of cognitive impairment, seizures, muscle weakness or spasticity, dystonia, and choreoathetosis. Endocrine disorders secondary to pituitary insufficiency are common, with diabetes insipidus being the most frequently observed finding. Hypothalamic dysfunction may also manifest as autonomic instability [Levey et al., 2010].
Non-syndromic, non-chromosomal HPE is typically inherited in an autosomal dominant manner with incomplete penetrance and variable expressivity [Dubourg et al., 2007; Solomon et al., 2010]. HPE is a multifactorial disorder, which is best modeled by a combination of susceptibility genes and environmental factors interacting to produce a wide range of phenotypes. At least 12 genes have been shown to be associated with non-syndromic, non-chromosomal HPE, the first of which was SHH (Sonic Hedgehog). Since this discovery, several other genes involved in the SHH pathway have been linked to HPE [Roessler et al., 1996; Roessler and Muenke, 2010]. Like SHH, TGIF (Transforming Growth Factor Beta-Induced Factor; OMIM 602630) was initially found to be associated with HPE through a positional candidate gene approach based on patients with HPE and known cytogenetic anomalies containing the HPE4 locus on chromosome 18, whose minimal critical region includes TGIF [Overhauser et al., 1995; Gripp et al., 2000]. Subsequent studies have shown that among patients with non-syndromic HPE and normal karyotypes, the 4 most commonly mutated HPE-associated genes are (in order of decreasing frequency) SHH, ZIC2, SIX3, and TGIF. In prospective studies, a mutation in at least one of these genes is found in approximately 25% of such probands, although mutations in TGIF occur in less than 2% [Pineda-Alvarez et al., 2010; Solomon et al., 2010].
TGIF maps to 18p11.3, and the protein product is known to be expressed in the developing forebrain and in midline facial structures in many species [Gripp et al., 2000; Jin et al., 2006; Knepper et al., 2006]. The role of TGIF in the pathogenesis of HPE is not well-delineated. However, evidence regarding key biological properties of TGIF suggests mechanisms by which alterations of TGIF results in HPE. The TGIF protein can inhibit retinoid signaling by blocking retinoid receptor response element (RXR) binding to the retinoid receptor [Bartholin et al., 2006]. This has been an area of particular interest, as retinoic acid is known to be involved in the development and patterning of the central nervous system [Maden, 2003]. Animal studies indicate that embryonic exposure to retinoic acid may lead to craniofacial malformations consistent with HPE. Thus, it is hypothesized that interference with retinoid signaling is one mechanism by which alterations of the TGIF gene may contribute to the development of human HPE [Sulik et al., 1995]. TGIF is also known to interact with intracellular Smad proteins to repress responses to the TGF-β family of growth and differentiation factors. This is of interest in the context of the pathogenesis of HPE, as Smad2 is part of the Nodal signaling pathway involved in neural axis development. Although mutations in TGIF decrease the protein's ability to bind Smad2, it has been unclear precisely how decreased TGIF activity affects the NODAL pathway [Roessler et al., 1996; Gripp et al., 2000]. However, studies in animal models have shown that Tgif function is required for both normal gastrulation and for regulation of the transcriptional response to Nodal signaling in early embryogenesis [Powers et al., 2010].
Recent studies involving relatively large cohorts of patients with mutations in HPE-associated genes have suggested that in select patients, the phenotype may be predicted by the mutated gene. For example, patients with ZIC2 mutations tend to have distinct craniofacial anomalies regardless of the severity of brain malformation [Solomon et al., 2009b]. Here, we report on all known patients with features of HPE and evidence of TGIF alterations in an attempt to understand if such a correlation can be found within this cohort.
Subjects and Methods
Patient Recruitment, Mutation Screening, and Clinical Assessment
Our laboratory has collected blood samples from approximately 1,000 patients with HPE-spectrum disorders and their family members over the course of 20 years. Under our National Human Genome Research Institute (NHGRI) IRB-approved protocol, and with appropriate consent obtained from participants, samples were tested for variations in the 4 most common HPE-associated genes (SHH, ZIC2, SIX3, and TGIF). The strategy for mutation testing of TGIF by dideoxynucleotide sequencing of exons has been described previously [Gripp et al., 2000; El-Jaick et al., 2007]. Clinical information, photographs, and neuroimaging results for patients with TGIF mutations were reviewed when available. Additional patients were ascertained through collaborations with diagnostic testing centers. Three patients were examined at the National Institutes of Health as part of our NHGRI IRB-approved clinical protocol on HPE.
Literature Review
A Medline search was conducted to find previously reported patients with HPE and mutations or haploinsufficiency of TGIF. Keywords for this search included: ‘TGIF’, ‘holoprosencephaly’, ‘HPE’, ‘18p’, ‘18p deletion’, ‘monosomy 18p’, and ‘18p11.3’. Referenced patients were included in this analysis if there was a confirmed mutation affecting TGIF. Also included in the analysis were patients with clinical features of HPE and cytogenetic anomalies affecting the short arm of chromosome 18 (18p) without additional chromosomal abnormalities and with clear evidence that the deletion includes the TGIF locus at 18p11.3 [Münke et al., 1988; Münke, 1989; Roessler and Muenke, 1998; Aguilella et al., 2003; Bendavid et al., 2006, 2009; El-Jaick et al., 2007; Richieri-Costa and Ribeiro, 2008; Sepulveda, 2009; Rosenfeld et al., 2010]. Cases involving deletions of 18p were only included in the final statistical analysis of phenotypes if clinical findings were consistent with HPE-spectrum anomalies. Cases were excluded if other chromosomal abnormalities were present beyond deletion of all or part of chromosome 18p, such as aberrations involving 18q or other chromosomes.
Results
Patients (General Characteristics)
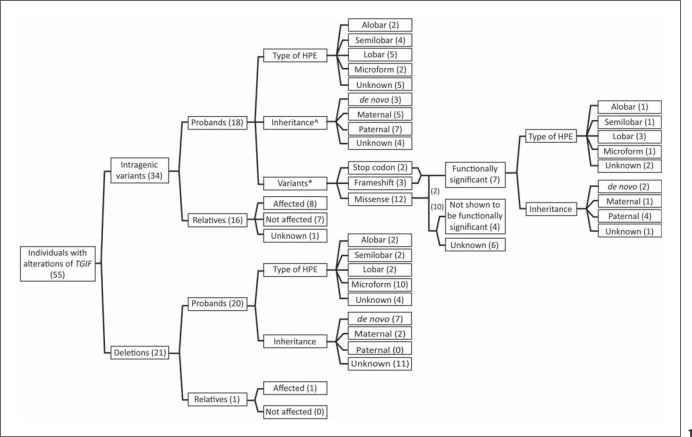
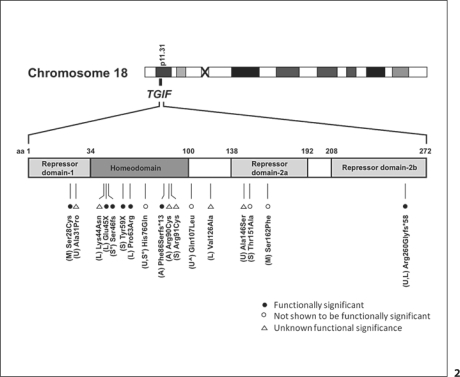
We identified 18 probands with molecularly-determined intragenic sequence mutations in TGIF, and 16 affected relatives with mutations. We also identified 20 probands with cytogenetic deletions of all or part of chromosome 18p (which include deletion of TGIF) and findings consistent with HPE-spectrum anomalies, and 1 affected relative with a deletion. Five probands with intragenic mutations and 2 probands with deletions are newly presented here. In addition, we present new clinical details for 5 previously reported cases (primarily reported in the context of mutation discovery). See table 1 for patient details, figure 1 for the breakdown of patients, figure 2 for mutation distribution, and online supplementary table 1 (for all online supplementary material, see www.karger.com/doi/10.1159/000328203) for additional patient information.
Table 1.
All known patients with either mutations in or deletions of TGIF
| Patienta | HPE typeb | Inheritance | DNA alteration | Predicted protein alteration | Predicted functional alterationc | Gender | Proband (or relationship to proband) | Reference |
|---|---|---|---|---|---|---|---|---|
| 1a | mic | pat | c.83C>G | p. Ser28Cys | decreased transcriptional repression | F | proband | Gripp et al., 2000 |
| 1b | mic | pat | c.83C>G | p. Ser28Cys | decreased transcriptional repression | M | father | Gripp et al., 2000 |
| 1c | mic | U | c.83C>G | p. Ser28Cys | decreased transcriptional repression | M | paternal grandfather | Gripp et al., 2000 |
| 2a | U | pat | c.91G>C | p. Ala31Pro | U | F | proband | this report |
| 2b | none | U | c.91G>C | p. Ala31Pro | U | M | father | this report |
| 3a | L | mat | c.132G>T | p. Lys44Asn | U | M | proband | Richieri-Costa and Ribeiro, 2008 |
| 3b | none | U | c.132G>T | p. Lys44Asn | U | F | mother | Richieri-Costa and Ribeiro, 2008 |
| 4 | L | de novo | c.133G>T | p. Glu45X | protein truncation with no transcriptional repression | M | proband | El-Jaick et al., 2007 |
| 5 | U | U | c.228C>A | p. His76Gln | likely no functional alteration | U | proband | El-Jaick et al., 2007 |
| 6a | S | pat/mat | c.l40_141delTG (pat); c.228C>A (mat) | p. Ser46fs (pat); p. His76Gln (mat) | paternal mutation results in truncation and loss of repression; maternal mutation likely causes no functional alteration | F | proband | El-Jaick et al., 2007 |
| 6b | none | U | c.228C>A | p. His76Gln | likely no functional alteration | F | mother | El-Jaick et al., 2007 |
| 6c | none | U | c.140 141delTG | p. Ser46fs | protein truncation with no transcriptional repression | M | father | El-Jaick et al., 2007 |
| 7a | S | pat | c.177C>G | p. Tyr59X | protein truncation with no transcriptional repression | F | proband | Aguilella et al., 2003 |
| 7b | mic | U | c.177C>G | p. Tyr59X | protein truncation with no transcriptional repression | M | father | Aguilella et al., 2003 |
| 8 | L | de novo | c.187C>G | p. Pro63Arg | likely misfolded with no transcriptional repression | U | proband | Gripp et al., 2000 |
| 9a | A | mat | c.257delT | p. Phe86Serfs*13 | likely protein truncation | F | proband | this report |
| 9b | none | U | c.257delT | p. Phe86Serfs*13 | likely protein truncation | F | mother | this report |
| 10 | A | de novo | c.268C>T | p. Arg90Cys | U | M | proband | Chen et al, 2002 |
| 11a | S | pat | c.271C>T | p. Arg91Cys | U | F | proband | this report |
| 11b | A | pat | not available; likely same as 11a | not available; likely same as 11a | U | F | sibling | this report |
| 11c | none | U | c.271C>T | p. Arg91Cys | U | M | father | this report |
| 12a | U | mat | not available; likely same as 12b | not available; likely same as 12b | likely no functional alteration | F | proband | Aguilella et al, 2003 |
| 12b | U | U | c.320A>T | p. Glnl07Leu | likely no functional alteration | F | mother | Aguilella et al, 2003; El-Jaick et al, 2007 |
| 13a | L | mat | c.377T>C | p. Vall26Ala | U | M | proband | Chen etal, 2006 |
| 13b | S | mat | not available; likely same as 13a | not available; likely same as 13a | U | U | sibling | Chen etal, 2006 |
| 13c | L | mat | not available; likely same as 13a | not available; likely same as 13a | U | U | sibling | Chen etal, 2006 |
| 13d | mic | U | c.377T>C | p. Vall26Ala | U | F | mother | Chen etal, 2006 |
| 14 | U | U | c.436G>T | p. Alal46Ser | U | M | proband | this report |
| 15 | S | U | c.451A>GinrGiF; (SHH: c.l283_1291del) | p. Thrl51ArainTGIF, (SHH: p.378_380del) | likely no functional alteration | F | proband | Nannietal, 1999; Gripp et al, 2000 |
| 16a | mic | pat | c.485C>T | p. Serl62Phe | likely no functional alteration | M | proband | Gripp et al, 2000; El-Jaick et al, 2007 |
| 16b | none | U | c.485C>T | p. Serl62Phe | likely no functional alteration | M | father | Gripp et al, 2000 |
| 17a | U | pat | c.778delC | p. Arg260Glyfs*58 | likely misfolded with absent TGF-β-dependent transcriptional repression and decreased RXR-dependent transcriptional repression | M | proband | El-Jaick et al, 2007 |
| 17b | mic | U | c.778delC | p. Arg260Glyfs*58 | likely misfolded with TGF-β and decreased RXR-dependent transcriptional repression | M | father | El-Jaick et al, 2007 |
| 18 | L | U | c.778delC | p. Arg260Glyfs*58 | likely misfolded with TGF-β and decreased RXR-dependent transcriptional repression | F | proband | this report (no evidence of relationship with family 17, and the 2 families are of different ethnicities) |
| 19 | A | U | gene deletion (FISH) | N/A | predicted null | M | proband | Bendavid et al., 2006 |
| 20 | S | U | gene deletion (MLPA) | N/A | predicted null | U | proband | Bendavid et al., 2009 |
| 21 | S | mat (both SHH mutation and 18p deletion) | del(18)(pll.23→pter) (SHH: C.1270OG) | N/A in TGIF; (SHH: p. Pro424Ala) | predicted null in TGIF | F | proband | Nannietal., 1999 |
| 22a | mic | mat | del(18)(pll.3→18pter) | N/A | predicted null | M | proband | Portnoi et al., 2007 |
| 22b | mic | U | del(18)(pll.3→18pter) | N/A | predicted null | F | mother | Portnoi et al., 2007 |
| 23 | U | U | del(18)(pll.l→18pter) | N/A | predicted null | F | proband | Kuchle et al., 1991 |
| 24 | mic | de novo | del(18)(pll→18pter) | N/A | predicted null | F | proband | Morales-Peralta and Lanti-gua, 1994 |
| 25 | U | U | del(18)(pll→18pter) | N/A | predicted null | M | proband | Boudailliez et al., 1983 |
| 26 | U | de novo | del(18)(p)(l/2ofl8p) | N/A | predicted null | F | proband | Faust et al., 1976 |
| 27 | L | U | del(18)(p)(3/4ofl8p) | N/A | predicted null | F | proband | Faust et al., 1976 |
| 28 | mic | de novo | del(18)(p) | N/A | predicted null | F | proband | Aughton et al., 1991 |
| 29 | mic | de novo | del(18)(p) | N/A | predicted null | M | proband | Dolanetal., 1981 |
| 30 | L | U | arr(18): 140,284-14,065,199 | N/A | predicted null | F | proband | Rosenfeld et al., 2010 |
| 31 | mic | U | arr(18): chr 18: 5,982-14,065,199 | N/A | predicted null | F | proband | Rosenfeld et al., 2010 |
| 32 | mic | de novo | arr(18): chr 18: 5,982-14,065,199 | N/A | predicted null | F | proband | Rosenfeld et al., 2010 |
| 33 | mic | U | arr(18): 102,328-15,079,388 | N/A | predicted null | M | proband | Rosenfeld et al., 2010 |
| 34 | U | de novo | arr(18): 140,284-10,600,909 | N/A | predicted null | F | proband | Rosenfeld et al., 2010 |
| 35 | mic | U | arr(18): 5,982-4,974,551 | N/A | predicted null | F | proband | Rosenfeld et al., 2010 |
| 36 | A | de novo | arr(18): 5,982-4,974,551 | N/A | predicted null | F | proband | Sepulveda, 2009 |
| 37 | mic | U | del(18)(p11.2→pter) | N/A | predicted null | M | proband | this report |
| 38 | mic | U | gene deletion (aCGH) | N/A | predicted null | M | proband | this report |
Some mutations have been shown to be loss-of-function, but the functional effects of others (such as p. His76Gln and p. Glnl07Leu) are not known, and may in fact be rare familial variants, A = Alobar; L = lobar; mat = maternal; mic = microform; N/A = not applicable; pat = paternal; S = semilobar; U = unknown.
Each family is listed with a different number; individuals within each family are given a different letter.
The form of HPE is described as 'unknown' for patients with no available neuroimaging or with insufficient information for classification.
Functional data derived from El-Jaick et al. [2007].
Fig. 1.
Schematic flow-chart of the breakdown of probands and relatives with TGIF alterations. * Two probands (#17a, #18a) have the same functionally significant variant; 2 patients (#5a, #6a) have the same variant not shown to be functionally significant. ⁁ Patient #6a has 2 variants, 1 maternal, and 1 paternal, though only the paternal variant is thought to be of functional significance.
Fig. 2.
Schematic representation of the TGIF coding region (cDNA). The mutations found in probands are depicted with the type of HPE noted in parentheses. Amino acids are labeled from left to right. Functional domains are shaded, including an amino-terminal repression domain, 2 carboxyl-terminal repression domains, and a homeodomain. Domains are based on Wotton et al. [1999b] and Mukherjee and Bürglin [2007]. Functional significance is indicated as based on El-Jaick et al. [2007]. ° Two variants found in the same patient; ⁁ variant found in proband's mother, proband's sample not available. A = Alobar; S = semilobar; L = lobar; M = microform; U = unknown; fs = frameshift.
Of the 7 probands with functionally significant intragenic mutations for whom gender was known, 5 (71%) were female and 2 (29%) were male. Among the 19 probands with whole-gene TGIF deletions for whom gender was known, 12 (63%) were female and 7 (37%) were male. There was not a statistically significant difference in gender distribution between probands with intragenic mutations and probands with deletions (χ2(1) = 0.155, p = 0.6942, no continuity correction applied).
Inheritance
Of the 7 functionally significant intragenic mutations in which inheritance was known for the probands, 2 (29%) were de novo, 1 (14%) was maternally-inherited, and 4 (57%) were paternally-inherited. One proband had a variation inherited from each parent, though only the paternal variation has evidence for abnormal gene function [El-Jaick et al., 2007]. Of the 9 probands with a TGIF deletion for whom inheritance was known, 7 (78%) were de novo, 2 (22%) were maternally-inherited, and none was paternally-inherited. Mutations were more likely to be de novo in patients with deletions versus intragenic mutations, and there was a statistically significant difference in the overall distribution of inheritance between patients with intragenic mutations and patients with deletions (χ2(2) = 6.97, ppermutated = 0.0356).
HPE Type
Of the 13 probands with intragenic mutations and known HPE type, 2 (15%) had alobar, 4 (30%) had semilobar, 5 (38%) had lobar, and 2 (15%) had microform HPE. Among the 7 probands with functionally significant intragenic mutations and known HPE type, 1 (14%) had alobar, 2 (29%) had semilobar, 3 (43%) had lobar, and 1 (14%) had microform HPE. Among the 16 probands with deletions and known HPE type, 2 (12%) had alobar, 2 (12%) had semilobar, 2 (12%) had lobar, and 10 (65%) had microform HPE. There was not a statistically significant difference in the distribution of HPE types between patients with functionally significant intragenic mutations and those with deletions (χ2(3) = 5.166, ppermutated = 0.1738).
We compared the overall presence of frank HPE (structural brain anomalies, including alobar, semilobar, or lobar types, in contrast to microform HPE) between probands with intragenic mutations versus those with whole gene deletions. Of the probands with known HPE type, 6 of 7 probands (86%) with functionally significant intragenic mutations had frank HPE, as compared to 6 of 16 probands (38%) with TGIF deletions. This is likely a selection bias, as reference laboratories typically sequence TGIF only when HPE is present, whereas testing (such as a microarray) to look for any genomic deletion is commonly done in a wider variety of clinical situations. Though there were more probands with frank HPE in the intragenic mutation group than in the deletion group, this difference was not statistically significant (by two-tailed Fisher's exact test, p = 0.0686). In an analysis of all patients in whom imaging was available, 14 of 27 patients (52%) (including both probands and relatives with functionally significant mutations) with intragenic mutations had structural brain anomalies, while 6 of 17 patients (35%) with deletions had structural brain anomalies. This difference in the prevalence of structural brain anomalies in the mutation group versus the group with deletions likely reflects ascertainment bias, as there was a greater proportion of probands compared to relatives in the deletion group. Patients with intragenic mutations in TGIF had a lower prevalence of structural brain anomalies compared to patients with intragenic mutations affecting SIX3 or ZIC2, though the difference was only significant compared to patients with mutations in ZIC2 (table 2) [Lacbawan et al., 2009; Solomon et al., 2009b].
Table 2.
Prevalence of structural brain anomalies according to the altered HPE-associated gene (among both probands and affected relatives)
| Gene | TGIF (n = 13)a n (%) | SHH(n = 103) n (%) | ZIC2 (n = 101) n (%) | SIX3 (n = 92) n (%) |
|---|---|---|---|---|
| Frank HPE (alobar, semilobar, lobar) | 6 (46) | 47 (46) | 88 (87) | 59 (64) |
| No frank HPE (microform or unaffected) | 7(54) | 56 (54) | 13(13) | 33 (36) |
| p value | N/A | 0.795 | 0.0013* | 0.3450 |
Frank HPE refers to the presence of structural brain anomalies (alobar, semilobar, lobar, or MIHV type HPE), in contrast to microform HPE. As there is not an available reference for patients with deletions of each gene, only patients with intragenic mutations were considered.
Significant p value (calculated by χ2 test with continuity correction); N/A = not applicable.
Number of probands + affected relatives with functionally significant intragenic mutations.
We next compared the prevalence of classic HPE types (alobar, semilobar, and lobar HPE; MIHV type was excluded due to the lack of cases) within our group of probands with TGIF mutations to those of general HPE cohort studies, specifically 5 previous studies that examined the prevalence of each HPE subtype. Two of these studies included liveborn patients and fetuses diagnosed with HPE [Muenke Lab; Lazaro et al., 2004; Ming and Muenke, 2002]. One study focused only on liveborn patients with HPE [Orioli and Castilla, 2007], and the final 2 specifically examined patients with known mutations in either ZIC2 or SIX3 [Lacbawan et al., 2009; Solomon et al., 2009b]. Probands with intragenic TGIF mutations included a greater proportion of patients with microform HPE when compared to the cohort of patients with mutations in ZIC2. Probands with TGIF deletions had a more even distribution between HPE subtypes compared to the other cohort studies (table 3) [Solomon et al., 2009b].
Table 3.
Comparison of HPE subtypes between patients with functionally significant mutations in or deletions of TGIF and previously studied cohorts of patients with non-syndromic, non-chromosomal HPE
| HPE type | Intragenic TGIF mutations (probands) n (%) | TGIF deletions (probands) n (%) | NIH, Muenke Laba n (%) | Lazaro et al., 2004a n (%) | Orioli and Castilla, 2007b n (%) | Solomon et al, 2009b (ZIC2a n (%) | Lacbawan et al, 2009 (SIX3)a n (%) |
|---|---|---|---|---|---|---|---|
| Alobar | 1(17) | 2(33) | 10(13) | 15 (22) | 33 (40) | 27 (34) | 15 (37) |
| Semilobar | 2(33) | 2(33) | 45 (60) | 31 (45) | 36 (43) | 42 (53) | 20 (49) |
| Lobar | 3(50) | 2(33) | 20 (27) | 23 (33) | 14(17) | 10(13) | 6(15) |
| Total | 6 | 6 | 69 | 69 | 83 | 79 | 41 |
| Comparison vs. TGIF cohort | N/A | N/A | mutations: p = 0.4083 deletions: p = 0.3211 | mutations: p = 0.7127 deletions: p = 0.7804 | mutations: p = 0.1269 deletions: p = 0.5961 | mutations: p = 0.0491* deletions: p = 0.3476 | mutations: p = 0.1167 deletions: p = 0.5073 |
Statistically significant differences; N/A = not applicable.
These cohorts include both living and deceased patients (liveborn infants and fetuses) with non-chromosomal, non-syndromic HPE.
This cohort includes only liveborn patients with HPE, including chromosomal and syndromic cases.
Additional Clinical Findings
The availability of clinical data for patients was highly variable, though 25 patients with intragenic mutations and 18 patients with TGIF deletions had adequate data for analysis of findings (table 4). For patients for whom clinical information was known, the most commonly reported manifestations in patients with intragenic TGIF mutations included (in order of decreasing prevalence) microcephaly (10/25 patients), hypotelorism (9/25), and cleft lip and/or palate (9/25). The most commonly reported findings in patients with deletions of TGIF were microcephaly (9/18), cleft lip and/or palate (7/18), midface hypoplasia (7/18), and single maxillary central incisor (6/18). As an aggregate, patients had craniofacial findings typical of non-chromosomal, non-syndromic HPE [Lacbawan et al., 2009; Solomon et al., 2009a, 2010]. There was no significant difference in the prevalence of the above findings between the 2 groups. However, there was a statistically significant difference in the presence or absence of extra-neuronal/craniofacial findings, which were more common in the TGIF deletion group. This is likely due to the fact that other genes on 18p were also deleted in these cases.
Table 4.
Clinical manifestations of patients with TGIF alterations (including probands and mutation or deletion-positive relatives)
| Clinical feature | Patients (n = 25) with intragenic TGIF mutations n (%) | Patients (n = 18) with TGIF deletions n (%) | p valuea |
|---|---|---|---|
| Microcephaly | 10 (40) | 9(50) | 0.5496 |
| Hypotelorism | 9(36) | 3(17) | 0.1911 |
| Cleft lip and/or palate | 9(36) | 7(39) | 0.8994 |
| Midface hypoplasia | 8(32) | 7(39) | 0.7497 |
| Single maxillary central incisor | 2(8) | 6(33) | 0.0517 |
| Hypopituitarism | 4(16) | 5(28) | 0.4554 |
| Extra-neuronal/craniofacial findings | 0 | 6(33) | 0.0030* |
| Total | 25 | 18 | N/A |
Significant; N/A = not applicable.
By 2-tailed Fisher's exact test or χ2 when appropriate.
TGIF Variants
Among the 18 families presented here, we identified 19 total sequence-based mutations (fig. 2; family #6 had 2 variants; families #5 and #6, and families #17 and #18 each shared a variant). Eighty-nine percent (17/19) of the variants were unique. Two unrelated families (#5, #6) had the same missense variation (c.228C>A, p.His76Gln), 2 unrelated families both had another missense variation in common (c.271C>T, p.Arg91Cys), and 2 unrelated families (#17, #18) carried the same frameshift mutation (c.778delC, p.Arg260Glyfs*58). While the frameshift mutation is certain to have functional consequences, it is less clear as to whether the p.His76Gln variation is truly associated with HPE or is instead a rare variant [El-Jaick et al., 2007].
Of the 17 different variations amongst 18 probands, 12 (71%) were missense mutations, 3 (18%) were frameshift mutations, and 2 (12%) were nonsense mutations. Variants are most likely to be missense (χ2(2) = 10.705, p = 0.004). As mentioned above, our understanding of the functional effects of each variant is incomplete. Based on previous functional studies in which 11 TGIF alterations from patients with HPE were examined using a cell-based assay, 7 mutations are believed to be pathogenic as evidenced by altered TGIF protein function (table 1) [El-Jaick et al., 2007].
Of note, some patients with variations in TGIF also had mutations in other HPE-associated genes. If these were all genuinely pathogenic alterations, this would be consistent with the ‘multi-hit hypothesis of HPE’, which alludes to the idea that more than one HPE-associated gene may require disruption in order to result in HPE [Ming and Muenke, 2002; Lacbawan et al., 2009; Solomon et al., 2009b]. One proband (patient 15) and her clinically unaffected mother were each found to have an in-frame deletion in SHH (c.1132_1140del, p.378_380del); the proband was also found to have a missense variant in TGIF (c.451A>G, p.Thr151Ala) [Nanni et al., 1999; Gripp et al., 2000; Ming and Muenke, 2002]. As the SHH alteration has never been shown to be pathogenic by functional assays, it is entirely possible that neither alteration actually contributes to the phenotypes [Roessler et al., 2009]. Another patient who had semilobar HPE (patient 21) had a maternally-inherited SHH variation (c.1270C>G, p.Pro424Ala), as well as a deletion of TGIF (del(18)(p11.23→pter)), resulting from a maternal translocation, with maternal chromosome analysis revealing 46,XX,t(1;18)(q43;p11.3). Significant family history included multiple miscarriages and congenital anomalies most likely associated with the maternal translocation [Moog et al., 2001; Ming and Muenke, 2002]. In this instance, the TGIF deletion is clearly likely to result in an abnormal phenotype, and the consequence of the SHH variant is unclear and is felt to be unlikely to be pathogenic [Roessler et al., 2009].
Mutation Location and Conservation
Of the 17 unique TGIF variants, 9 (53%) are in the homeodomain, 2 (12%) are in the repressor domain-1, 3 (18%) are in the repressor domain-2a, 1 (6%) is in the repressor domain-2b, and 2 (12%) are not in a known functional domain. Of the 9 mutations within the homeodomain, 5 are missense mutations, one of which is known to be functionally significant (p.Pro63Arg) and one of which is not likely to be functionally significant (p.His76Gln). The residue of the functionally significant variant is highly conserved amongst a wide spectrum of species. The remaining 3 missense mutations in the homeodomain with unknown functional significance have the same high conservation as the functionally significant variant. Specifically, we examined common chimpanzee, Rhesus macaque, domestic sheep, horse, rat, crab-eating macaque, black-capped squirrel monkey, gray short-tailed opossum, Xenopus laevis, zebrafish, green pufferfish, and Drosophila melanogaster using a publicly available database (COBALT, Constraint-based Multiple Alignment Tool; see online suppl. table 2 for further details.) This suggests that these are functionally significant alterations; however, further study is required prior to assigning pathogenicity. In addition, p.Ala31Pro is of interest because it is near the carboxyl terminus-binding protein (CtBP), and could potentially influence CtBP binding, though this has yet to be studied [Melhuish and Wotton, 2000].
Discussion
We have previously reported comprehensive analyses of patients with HPE and mutations in SIX3 and ZIC2 [Lacbawan et al., 2009; Solomon et al., 2009b]. Here, we described 38 probands with either intragenic mutations or cytogenetic anomalies affecting TGIF, and 17 relatives with the same genetic change. Mutations in TGIF account for a much smaller proportion of HPE than the other genes commonly associated with HPE, and are estimated to occur in less than 2% of probands with non-chromosomal, non-syndromic forms of HPE. Nevertheless, this is the largest known analysis of a cohort of patients with mutations affecting this gene, and our findings allow for some conclusions that should be helpful for clinicians encountering patients with HPE in general, and specifically, when counseling families of patients with TGIF variants.
First, the case of TGIF clearly demonstrates a common problem in many genetic disorders: detected genetic variants may be of unclear functional significance. Unlike mutations in ZIC2, which are often null alleles, variants in TGIF are frequently missense variants, and thus present a diagnostic and counseling dilemma. Use of publicly available databases and software (for example, those that evaluate the evolutionary conservation of the residue in question or that predict the consequence of a protein alteration) may be used, though information gathered from these sources cannot substitute for basic scientific analysis. In addition, family studies are always indicated since they may provide some clarity to the clinical scenario, given that de novo variants are presumably more significant. However, until a functional assay becomes commonly available, it will be difficult to accurately assign pathogenicity to each variant. An additional limitation in interpreting these results is that, while our laboratory and others sequence TGIF exons and flanking sequences, this methodology has the potential to miss small exonic deletions or variations within intronic sequences that may be functionally significant. Clinicians and genetic testing facilities must take this into account when discussing findings with affected families. Furthermore, efforts to establish molecular databases need to be supported in an effort to tabulate rare variants seen in different populations.
Second, it is interesting to note that over half of the individual mutations occurred within the homeodomain of the TGIF protein, a finding similar to that reported in patients with mutations affecting SIX3 [Lacbawan et al., 2009]. Like mutations within the SIX3 homeodomain, all mutations within the TGIF homeodomain (with the possible exception of p.His76Gln) appear to lead to decreased protein function [El-Jaick et al., 2007]. This supports molecular studies in which the homeodomain was found to be not only essential for DNA binding, but also for TGIF to function as a transcriptional repressor [Wotton et al., 1999a]. In addition, the clustering of mutations in the homeodomain suggests that either this region is more prone to sequence variations, that sequence variations in other regions of the gene may be lethal, or perhaps that non-homeodomain variants do not actually produce a HPE-related phenotype.
Third, we see a wide range of clinical severity in patients with TGIF mutations, ranging from very subtle manifestations typically only ascertained following the birth of a severely affected relative, to having profound sequelae of HPE incompatible with life. Further, craniofacial features appear consistent with the spectrum of midline deficits seen in patients with HPE in general [Solomon et al., 2010]. It is not surprising that patients with microdeletions of the TGIF gene or larger 18p deletions including the TGIF locus tend to display additional manifestations, such as congenital anomalies of the heart and digits not usually observed in patients with intragenic mutations. This is likely due to deletion of additional genes near the TGIF locus. At present, it is difficult to determine genes that could contribute to these manifestations, as findings may vary greatly among these patients [Turleau, 2008]. One attractive candidate gene is TWSG1, located at 18p11.22, a gene demonstrated in animal models to play a role in forebrain, foregut, and skeletal development [Nosaka et al., 2003; Petryk et al., 2004]. However, recent human studies have demonstrated minimal evidence for involvement of this gene in human HPE [Kauvar et al., 2011]. More broadly, information about the prevalence of and difference between findings among the 2 groups described here may be important to clinicians and affected families.
Fourth, similar to the case with SHH, but in contrast to ZIC2 and SIX3, over half of all patients (including both probands and relatives) with TGIF mutations are relatively mildly affected. These mildly affected patients may be described as either microform or non-penetrant carriers. However, we suspect that careful physical examination by experienced clinical geneticists often reveals subtle findings in patients previously labeled as ‘unaffected’. The prevalence of such mild clinical findings in patients with intragenic mutations in SHH, SIX3, and ZIC2 is 54, 36 and 13%, respectively [Lacbawan et al., 2009; Solomon et al., 2009b 2010]. These findings indicate that mutations in TGIF may result in less severe phenotypes compared to patients with mutations in either SIX3 or ZIC2.
An explanation for the generally mild phenotypes observed in patients with mutations in TGIF remains to be determined. It is interesting to note that among those with complete deletion of TGIF (i.e. partial monosomy 18p), less than half of the patients demonstrate findings of HPE. Our findings are consistent with earlier studies that have estimated that only ∼10% of all patients with 18p deletions (including deletion of TGIF) have HPE [Roessler and Muenke, 1998; Turleau, 2008]. This is in contrast to deletions of 2p21 (which includes SIX3), where virtually all patients have HPE, and deletions of 7q36 (which includes SHH), where approximately half of the patients have HPE. An explanation for these differences may involve nearby genes that also play a role in HPE pathogenesis.
The wide spectrum of severity and the difficulties in making precise genotype-phenotype correlations point to a complex model of HPE pathogenesis that demands further research. Functional analysis of Tgif in murine models has been inconclusive, although several findings point to this complex pathogenesis. For example, mouse models with homozygous disruption of Tgif fail to produce findings of HPE. In contrast, mice with decreased expression or knockdown of other HPE-associated genes (Zic2, Shh, Six3) typically display a strong HPE phenotype [Hayhurst and McConnell, 2003; Schachter and Krauss, 2008]. This suggests that, in addition to alterations in TGIF, other genetic modifiers (on 18p or other chromosomal regions) or environmental factors are necessary to result in HPE. TSWG1, as described above, was initially hypothesized to be one such genetic modifier, as it has been linked to forebrain development, although recent mutation analysis of the coding region of TWSG1 suggests that this gene does not play a significant role in human HPE [Petryk et al., 2004; Rosenfeld et al., 2010; Kauvar et al., 2011]. Another possibility may relate to the fact that mice with Tgif mutations have increased susceptibility to the teratogen retinoic acid [Bartholin et al., 2006]. Additionally, while homozygous mutations in Tgif1 (the mouse homologue to HPE-associated TGIF1 in humans) fail to create a pathogenic phenotype, mice with mutations in both Tgif1 and Tgif2 (a gene similar in structure to Tgif1) fail to undergo gastrulation, indicating a significant but incompletely defined role of this gene in mammalian embryogenesis [Powers et al., 2010].
Supplementary Material
Acknowledgements
We are extremely grateful to the patients and families presented in this report and to all other research participants, as well as to the Carter Centers for Brain Research in Holoprosencephaly and Related Malformations. This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
References
- Aguilella C, Dubourg C, Attia-Sobol J, Vigneron J, Blayau M, et al. Molecular screening of the TGIF gene in holoprosencephaly: identification of two novel mutations. Hum Genet. 2003;112:131–134. doi: 10.1007/s00439-002-0862-8. [DOI] [PubMed] [Google Scholar]
- Aughton DJ, AlSaadi AA, Transue DJ. Single maxillary central incisor in a girl with del(18p) syndrome. J Med Genet. 1991;28:530–532. doi: 10.1136/jmg.28.8.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkovich AJ, Quint DJ. Middle interhemispheric fusion: an unusual variant of holoprosencephaly. AJNR Am J Neuroradiol. 1993;14:431–440. [PMC free article] [PubMed] [Google Scholar]
- Bartholin L, Powers SE, Melhuish TA, Lasse S, Weinstein M, Wotton D. TGIF inhibits retinoid signaling. Mol Cell Biol. 2006;26:990–1001. doi: 10.1128/MCB.26.3.990-1001.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendavid C, Haddad BR, Griffin A, Huizing M, Dubourg C, et al. Multicolour FISH and quantitative PCR can detect submicroscopic deletions in holoprosencephaly patients with a normal karyotype. J Med Genet. 2006;43:496–500. doi: 10.1136/jmg.2005.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendavid C, Rochard L, Dubourg C, Seguin J, Gicquel I, et al. Array-CGH analysis indicates a high prevalence of genomic rearrangements in holoprosencephaly: an updated map of candidate loci. Hum Mutat. 2009;30:1175–1182. doi: 10.1002/humu.21016. [DOI] [PubMed] [Google Scholar]
- Boudailliez B, Morichon-Delvallez N, Goldfarb A, Pautard JC, Lenaerts C, Piussan C. Solitary upper incisor, hypopituitarism and monosomy 18p chromosome aberration. J Genet Hum. 1983;31:39–42. [PubMed] [Google Scholar]
- Chen CP, Chern SR, Du SH, Wang W. Molecular diagnosis of a novel heterozygous 268C→T (R90C) mutation in TGIF gene in a fetus with holoprosencephaly and premaxillary agenesis. Prenat Diagn. 2002;22:5–7. doi: 10.1002/pd.202. [DOI] [PubMed] [Google Scholar]
- Chen M, Kuo SJ, Liu CS, Chen WL, Ko TM, et al. A novel heterozygous missense mutation 377T>C (V126A) of TGIF gene in a family segregated with holoprosencephaly and moyamoya disease. Prenat Diagn. 2006;26:226–230. doi: 10.1002/pd.1385. [DOI] [PubMed] [Google Scholar]
- Dolan LM, Willson K, Wilson WG. 18p– syndrome with a single central maxillary incisor. J Med Genet. 1981;18:396–398. doi: 10.1136/jmg.18.5.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg C, Bendavid C, Pasquier L, Henry C, Odent S, David V. Holoprosencephaly. Orphanet J Rare Dis. 2007;2:8–21. doi: 10.1186/1750-1172-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Jaick KB, Powers SE, Bartholin L, Myers KR, Hahn J, et al. Functional analysis of mutations in TGIF associated with holoprosencephaly. Mol Genet Metab. 2007;90:97–111. doi: 10.1016/j.ymgme.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust J, Habedank M, Nieuwenhuijsen C. The 18p– syndrome. Report of four cases. Eur J Pediatr. 1976;123:59–66. doi: 10.1007/BF00497681. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, et al. Mutations in TGIF cause holoprosencephaly and link NODAL signaling to human neural axis development. Nat Genet. 2000;25:205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Barnes PD. Neuroimaging advances in holoprosencephaly: Refining the spectrum of the midline malformation. Am J Med Genet C Semin Med Genet. 2010;154C:120–132. doi: 10.1002/ajmg.c.30238. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Barkovich AJ, Stashinko EE, Kinsman SL, Delgado MR, Clegg NJ. Factor analysis of neuroanatomical and clinical characteristics of holoprosencephaly. Brain Dev. 2006;28:413–419. doi: 10.1016/j.braindev.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Barnes PD, Clegg NJ, Stashinko EE. Septopreoptic holoprosencephaly: a mild subtype associated with midline craniofacial anomalies. AJNR Am J Neuroradiol. 2010;31:1596–1601. doi: 10.3174/ajnr.A2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst M, McConnell SK. Mouse models of holoprosencephaly. Curr Opin Neurol. 2003;16:135–141. doi: 10.1097/01.wco.0000063761.15877.40. [DOI] [PubMed] [Google Scholar]
- Jin JZ, Gu S, McKinney P, Ding J. Expression and functional analysis of Tgif during mouse midline development. Dev Dyn. 2006;235:547–553. doi: 10.1002/dvdy.20642. [DOI] [PubMed] [Google Scholar]
- Kauvar EF, Hu P, Pineda-Alvarez DE, Solomon BD, Dutra A, et al. Minimal evidence for a direct involvement of twisted gastrulation homolog 1 (TWSG1) gene in human holoprosencephaly. Mol Genet Metab. 2011;102:470–480. doi: 10.1016/j.ymgme.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knepper JL, James AC, Ming JE. TGIF, a gene associated with human brain defects, regulates neuronal development. Dev Dyn. 2006;235:1482–1490. doi: 10.1002/dvdy.20725. [DOI] [PubMed] [Google Scholar]
- Küchle M, Kraus J, Rummelt C, Naumann GO. Synophthalmia and holoprosencephaly in chromosome 18p deletion defect. Arch Ophthalmol. 1991;109:136–137. doi: 10.1001/archopht.1991.01080010138045. [DOI] [PubMed] [Google Scholar]
- Lacbawan F, Solomon BD, Roessler E, El-Jaick K, Domené S, et al. Clinical spectrum of SIX3-associated mutations in holoprosencephaly: correlation between genotype, phenotype, and function. J Med Genet. 2009;46:389–398. doi: 10.1136/jmg.2008.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro L, Dubourg C, Pasquier L, Le Duff F, Blayau M, et al. Phenotypic and molecular variability of the holoprosencephalic spectrum. Am J Med Genet A. 2004;129A:21–24. doi: 10.1002/ajmg.a.30110. [DOI] [PubMed] [Google Scholar]
- Leoncini E, Baranello G, Orioli IM, Annerén G, Bakker M, et al. Frequency of holoprosencephaly in the International Clearinghouse Birth Defect Surveillance Systems: searching for population variations. Birth Defects Res A Clin Mol Teratol. 2008;82:585–591. doi: 10.1002/bdra.20479. [DOI] [PubMed] [Google Scholar]
- Levey EB, Stashinko E, Clegg NJ, Delgado MR. Management of children with holoprosencephaly. Am J Med Genet C Semin Med Genet. 2010;154C:183–190. doi: 10.1002/ajmg.c.30254. [DOI] [PubMed] [Google Scholar]
- Maden M. Retinoid signaling in the development of the central nervous system. Nat Rev Neurosci. 2003;3:843–853. doi: 10.1038/nrn963. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Shiota K. Holoprosencephaly in human embryos: epidemiologic studies in 150 cases. Teratology. 1977;16:261–272. doi: 10.1002/tera.1420160304. [DOI] [PubMed] [Google Scholar]
- Melhuish TA, Wotton D. The interaction of the carboxyl terminus-binding protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF. J Biol Chem. 2000;275:39762–39766. doi: 10.1074/jbc.C000416200. [DOI] [PubMed] [Google Scholar]
- Ming JE, Muenke M. Multiple hits during early embryonic development: digenic diseases and holoprosencephaly. Am J Hum Genet. 2002;71:1017–1032. doi: 10.1086/344412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog U, De Die-Smulders CE, Schrander-Stumpel CT, Engelen JJ, Hamers AJ, et al. Holoprosencephaly: the Maastricht experience. Genet Couns. 2001;12:287–298. [PubMed] [Google Scholar]
- Morales-Peralta E, Lantigua A. Deletion 18p associated with a single maxillary incisor: a case study. Rev Brasil Genet. 1994;17:341–343. [Google Scholar]
- Mukherjee K, Bürglin Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J Mol Evol. 2007;65:137–153. doi: 10.1007/s00239-006-0023-0. [DOI] [PubMed] [Google Scholar]
- Münke M. Clinical, cytogenetic and molecular approaches to the genetic heterogeneity of holoprosencephaly. Am J Med Genet. 1989;34:237–245. doi: 10.1002/ajmg.1320340222. [DOI] [PubMed] [Google Scholar]
- Münke M, Page DC, Brown LG, Armson BA, Zackai EH, et al. Molecular detection of a Yp/18 translocation in a 45,X holoprosencephalic male. Hum Genet. 1988;80:219–223. doi: 10.1007/BF01790089. [DOI] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Bocian M, Steinhaus K, Bianchi DW, et al. The mutational spectrum of the sonic hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum Mol Genet. 1999;8:2479–2488. doi: 10.1093/hmg/8.13.2479. [DOI] [PubMed] [Google Scholar]
- Nosaka T, Morita S, Kitamura H, Nakajima H, Shibata F, et al. Mammalian twisted gastrulation is essential for skeleto-lymphogenesis. Mol Cell Biol. 2003;23:2969–2980. doi: 10.1128/MCB.23.8.2969-2980.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli IM, Castilla EE. Clinical epidemiologic study of holoprosencephaly in South America. Am J Med Genet A. 2007;143A:3088–3099. doi: 10.1002/ajmg.a.32104. [DOI] [PubMed] [Google Scholar]
- Overhauser J, Mitchell HF, Zackai EH, Tick DB, Rojas K, Muenke M. Physical mapping of the holoprosencephaly critical region in 18p11.3. Am J Hum Genet. 1995;57:1080–1085. [PMC free article] [PubMed] [Google Scholar]
- Petryk A, Anderson RM, Jarcho MP, Leaf I, Carlson CS, et al. The mammalian twisted gastrulation gene functions in foregut and craniofacial development. Dev Biol. 2004;267:374–386. doi: 10.1016/j.ydbio.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Pineda-Alvarez DE, Dubourg C, David V, Roessler E, Muenke M. Current recommendations for the molecular evaluation of newly diagnosed holoprosencephaly patients. Am J Med Genet C Semin Med Genet. 2010;154C:93–101. doi: 10.1002/ajmg.c.30253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plawner LL, Delgado MR, Miller VS, Levey EB, Kinsman SL, et al. Neuroanatomy of holoprosencephaly as a predictor of function: beyond the face predicting the brain. Neurology. 2002;50:1058–1066. doi: 10.1212/wnl.59.7.1058. [DOI] [PubMed] [Google Scholar]
- Portnoï MF, Gruchy N, Marlin S, Finkel L, Denoyelle F, et al. Midline defects in deletion 18p syndrome: clinical and molecular characterization of three patients. Clin Dysmorphol. 2007;16:247–252. doi: 10.1097/MCD.0b013e328235a572. [DOI] [PubMed] [Google Scholar]
- Powers SE, Taniguchi K, Yen W, Melhuish TA, Shen J, et al. Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation. Development. 2010;137:249–259. doi: 10.1242/dev.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richieri-Costa A, Ribeiro LA. Variable phenotypic manifestations of a K44N mutation in the TGIF gene. Brain Dev. 2008;30:203–205. doi: 10.1016/j.braindev.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Roessler E, Muenke M. Holoprosencephaly: a paradigm for the complex genetics of brain development. J Inherit Metab Dis. 1998;21:481–497. doi: 10.1023/a:1005406719292. [DOI] [PubMed] [Google Scholar]
- Roessler E, Muenke M. The molecular genetics of holoprosencephaly. Am J Med Genet C Semin Med Genet. 2010;154C:52–61. doi: 10.1002/ajmg.c.30236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- Roessler E, El-Jaick KB, Dubourg C, Vélez JI, Solomon BD, et al. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum Mutat. 2009;10:E921–E935. doi: 10.1002/humu.21090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Ballif BC, Martin DM, Aylsworth AS, Bejjani BA, et al. Clinical characterization of individuals with deletions of genes in holoprosencephaly pathways by aCGH refines the phenotypic spectrum of HPE. Hum Genet. 2010;127:421–440. doi: 10.1007/s00439-009-0778-7. [DOI] [PubMed] [Google Scholar]
- Schachter KA, Krauss RS. Murine models of holoprosencephaly. Curr Top Dev Biol. 2008;84:139–170. doi: 10.1016/S0070-2153(08)00603-0. [DOI] [PubMed] [Google Scholar]
- Sepulveda W. Monosomy 18p presenting with holoprosencephaly and increased nuchal translucency in the first trimester: report of 2 cases. J Ultrasound Med. 2009;28:1077–1080. doi: 10.7863/jum.2009.28.8.1077. [DOI] [PubMed] [Google Scholar]
- Solomon BD, Lacbawan F, Jain M, Domené S, Roessler E, et al. A novel SIX3 mutation segregates with holoprosencephaly in a large family. Am J Med Genet A. 2009a;149A:919–925. doi: 10.1002/ajmg.a.32813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Lacbawan F, Mercier S, Clegg NJ, Delgado MR, et al. Mutations in ZIC2 in human holoprosencephaly: description of a novel ZIC2-specific phenotype and comprehensive analysis of 157 individuals. J Med Genet. 2009b;47:513–524. doi: 10.1136/jmg.2009.073049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Mercier S, Vélez JI, Pineda-Alvarez DE, Wyllie A, et al. Analysis of genotype-phenotype correlations in human holoprosencephaly. Am J Med Genet C Semin Med Genet. 2010;154C:133–141. doi: 10.1002/ajmg.c.30240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stashinko EE, Clegg NJ, Kammann HA, Sweet VT, Delgado MR, et al. A retrospective study of perinatal risk factors of 104 living children with holoprosencephaly. Am J Med Genet A. 2004;128A:114–119. doi: 10.1002/ajmg.a.30070. [DOI] [PubMed] [Google Scholar]
- Sulik KK, Dehart DB, Rogers JM, Chernoff N. Teratogenicity of low doses of all-trans retinoic acid in presomite mouse embryos. Teratology. 1995;51:398–403. doi: 10.1002/tera.1420510605. [DOI] [PubMed] [Google Scholar]
- Turleau Monosomy 18p. Orphanet J Rare Dis. 2008;3:4–8. doi: 10.1186/1750-1172-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Lo RS, Lee S, Massague JA. Smad transcriptional corepressor. Cell. 1999a;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- Wotton D, Lo RS, Swaby LA, Massagué J. Multiple modes of repression by the Smad transcriptional corepressor TGIF. J Biol Chem. 1999b;274:37105–37110. doi: 10.1074/jbc.274.52.37105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.