Abstract
The chromosome region 22q11.2 has long been recognized to be susceptible to genomic rearrangement. More recently, this genomic instability has been shown to extend distally (involving LCR22E–H) to the commonly deleted/duplicated region. To date, 21 index cases with ‘distal’ 22q11.2 duplications have been reported. We report on the clinical and molecular characterization of 16 individuals with distal 22q11.2 duplications identified by DNA microarray analysis. Two of the individuals have been partly described previously. The clinical phenotype varied among the patients in this study, although the majority displayed various degrees of developmental delay and speech disturbances. Other clinical features included behavioral problems, hypotonia, and dysmorphic facial features. Notably, none of the patients was diagnosed with a congenital heart defect. We found a high degree of inherited duplications. Additional copy number changes of unclear clinical significance were identified in 5 of our patients, and it is possible that these may contribute to the phenotypic expression in these patients as has been suggested recently in a 2-hit ‘digenic’ model for 16p12.1 deletions. The varied phenotypic expression and incomplete penetrance observed for distal 22q11.2 duplications makes it exceedingly difficult to ascribe pathogenicity for these duplications. Given the observed enrichment of the duplication in patient samples versus healthy controls, it is likely that distal 22q11.2 duplications represent a susceptibility/risk locus for speech and mild developmental delay.
Key Words: 22q11.2 distal duplication, Array comparative genomic hybridization, Behavioral disorders, Copy number variations, Developmental delay, Low copy repeats, SNP array, Speech delay
Introduction
The chromosome region 22q11.2 has long been recognized as a hotspot for genomic rearrangement and related disorders, such as 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome, OMIM 188400/OMIM 192430) [DiGeorge and Harley, 1965; Shprintzen et al., 1978], der(22) t(11;22) syndrome (OMIM 609029) and cat-eye syndrome (OMIM 115470) [Guanti, 1981; Edelmann et al., 1999a]. Der(22) syndrome and cat-eye syndrome are rare conditions characterized by increased copy-number of the most centromeric part of 22q11 [Zackai and Emanuel, 1980; McDermid and Morrow, 2002], whereas the microdeletions of 22q11.2 occur more often, with an estimated frequency of 1 in 4,000–6,000 live births [Yamagishi, 2002; Botto et al., 2003]. The 22q11 deletion syndrome is characterized by congenital heart defects, immune deficiency, transient neonatal hypocalcemia, velopharyngeal insufficiency and a distinctive facial appearance but also by learning disabilities and behavioral anomalies. The phenotype is variable with multiple organ systems being affected [Shprintzen et al., 1981; Scambler et al., 1992; Robin and Shprintzen, 2005]. Although the majority of the 22q11.2 deletions are de novo, some deletions (6–25%) are inherited from mildly affected or normal parents [Leana-Cox et al., 1996; McDonald-McGinn et al., 1997, 2001; Ryan et al., 1997; Matsuoka et al., 1998].
The genomic region of chromosome band 22q11.2 contains several large segmental duplications/low copy repeats (LCRs) that function as mediators of non-allelic homologous recombination (NAHR) and predispose the genomic region to chromosomal rearrangements [Edelmann et al., 1999b; Shaikh et al., 2000; Ensenauer et al., 2003]. Eight LCR clusters (LCR22A–H) have been identified in the 22q11.2 genomic region [Edelmann et al., 1999c; Shaikh et al., 2000, 2007]. The modules that build these LCR show significant (97–98%) sequence identity to each other, although the LCR22s differ between each other in content and organization of the modules [Shaikh et al., 2000]. Most (>85%) individuals with proximal (involving LCR22A–D) 22q11 deletions (i.e. 22q11.2 deletion syndrome) have a 3-Mb deletion [Morrow et al., 1995; Shaikh et al., 2000; Emanuel, 2008] with breakpoints in LCR22s A and D [Edelmann et al., 1999b; Babcock et al., 2003], the largest and most complex of the LCR22s. Deletions mediated by distal LCR22s (LCR22E–H) have also been described, although these deletions are found less frequently than the common proximal 22q11 deletions [Rauch et al., 1999; Saitta et al., 1999; Mikhail et al., 2007; Ben-Shachar et al., 2008]. This may be due to differences in the rates of genomic rearrangement mediated by the various LCR clusters (due to underlying sequence identity/motif organization differences) [Shaikh et al., 2007] or the wider phenotypic spectrum associated with distal deletions. A systematic assessment of atypical 22q11.2 deletions showed that atypical congenital heart defects and mild dysmorphism are recognizable features in patients with distal 22q11.2 deletions but very uncommon in patients with conotruncal heart defects [Rauch et al., 2005].
The proximal breakpoint for most reported distal 22q11.2 deletions has been shown to lie within LCR22D, whereas the distal breakpoints appear to be spread across the other LCR22 clusters [Saitta et al., 1999; Shaikh et al., 2007]. In 2007, detailed molecular analysis reported by Shaikh et al. showed that the distal deletion breakpoints mapped to a BCRL (breakpoint cluster region-like) module, suggesting that homologous sequences within this module may represent a rearrangement hotspot. The BCRL module is present in all LCR22s except for in LCR22B [Shaikh et al., 2007]. The orientation of these BCRL modules may predict between which LCR22s it is likely that NAHR will occur, since it has been proposed that modules within the LCRs that have a direct orientation with respect to one another are likely to mediate rearrangements [Shaffer and Lupski, 2000].
To date, about 50 index cases with proximal (involving LCR22A–D) 22q11.2 duplications have been reported [Edelmann et al., 1999c; Ensenauer et al., 2003; Hassed et al., 2004; Portnoi et al., 2005; Sparkes et al., 2005; Alberti et al., 2007; Ou et al., 2008; Wentzel et al., 2008; Yu et al., 2008]. The phenotypes of the patients with proximal 22q11.2 microduplications are diverse, with symptoms ranging from mild learning disability and mild dysmorphic facial features to severe mental retardation and multiple congenital malformations [Ensenauer et al., 2003]. Other phenotypic features observed include speech delay, behavioral problems, hearing loss, growth delay, urogenital abnormalities, muscular hypotonia, congenital heart malformation, seizures and bladder exstrophy [Wentzel et al., 2008; Portnoi, 2009; Draaken et al., 2010; Lundin et al., 2010]. Many families in which patients inherited the duplication from mildly affected or asymptomatic parents have been reported. The paucity of reported proximal 22q11.2 microduplication cases may, in part, be explained by the absence of a defined phenotype and the wide range of and sometimes mild symptoms with the possibility of incomplete penetrance [Wentzel et al., 2008]. Therefore, mild cases with microduplications within proximal 22q11.2 may not be subject to testing. In addition, limitations in the previously used molecular techniques made it difficult to detect extra copies of such small regions [Sparkes et al., 2005].
Twenty-two index cases with 22q11.2 duplications involving the distal LCR22s have been reported so far [Descartes et al., 2008; Ou et al., 2008; Coppinger et al., 2009; Shimojima et al., 2010]. Similar to the proximal duplications, there seems to be a high rate of familial transmission [Ensenauer et al., 2003; Hassed et al., 2004; Portnoi et al., 2005; Ou et al., 2008] and the phenotypes vary among family members carrying the duplications [Ensenauer et al., 2003; Yobb et al., 2005; Ou et al., 2008]. In the study by Coppinger et al., the clinical picture varied with no clearly definable collection of phenotypic features shared among the patients and no correlation could be recognized between the severity of the clinical features and the size or location of the duplications [Coppinger et al., 2009].
In this present study, we report on the clinical and molecular characterization of 16 individuals with duplications of chromosome region 22q11.21–q11.23 (i.e. distal 22q11.2 duplications).
Material and Methods
Array analysis was performed on 11,463 patients referred to different European and Australian clinical genetics centers for a variety of neurodevelopmental phenotypes. Six patients were recruited from Nijmegen (the Netherlands), 6 patients from Melbourne (Australia), 2 patients from Oxford (England), 1 patient from Pavia (Italy) and 1 patient from Stockholm (Sweden). Two of the patients have been published previously elsewhere (table 1) [Bruno et al., 2009; Wincent et al., 2011].
Table 1.
Molecular details and phenotypic features of individuals with a distal 22q11.2 duplication
| Case | dup chr22 band | Seg dup | bp start; stop (NCBI36/hg18) | Size (Mb) | Platform | Origin | Additional aberrations | Age of patient when investigated | Sex | Developmental delay (mild/moderate/severe) | Autism/behavioral problems | Speech or language problems | Dys- morphic features | Heart defect | Hypotonia | Seizures | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | q11.21q11.22 | C-D/E | 19388824; 20784027 | 1.39 | 180K Agilent | de novo | 14 months | M | no | no | no | yes | no | no | no | Beekwith-Wiedeman-like facial features, macroglossia, facial hemangiomata, polyhydramnios, enlarged placenta | |
| 2 | q11.21q11.22 | D-E | 20128873; 21284860 | 1.15 | 180K Agilent | maternally inherited (healthy mother) | dup(6)(p22.3), 170 kb (1 gene), paternally inherited | 2 years 6 months | F | mild | no | yes | yes | no | yes | no | high broad forehead, downslanting low-set ears with flat upper helices, coarse nose, broad mouth, full lips, retrognathia |
| 3 | q11.21q11.22 | D-D/E | 20160123; 21039509 | 0.88 | Illumina-12-300K | maternally inherited (affected mother) | del(16)(q24.1q24.2), 1.57 Mb (20 genes), paternally inherited | F | yes | ? | ? | ? | ? | ? | ? | ||
| 4 | q11.21q11.22 | D-D/E | 20253470; 20982454 | 0.73 | 250K NSP Affymetrix | maternally inherited | 16 years | F | mild | behavioral problems, auto mutilation | ? | yes | no | yes neonatal | no | cervical syringomyelia, Chiari I malformation, retrocerebellar cysts, hirsutism, coarse facies | |
| 5 | q11.21q11.22 | E-F | 21328248; 21984237 | 0.65 | 180K Agilent | maternally inherited (healthy mother) | 2 years | M | mild | yes | yes | yes | no | no | no | ||
| 6 | q11.22q11.23 | E-F | 21392613; 21984436 | 0.59 | Illumina-12-300K | unknown | F | severe | autism | ? | no | ? | ? | epilepsia | absence of septum pellucidum | ||
| 7 | q11.22q11.23 | E-H | 21322838; 23319936 | 1.99 | Illumina CytoSNP-12 | unknowna | 9 years | F | no | yes | expressive language impairment | no | no | no | no | peculiar gait | |
| 8 | q11.22q11.23 | E-H | 21328284; 23326964 | 1.99 | 180K Agilent | unknown | del(4)(ql2), 77 kb (3 genes), origin unknown | 3 years 6 months | F | moderate | no | yes | yes | no | lax ankle joints | yes, rolandic epilepsy | brachycephaly and fetal pads, (may have downslanting palpebral fissures), MRI showed enlarged Sylvian fissure |
| 9b | q11.22q11.23 | E-H | 21362255; 23325382 | 1.96 | 244K Agilent | paternally inherited (healthy father) | 0–4 years | ? | moderate | ? | ? | no | ? | ? | yes | frontomedial polymicrogyri and corpus callosum agenesis, normal growth | |
| 10 | q11.23 | F-H | 21998015; 23322070 | 1.32 | 180K Agilent | maternally inherited (healthy mother) | 35 years | M | moderate | autism | yes | no | no | yes | no | at —25 years progressive decline in functioning: loss of speech, progressive spasticity (now wheelchair dependent), problems with swallowing/eating (PEG); high suspicion of mitochondrial disorder, tests still ongoing | |
| 11 | q11.23 | F-H | 22003184; 23307286 | 1.30 | Illumina-12-300K | maternally inherited (healthy mother) | 4 years | F | yes | ? | ? | ? | ? | ? | ? | consanguine parents (first cousins); the patient's genome showed 5% homozygosity | |
| 12 | q11.23 | F-H | 22003184; 23409925 | 1.41 | Illumina-12-300K | paternally inherited | M | ? | ? | ? | ? | ? | ? | ? | IUGR/failure to thrive, central sleep apnea | ||
| 13 | q11.23 | F-H | 22014324; 23321952 | 1.31 | 244K Agilent | maternally inherited | ? | yes | ? | ? | yes | ? | ? | ? | |||
| 14 | q11.23 | F-H | 22017292; 23284714 | 1.27 | 250K NSP Affymetrix | unknown | del(16)(pl3.2), 250 kb (1 gene), origin unknown | 11 years | M | moderate | no | yes | no | no | mild | yes | MRI normal |
| 15 | q11.23 | F-H | 22038020; 23409925 | 1.37 | Illumina-12-300K | paternally inherited (healthy father) | dup(4)(q35.2), 1.94 Mb (7) genes paternally inherited | M | ? | ? | speech delay | ? | ? | ? | ? | ||
| 16c | q11.23 | F-H | 21998015; 23308071 | 1.31 | 244K Agilent | de novo | 4 years | M | mild | no | severe speech and language disturbance | no | no | no | no | ||
| Summary | 12/14 | 5/10 | 8/9 | 6/12 | 0/9 | 5/9 | 4/11 | ||||||||||
Not in father and two siblings. Mother unavailable.
Case 10, Bruno et al. [2009].
Case 34, Wincent et al. [2011].
In order to find etiological causes for the patients’ phenotypes, DNA samples isolated from peripheral blood lymphocytes were analyzed with genome-wide DNA microarrays (molecular karyotyping). The arrays used encompassed 38K BAC, 180K Agilent, 244K Agilent, Illumina-12-300K or 250K NSP Affymetrix (table 1). If possible, the patients initially analyzed by the 38K BAC array or the 250K NSP array were reanalyzed with the 244K/180K Agilent array in order to refine the breakpoints. If available, parental samples were investigated for inheritance by molecular karyotyping or multiplex ligation probe amplification (MLPA). Phenotypic data on patients and parents were collected from the referring physicians.
244K/180K oligonucleotide arrays with complete genome coverage produced by Agilent Technologies (Palo Alto, Calif., USA) were used. Experiments were performed according to the manufacturer's protocol. After hybridization and washing, the slides were scanned on an Agilent Microarray Scanner. Captured images were analyzed with Feature Extraction Software v. 9.1 and ADM2 algorithm in the DNA analytics software (V4.0) (Agilent Technologies). Genomic start and stop positions of the duplications were determined by visual inspection of the numerical normalized log2 ratio values in the table view of the DNA analytics software package. Genomic positions are according to the NCBI36/hg18 build in UCSC.
MLPA was used in some of the cases for confirmation of gene dose imbalances and for investigation of parental samples. Synthetic MLPA probes were designed as described previously [Barbaro et al., 2007]. The MLPA reaction was performed according to the manufacturer's standard protocol and reagents (MRC Holland, Amsterdam, the Netherlands).
Results
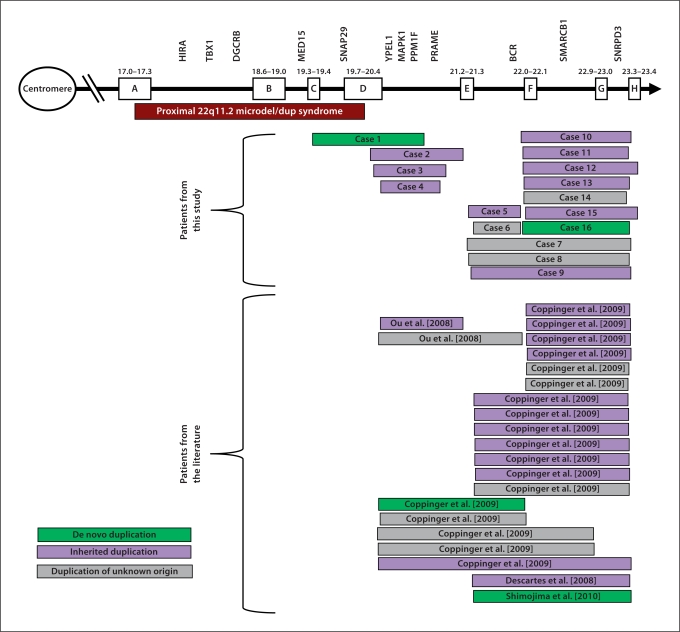
Among 11,463 patients with idiopathic mental retardation, brain malformations, autism spectrum disorders, and/or speech delay, we identified 16 individuals with duplications of chromosome band 22q11.21–q11.23. One duplication was flanked proximally by LCR-C, 3 by LCR-D, 5 by LCR-E and 7 by LCR-F. The distal breakpoints were in 1 case flanked by LCR-E, in 2 cases by LCR-F, and in 10 cases by LCR-H. In 3 cases the distal breakpoint was located between LCR-D and LCR-E (table 1, fig. 1).
Fig. 1.
Schematic overview of distal 22q11.2 duplications from this study and from the literature.
In 2 cases the duplication was de novo, in 10 cases it was inherited and in 4 cases parental samples were not available. The parents of cases 2, 5, 9, 11 and 15 had duplications but were reported to be clinically healthy. The duplication was also identified in the apparently unaffected maternal grandmother of case 5. However, the mother of case 3, who had the duplication, had learning and behavioral problems. The clinical status of the other parents carrying duplications was unavailable. Case 1 was initially referred to the genetic center to exclude Beckwith-Wiedemann syndrome because during pregnancy a large tongue, a large placenta and polyhydramnion were noted. After birth he showed facial hemangioma and a large tongue. However, methylation of LIT1 and H19 were normal and since the boy did not demonstrate any other BWS characteristics (such as overgrowth, asymmetry, organomegaly, hypoglycaemia, ear creases or umbilical hernia), he was no longer suspected of BWS. The de novo duplication identified in this patient, partly overlapped that of a previously reported de novo duplication identified in patient 10 in the study by Coppinger et al. [2009] (fig. 1). Case 16, a boy with a severe speech and language disturbance, is to the best of our knowledge the first reported patient with a de novo duplication involving LCRF–H.
There have been no previous reports of cases with duplications flanked by LCRE–F, but 2 such duplications were identified among our patients. Case 5, with a maternally inherited duplication, had mild developmental delay and behavioral problems. The duplication was also identified in the maternal grandmother. Case 6 had a severe developmental delay, autism, epilepsy and absence of septum pellucidum. The origin of her duplication was unknown. One paternally inherited D–E duplication has previously been described in a girl with developmental delay and dysmorphic features (patient 7 in the study by Ou et al.) [Ou et al., 2008]. We identified a maternally inherited D–E duplication in a girl (case 2) with a mild developmental delay, friendly behavior and dysmorphic facial features including a high broad forehead, downslanting low-set ears with flat upper helices, coarse nose, broad mouth, full lips, and retrognathia.
In 5 cases (31%), additional copy number changes of unclear significance were identified. A paternally inherited 170-kb duplication of chromosome 6p22.3 (chr6: 15387469–15557420) was found in case 2. The duplication comprised 2 exons of JARID2 which is an ortholog of the mouse jumonji gene, which encodes a nuclear protein essential for mouse embryogenesis. Case 3 showed a paternally inherited 1.57-Mb deletion in 16q24.1–q24.2 (chr16: 85271219–86844155) involving approximately 20 genes among which were MAP1LC3B, JPH3, and SLC7A5. Case 8 had a 77-kb deletion of chromosome 4q12 (chr4: 56985292–57062624) comprising the genes PPAT, PAICS, and SRP72. In case 14, a 250-kb deletion of 16p13.2 (SNP_A-4205445→SNP_A-2004534) of unknown origin involving GRIN2A, that encodes an NMDA receptor subunit, was identified. Submicroscopic deletions, point mutations and translocation in 16p13 encompassing GRIN2A have recently been associated with neurodevelopmental phenotypes [Endele et al., 2010; Reutlinger et al., 2010]. This deletion may therefore also contribute to the phenotype of this patient. Case 15 had a paternally inherited 1.94-Mb duplication of chromosome 4q35.2 (chr4: 187452010–189387111) comprising 7 genes.
The phenotypes of the cases are summarized in table 1. The clinical phenotype varied among the individuals in this study, although a majority of cases displayed various degrees of developmental delay, ranging from mild to severe, and speech disturbances. Other clinical features present in more than 5 cases included behavioral problems, hypotonia and dysmorphic facial features. Notably, none of the cases in our study had a diagnosed congenital heart defect.
Discussion
In the present study, we report on the clinical and molecular characterization of 16 individuals with distal duplications of chromosome band 22q11.21–q11.23 (involving LCR22E–H and located distal to the region typically deleted in DiGeorge syndrome). Distal 22q11.21–q11.23 duplications are rare and to date only 22 cases have been reported. We identified in this study 16 such duplications among 11,463 patients tested which results in an estimated frequency of around 0.1%. The estimated frequency is slightly elevated in our patient population compared to the study by Coppinger et al. [2009], who identified 18 distal duplications among 22,096 patients tested. This may be due to patient selection, since a patient cohort with speech delay, brain malformations and autism spectrum disorders were included in our study. In concordance with the findings by Coppinger et al. [2009], many of the duplications in our cohort were flanked by LCRE–H and LCRF–H. Since BCRL-E, BCRL-F, and BCRL-H are in the same orientation [Shaikh et al., 2007], our findings of E–H-, E–F- and F–H-mediated duplications support the hypothesis by Shaikh et al. that BCRL motifs in the same orientation facilitate NAHR [Shaikh et al., 2007]. More surprising was that 3 of our distal breakpoints were not flanked by known LCR22s, but all resided in a region of ∼20.9–21.0 Mb on chromosome 22q (fig. 1). This could indicate an additional locus that predisposes to NAHR (the proximal breakpoints lie within LCR22s likely excluding a replication-based mechanism).
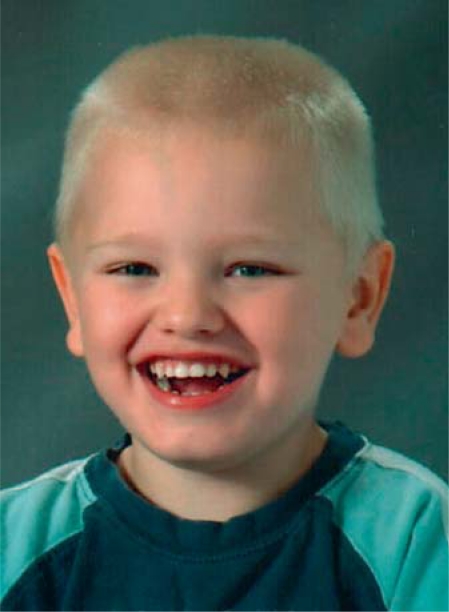
Case 16 in our study, with a de novo duplication involving LCRF–H, has a phenotype that is highly concordant with that of patient 14 in the study by Coppinger et al.[2009] (in that article, the patient L photo in fig. 1 corresponds to patient 14 in table 2 [pers. commun.]). Both cases had speech impairment and they have a similar facial appearance (fig. 2), although neither shows evident facial dysmorphic features. Case 16 had very mild developmental delay but severe speech and language disturbance. It is noteworthy that 6 of the 10 patients with E/F–H duplications in our study had a speech delay. Case 7 had expressive language impairment with articulation difficulties. She was also reported to be shy and oversensitive and had problems in maintaining friendships. Her father, who did not carry the duplication, had literacy problems and a family history of dyslexia. Case 7 and her 2 older brothers, neither of whom carried the duplication, all attended mainstream education and had performance IQ in the normal range. The oldest brother had some expressive vocabulary problems but was otherwise linguistically normal and the younger of the brothers had some difficulties in following and giving simple instructions but had no overt language difficulties. Case 8 used approximately 20 words and she did not put 2 words together at the age of 3.5 years. Case 10 spoke his first words at 4 years of age and although was later able to speak, he has had a severe decline in functioning since age 25 and can no longer speak. Furthermore, he had a gastrostomy because of severe difficulties with swallowing, and he is now wheelchair dependent due to progressive spasticity. There is a high suspicion of a mitochondrial disorder, although this could not be confirmed by genetic tests (TK2, POLG1, DGUOK, Twinkle, PDHA1 mutation analysis in fibroblasts).
Fig. 2.
Facial features of case 16.
So far, microduplications of distal chromosome 22q11.2 may be largely undetected as a result of an unspecific and/or mild phenotype leading to problems with ascertainment. In concordance with previous studies of both proximal and distal 22q11.2 duplications, we found a high degree of inherited duplications (in 83% of cases in this study where parental samples were available). Parents of a child with an inherited chromosome abnormality may sometimes show mild variations of the child's phenotype, which for example have been reported for the 22q11.21 microdeletion syndrome that predominantly has a de novo occurrence [Leana-Cox et al., 1996; McDonald-McGinn et al., 1997, 2001; Ryan et al., 1997; Matsuoka et al., 1998]. Unfortunately, we only had phenotypic data available on 7 of the parents from whom duplications were inherited. Six of these parents were reported to be healthy and 1 parent was affected.
The varied phenotypic expression and incomplete penetrance observed for distal 22q11.2 duplications makes it exceedingly difficult to ascribe pathogenicity for these duplications. Although the fact that all probands reported so far display a clinical phenotype might be due to ascertainment bias, distal 22q11.2 duplications are rarely reported as normal variants. The duplications reported in control samples in the Database of Genomic Variation [http://projects.tcag.ca/variation/] overlapping the duplications identified in our patients are smaller and do not cover all the genes. Given that distal 22q11.2 duplications, along with a growing number of recurrent genomic deletions and duplications [Itsara et al., 2009], appear to be enriched in individuals with neurodevelopmental and neurobehavioral phenotypes compared to control samples, it is likely that distal 22q11.2 duplications represent a susceptibility/risk locus for speech and mild developmental delay rather than causal variants. These copy number changes are insufficient to cause the observed phenotypic abnormality, and additional genetic, epigenetic or environmental factors may be required.
To this end, it is noteworthy that a digenic/multigenic model has recently been demonstrated for 16p12.1 deletions [Girirajan et al., 2010], which are inherited in the majority of cases and show considerable variability in expression. These deletions have been shown to cooccur with secondary pathogenic or ‘uncertain significance’ copy number change in approximately 24% of cases. The second hit could potentially be another copy number variant, a disruptive single-base-pair mutation in a functionally related gene, or an environmental event that influences the phenotype. We identified additional copy number changes of unclear clinical significance in 5 of our cases making a 2-hit event plausible. These additional copy number changes were in case 2, 3 and 15 inherited and in case 8 and 14 parental samples were not available. In case 15, the additional copy number change was inherited from the parent carrying the 22q11 duplication. However, in case 2 and 3, the additional change was inherited from the non-22q11.2 duplication carrier parent making these additional copy number changes good candidates for the ‘second hit’ event. Nevertheless, proving digenicity/multigenicity in individual cases is not feasible at present, and it is unlikely that copy number variations represent the additional ‘hit’ in the majority of cases. The study of individuals/families with distal 22q11.2 duplications by whole genome or exome sequencing, as has recently been demonstrated on a small number of idiopathic mental retardation cases [Vissers et al., 2010], may begin to shed light on how and to what extent the duplications contribute to phenotypic expression.
Conclusion
Although there are now more than 35 index cases with distal 22q11.2 microduplications (including the cases from this study) reported in the literature, extended investigations of families harboring these duplications are needed to provide insight into the mechanisms of pathogenicity of these duplications. There is an urgent need for ascertainment of risk figures for phenotypic abnormality in individuals with 22q11.2 distal duplications to help alleviate the current interpretational challenges for diagnostic (including prenatal) testing and counseling.
Acknowledgements
We are grateful to Margareta Lagerberg and Dr. Elham Sadighi Akha for performing array hybridizations. This work was supported by funds from the Swedish Research Council, the Karolinska Institutet, Frimurarna Barnahuset Foundation, Linnea och Josef Carlsson Foundation, Kronprinsessan Lovisa Foundation, Åke Wibergs foundation and by the Wellcome Trust 498 (075491/Z/04). S.J.L.K., Elham Sadighi Akha, H.S. and D.A.D.K. are supported by the NIHR Biomedical Research Centre, Oxford with funding from the Department of Health's NIHR Biomedical Research Centre's funding scheme. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health. S.J.L.K., Elham Sadighi Akha and D.F.N. are also supported by the Wellcome Trust (075491/Z/04). D.F.N. is an MRC career development fellow. Sample 7 was collected as part of the SLI Consortium (SLIC) and was funded by the Wellcome Trust.
References
- Alberti A, Romano C, Falco M, Cali F, Schinocca P, et al. 1.5 Mb de novo 22q11.21 microduplication in a patient with cognitive deficits and dysmorphic facial features. Clin Genet. 2007;71:177–182. doi: 10.1111/j.1399-0004.2007.00750.x. [DOI] [PubMed] [Google Scholar]
- Babcock M, Pavlicek A, Spiteri E, Kashork CD, Ioshikhes I, et al. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 2003;13:2519–2532. doi: 10.1101/gr.1549503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaro M, Oscarson M, Schoumans J, Staaf J, Ivarsson SA, Wedell A. Isolated 46,XY gonadal dysgenesis in two sisters caused by a Xp21.2 interstitial duplication containing the DAX1 gene. J Clin Endocrinol Metab. 2007;92:3305–3313. doi: 10.1210/jc.2007-0505. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, et al. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet. 2008;82:214–221. doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto LD, May K, Fernhoff PM, Correa A, Coleman K, et al. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics. 2003;112(1Pt1):101–107. doi: 10.1542/peds.112.1.101. [DOI] [PubMed] [Google Scholar]
- Bruno DL, Ganesamoorthy D, Schoumans J, Bankier A, Coman D, et al. Detection of cryptic pathogenic copy number variations and constitutional loss of heterozygosity using high resolution SNP microarray analysis in 117 patients referred for cytogenetic analysis and impact on clinical practice. J Med Genet. 2009;46:123–131. doi: 10.1136/jmg.2008.062604. [DOI] [PubMed] [Google Scholar]
- Coppinger J, McDonald-McGinn D, Zackai E, Shane K, Atkin JF, et al. Identification of familial and de novo microduplications of 22q11.21–q11.23 distal to the 22q11.21 microdeletion syndrome region. Hum Mol Genet. 2009;18:1377–1383. doi: 10.1093/hmg/ddp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descartes M, Franklin J, de Stahl TD, Piotrowski A, Bruder CE, et al. Distal 22q11.2 microduplication encompassing the BCR gene. Am J Med Genet A. 2008;146A:3075–3081. doi: 10.1002/ajmg.a.32572. [DOI] [PubMed] [Google Scholar]
- DiGeorge AM, Harley RD. The association of aniridia, Wilms's tumor, and genital abnormalities. Trans Am Ophthalmol Soc. 1965;63:64–69. [PMC free article] [PubMed] [Google Scholar]
- Draaken M, Reutter H, Schramm C, Bartels E, Boemers TM, et al. Microduplications at 22q11.21 are associated with non-syndromic classic bladder exstrophy. Eur J Med Genet. 2010;53:55–60. doi: 10.1016/j.ejmg.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Spiteri E, McCain N, Goldberg R, Pandita RK, et al. A common breakpoint on 11q23 in carriers of the constitutional t(11;22) translocation. Am J Hum Genet. 1999a;65:1608–1616. doi: 10.1086/302689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Morrow BE. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet. 1999b;64:1076–1086. doi: 10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, et al. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999c;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- Emanuel BS. Molecular mechanisms and diagnosis of chromosome 22q11.2 rearrangements. Dev Disabil Res Rev. 2008;14:11–18. doi: 10.1002/ddrr.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endele S, Rosenberger G, Geider K, Popp B, Tamer C, et al. Mutation in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, et al. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet. 2003;73:1027–1040. doi: 10.1086/378818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guanti G. The aetiology of the cat eye syndrome reconsidered. J Med Genet. 1981;18:108–118. doi: 10.1136/jmg.18.2.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassed SJ, Hopcus-Niccum D, Zhang L, Li S, Mulvihill JJ. A new genomic duplication syndrome complementary to the velocardiofacial (22q11 deletion) syndrome. Clin Genet. 2004;65:400–404. doi: 10.1111/j.0009-9163.2004.0212.x. [DOI] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leana-Cox J, Pangkanon S, Eanet KR, Curtin MS, Wulfsberg EA. Familial DiGeorge/velocardiofacial syndrome with deletions of chromosome area 22q11.2: report of five families with a review of the literature. Am J Med Genet. 1996;65:309–316. doi: 10.1002/(SICI)1096-8628(19961111)65:4<309::AID-AJMG12>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Lundin J, Söderhäll C, Lundén L, Hammarsjö A, White I, et al. 22q11.2 microduplication in two patients with bladder exstrophy and hearing impairment. Eur J Med Genet. 2010;53:61–65. doi: 10.1016/j.ejmg.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Matsuoka R, Kimura M, Scambler PJ, Morrow BE, Imamura S, et al. Molecular and clinical study of 183 patients with conotruncal anomaly face syndrome. Hum Genet. 1998;103:70–80. doi: 10.1007/s004390050786. [DOI] [PubMed] [Google Scholar]
- McDermid HE, Morrow BE. Genomic disorders on 22q11. Am J Hum Genet. 2002;70:1077–1088. doi: 10.1086/340363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn DM, LaRossa D, Goldmuntz E, Sullivan K, Eicher P, et al. The 22q11.2 deletion: screening, diagnostic workup, and outcome of results; report on 181 patients. Genet Test. 1997;1:99–108. doi: 10.1089/gte.1997.1.99. [DOI] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Tonnesen MK, Laufer-Cahana A, Finucane B, Driscoll DA, et al. Phenotype of the 22q11.2 deletion in individuals identified through an affected relative: cast a wide FISHing net! Genet Med. 2001;3:23–29. doi: 10.1097/00125817-200101000-00006. [DOI] [PubMed] [Google Scholar]
- Mikhail FM, Descartes M, Piotrowski A, Andersson R, Diaz de Stahl T, et al. A previously unrecognized microdeletion syndrome on chromosome 22 band q11.2 encompassing the BCR gene. Am J Med Genet A. 2007;143A:2178–2184. doi: 10.1002/ajmg.a.31882. [DOI] [PubMed] [Google Scholar]
- Morrow B, Goldberg R, Carlson C, Das Gupta R, Sirotkin H, et al. Molecular definition of the 22q11 deletions in velo-cardio-facial syndrome. Am J Hum Genet. 1995;56:1391–1403. [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Berg JS, Yonath H, Enciso VB, Miller DT, et al. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet Med. 2008;10:267–277. doi: 10.1097/GIM.0b013e31816b64c2. [DOI] [PubMed] [Google Scholar]
- Portnoi MF. Microduplication 22q11.2: a new chromosomal syndrome. Eur J Med Genet. 2009;52:88–93. doi: 10.1016/j.ejmg.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Portnoi MF, Lebas F, Gruchy N, Ardalan A, Biran-Mucignat V, et al. 22q11.2 duplication syndrome: two new familial cases with some overlapping features with DiGeorge/velocardiofacial syndromes. Am J Med Genet A. 2005;137:47–51. doi: 10.1002/ajmg.a.30847. [DOI] [PubMed] [Google Scholar]
- Rauch A, Pfeiffer RA, Leipold G, Singer H, Tigges M, Hofbeck M. A novel 22q11.2 microdeletion in DiGeorge syndrome. Am J Hum Genet. 1999;64:659–666. doi: 10.1086/302235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Zink S, Zweier C, Thiel CT, Koch A, et al. Systematic assessment of atypical deletions reveals genotype-phenotype correlation in 22q11.2. J Med Genet. 2005;42:871–876. doi: 10.1136/jmg.2004.030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reutlinger C, Helbig I, Gawelcyk B, Subero JI, Tönnies H, et al. Deletions in 16p13 including GRIN2A in patients with intellectual disability, various dysmorphic features, and seizure disorders of the rolandic region. Epilepsia. 2010;51:1870–1873. doi: 10.1111/j.1528-1167.2010.02555.x. [DOI] [PubMed] [Google Scholar]
- Robin NH, Shprintzen RJ. Defining the clinical spectrum of deletion 22q11.2. J Pediatr. 2005;147:90–96. doi: 10.1016/j.jpeds.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, et al. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet. 1997;34:798–804. doi: 10.1136/jmg.34.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitta SC, McGrath JM, Mensch H, Shaikh TH, Zackai EH, Emanuel BS. A 22q11.2 deletion that excludes UFD1L and CDC45L in a patient with conotruncal and craniofacial defects. Am J Hum Genet. 1999;65:562–566. doi: 10.1086/302514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scambler PJ, Kelly D, Lindsay E, Williamson R, Goldberg R, et al. Velo-cardio-facial syndrome associated with chromosome 22 deletions encompassing the DiGeorge locus. Lancet. 1992;339:1138–1139. doi: 10.1016/0140-6736(92)90734-k. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, O'Connor RJ, Pierpont ME, McGrath J, Hacker AM, et al. Low copy repeats mediate distal chromosome 22q11.2 deletions: sequence analysis predicts breakpoint mechanisms. Genome Res. 2007;17:482–491. doi: 10.1101/gr.5986507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima K, Imai K, Yamamoto T. A de novo 22q11.22q11.23 interchromosomal tandem duplication in a boy with developmental delay, hyperactivity and epilepsy. Am J Med Genet A. 2010;152A:2820–2826. doi: 10.1002/ajmg.a.33658. [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg RB, Lewin ML, Sidoti EJ, Berkman MD, et al. A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: velo-cardio-facial syndrome. Cleft Palate J. 1978;15:56–62. [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg RB, Young D, Wolford L. The velo-cardio-facial syndrome: a clinical and genetic analysis. Pediatrics. 1981;67:167–172. [PubMed] [Google Scholar]
- Sparkes R, Chernos J, Dicke F. Duplication of the 22q11.2 region associated with congenital cardiac disease. Cardiol Young. 2005;15:229–231. doi: 10.1017/S1047951105000466. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Wentzel C, Fernstrom M, Ohrner Y, Anneren G, Thuresson AC. Clinical variability of the 22q11.2 duplication syndrome. Eur J Med Genet. 2008;51:501–510. doi: 10.1016/j.ejmg.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Wincent J, Anderlid BM, Lagerberg M, Nordenskjold M, Schoumans J. High-resolution molecular karyotyping in patients with developmental delay and/or multiple congenital anomalies in a clinical setting. Clin Genet. 2011;79:147–157. doi: 10.1111/j.1399-0004.2010.01442.x. [DOI] [PubMed] [Google Scholar]
- Yamagishi H. The 22q11.2 deletion syndrome. Keio J Med. 2002;51:77–88. doi: 10.2302/kjm.51.77. [DOI] [PubMed] [Google Scholar]
- Yobb TM, Somerville MJ, Willatt L, Firth HV, Harrison K, et al. Microduplication and triplication of 22q11.2: a highly variable syndrome. Am J Hum Genet. 2005;76:865–876. doi: 10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Cox K, Friend K, Smith S, Buchheim R, et al. Familial 22q11.2 duplication: a three-generation family with a 3-Mb duplication and a familial 1.5-Mb duplication. Clin Genet. 2008;73:160–164. doi: 10.1111/j.1399-0004.2007.00938.x. [DOI] [PubMed] [Google Scholar]
- Zackai EH, Emanuel BS. Site-specific reciprocal translocation, t(11;22) (q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am J Med Genet. 1980;7:507–521. doi: 10.1002/ajmg.1320070412. [DOI] [PubMed] [Google Scholar]