Abstract
Branchio-oculo-facial syndrome (BOFS) is a rare disorder characterized by maldevelopment of the first and second branchial arches, skin defects, facial dysmorphism, auricular, ophthalmological and oral abnormalities. A high clinical variability has been reported. Recently, mutations in TFAP2A were found to underlie this condition. A small duplication on 5q13 was detected in 2 family members with mild BOFS features. Molecular cytogenetic delineation of the duplication demonstrated that only 7 genes are affected: LOC100289045, RGNEF, UTP15, ANKRA2, FUNDC2P1, BTF3 and FOXD1. The latter is expressed in the developing branchial arches and involved in cranio-facial development. Zebrafish embryos with combined inhibition of the expression of foxd1l and tfap2a show optic axis defects. We identified a novel locus associated with a mild BOFS-like phenotype. The functional in vivo experiments suggest an interaction between FOXD1 and TFAP2A.
Key Words: Branchial defects, Branchio-oculo-facial syndrome, FOXD1, TFAP2A, Zebrafish
Introduction
Branchio-oculo-facial syndrome (BOFS) (OMIM 113620) is characterized by abnormalities of the structures derived from the first and second branchial arches. The patients have facial dysmorphism: broad or bifurcated nasal tip, non-midline cleft lip, cleft palate, facial nerve palsy and ear abnormalities. Typical features are the branchial defects: sinuses or aplastic skin in the cervical area, supra-auricular sinuses and hemangiomatous lesions. Less frequently, individuals with BOFS can have genitourinary or limb abnormalities and dermoid cysts. Mental development is usually normal [Raveh et al., 2000; Milunsky et al., 2008]. Clinical variability has been reported between the different patients and within the affected families. The syndrome is inherited in an autosomal dominant manner and mutations in TFAP2A (activating enhancer-binding protein 2 alpha) underlie this condition [Milunsky et al., 2008]. TFAP2A is a transcription factor, located on chromosome 6p24. It is expressed during embryonic development in the branchial arches, the neural crest and the presumptive epidermis [Luo et al., 2003]. Homozygous knock-out mice die prenatally with severe developmental defects such as cranial neural tube and body wall closure defects, craniofacial and limb anomalies [Schorle et al., 1996; Zhang et al., 1996]. Chimeric null mice have an abnormal development of the maxillary and mandibular prominences. They display midline clefts, hypoplastic jaw and variable eye, limb and skin anomalies. The phenotype of these chimeric animals overlaps with the human BOFS [Nottoli et al., 1998].
Following gene identification, mutation screening confirmed the presence of alterations, mutations and deletions in all typical BOFS patients. It is, however, not known if TFAP2A is mutated in the atypical BOFS patients [Stoetzel et al., 2009].
We report the clinical and molecular data from the analysis of a family with mild BOFS features and identify a 1.1-Mb duplication on chromosome 5q13. The locus contains 7 genes. Based on the expression profile and zebrafish morpholino knockdown experiments, we hy- pothesize that the duplication of FOXD1 causes this OFS-like phenotype.
Subjects and Methods
Ethics Approval
The study was approved by the ‘Commissie Medische Ethiek’, Faculty of Medicine, Catholic University of Leuven.
Clinical Description
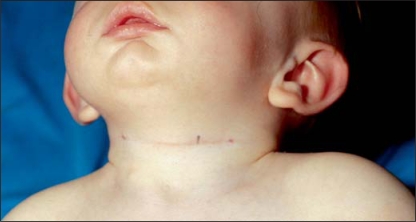
The proband is the first and only child of nonconsanguineous parents, born at term with a birth weight of 2440 g (p3) and a length of 49 cm (p50). After birth the boy had feeding difficulties and general hypotonia. Branchial cysts were present bilaterally (fig. 1). At the age of 9 months, he was seen at the genetic clinic and his weight was 6230 g (<p3), his length 68.5 cm (p9) and his head circumference 43.5 cm (p3). He had flat occiput, retrognatia, everted lower lip, broad nasal bridge and fifth finger clinodactyly. He had a pyloric stenosis treated by pylorotomy. His psychomotor development was mildly delayed. The mother presented with dysmorphic facial features: upslanting palpebral fissures, retrognatia, everted lower lip and white forelock. She functions at a borderline intelligence level.
Fig. 1.
Branchial anomalies in the affected child.
Patient Analysis
The patients were examined by experienced clinical geneticists and informed consent was provided.
Sequencing of TFAP2A
DNA was extracted following standard procedure. TFAP2A exons were amplified by PCR with primers extracted from a publicly available database: http://elxr.swmed.edu (online suppl. table 1, www.karger.com/doi/10.1159/000327707). For exon 4, the specific PCR product was further purified using the Qiagen gel extraction kit following the protocol supplied by the producer (Qiagen, Venlo, the Netherlands). The amplicons were subjected to Sanger sequencing with the BigDye terminator system. The resulting labeled fragments were separated and detected on an ABI 3100 sequencer.
Array CGH
Array CGH was performed on a DNA sample from the affected child using a Syndrome Plus oligonucleotide array (Oxford Gene Technology, Oxford, UK). This array contains 105,000 targets covering more than 200 syndrome-related regions and genes related with autism, heart, eye anomalies with at least 3-kb density and backbone targets spaced each 40 kb across the genome. Five hundred nanogram of patient and reference DNA (from a healthy male individual) were differentially labeled with 3-μl Cy5-dCTP and Cy3-dCTP (GE Healthcare, Chalfont St. Giles, United Kingdom) and 5-μl dNTP mix in 50-μl reaction using the Bioprime labeling kit (Invitrogen, Carlsbad, Calif., USA). After labeling, the samples were combined, precipitated and resuspended in 75-μl TE buffer (Promega, Leiden, the Netherlands). Further probe preparation, hybridization and washing were performed as recommended by the manufacturer.
Data were analyzed and visualized using Cytosure software (Oxford Gene Technology, Oxford, UK).
Confirmation of the array result and segregation analysis was done by quantitative PCR (qPCR) with 2 primer pairs located within the duplicated region and control primers in the p53 gene (online suppl. table 1). Samples from the patient, the mother and 3 control individuals were examined. The results were analyzed with the ΔΔCt method [Menten et al., 2006].
Zebrafish Experiments
A BLAT search with the human FOXD1 protein sequence in the UCSC genome browser identified the closest zebrafish ortholog. foxd1l showed the highest homology and was located in a syntenic genomic region.
Two BAC clones containing zebrafish foxd1l (CH73–374J10, CH1073–450F9) and a fosmid encompassing the human FOXD1 gene (WI22726L3) were obtained from the BACPAC CHORI resources (http://bacpac.chori.org/). Cultures were prepared according to the instructions of BACPAC CHORI. DNA was isolated by midi prep (Qiagen, Venlo, the Netherlands). Working solutions of 50–100–300 ng/μl have been prepared.
Knockdown of the tfap2a gene was done by splice block morpholino (Gene Tools, www.gene-tools.com) using the same sequence as described previously [O'Brien et al., 2004].
We designed a morpholino targeting the translation start site of foxd1l. The sequences of the morpholinos are shown in online supplementary table 1. The morpholinos were ordered from Gene Tools (Philomath, Oreg., USA) and reconstituted in distilled water. Working solutions were prepared with 1–2.5–5 ng/nl and 0.4–0.8–1.6 ng/nl concentrations for tfap2a and foxd1l morpholinos, respectively. A combined solution from both morpholinos was prepared with 2.5 ng/nl and 0.4 ng/nl for tfap2a and anti-foxd1l, respectively. All solutions for injections contained rhodamine as a tracer for injection.
Zebrafish adults were kept in standard conditions from the university aquatic facility. Embryos were raised in dishes containing no more than 30 embryos at 28.5°C and staged according to Kimmel et al. [1995].
Injections were done in over 70 embryos with 1 nl BAC, fosmid or morpholino solutions in the yolk immediately below the blastomeres at 1–4 cell stage. All experiments were repeated at least once and yielded consistent results. The embryos were screened at 4 h post fertilization and only embryos retaining the fluorescent dye and uniform for the injected amount were kept (over 50 embryos). Injections were done with tfap2a and foxd1l morpholinos separately and with a solution containing both tfap2a and foxd1l morpholinos.
The expression of the BAC and fosmid larvae was controlled using RT-qPCR (real-time quantitative PCR). For this purpose, RNA was extracted with TRIzol® (Invitrogen, Carlsbad, Calif., USA) from injected larvae and noninjected controls at 3 days post fertilization (dpf). After DNAseI (Roche Applied Science, Vilvoorde, Belgium) treatment, cDNA was synthesized using Superscript III (Invitrogen, Carlsbad, Calif., USA) with a mixture of random and poly-A primers, following the procedure described by the producer. RT-qPCR was done with expression primers within FOXD1, foxd1l and a control gene named gapdh. The primer sequences are shown in online supplementary table 1. The results were analyzed with the ΔΔCt method [Vandesompele et al., 2002].
Results
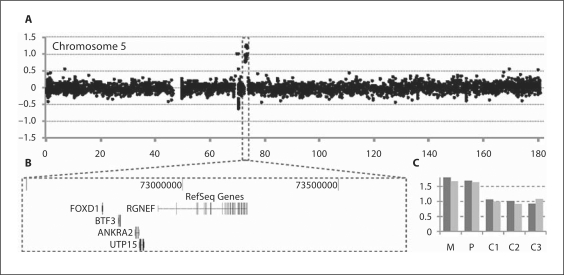
In a mother and child with BOFS-like phenotype, sequencing of the TFAP2A gene did not reveal any mutation. Genome-wide analysis for submicroscopic chromosomal deletions and duplications in the affected child by array CGH showed that the oligomeres located within a 1.1-Mb region on chromosome 5q13 had an increased log2 ratio compatible with duplication (chr5: 72556070–73668680 bp) (fig. 2A and B). qPCR confirmed the presence of the duplication in the child and showed that the affected mother was also a carrier (fig. 2C). Since the region was neither reported as copy variable in 4,000 normal individuals in a local database nor in the Database of Genomic Variants (http://projects.tcag.ca/variation/, version from August 2009), we consider this submicroscopic duplication as a candidate locus for the phenotype in mother and child.
Fig. 2.
Submicroscopic duplication detected by array CGH. A The partial array CGH result for chromosome 5 is displayed. The black dots represent the individual targets from the array, ranged according to their genomic position on the X axis and their log2 ratio on the Y axis. The duplicated probes are highlighted by a box. B A screenshot from Ensembl genome browser demonstrates the duplicated region. C qPCR confirmation of the array results with 2 primer pairs. Relative copy numbers as estimated by the ΔΔCt method are displayed for the patient (P), the mother (M) and 3 normal control individuals (C1, C2 and C3). Values around 1 and 1.5 correspond to a normal copy number and a heterozygous duplication, respectively.
The duplicated region encompasses 7 annotated genes: LOC100289045, RGNEF, UTP15, ANKRA2, FUNDC2P, BTF3 and FOXD1. The expression profiles collected in UniGene (http://www.ncbi.nlm.nih.gov/UniGene/), ZFIN (http://zfin.org/) and the Mouse Genome Database (http://www.informatics.jax.org/) showed that 6 genes have aubiquitous expression, whereas FOXD1 has a specific expressionin the brain, ear, eye, skin and branchial arches during the embryonic development. Hence, duplication of FOXD1 is a likely explanation for the observed phenotype.
To further investigate this hypothesis, we performed functional studies in zebrafish. Since the duplication results in an extra copy of FOXD1, the observed phenotype is likely due to overexpression of this gene during human development. To mimic this overexpression in zebrafish, 3 different vectors, 2 BACs, CH73–374J10, CH1073–450F9 containing the zebrafish foxd1l and a fosmid with the human FOXD1 gene, were injected in the zebrafish egg. Each vector was injected at 50–100–300 ng/μl. The injected embryos were staged every 12 h for the first 2 days and further on every 24 h until 5 dpf. The morphological changes were recorded at every stage. RT-qPCR on cDNA synthesized with RNA extracted at 3 dpf from the zebrafish injected with the fosmid containing the human FOXD1 gene and from non-injected controls was done. Primers specific to the human gene and control zebrafish gene were used. The experiment showed expression of the human gene. The phenotypes between the different injected embryos were highly variable making it impossible to draw conclusions from the effect of the overexpression. Other groups using the same technique have shown that the number of fish retaining the exogenous DNA is 1–5% to maximum 25% with a variable level of expression between the different lines [Nüsslein-Volhard, 2002]. Most likely, somatic mosaicism as a consequence of unequal distribution of the clones was occurring in the injected embryos which can explain the highly variable phenotypes.
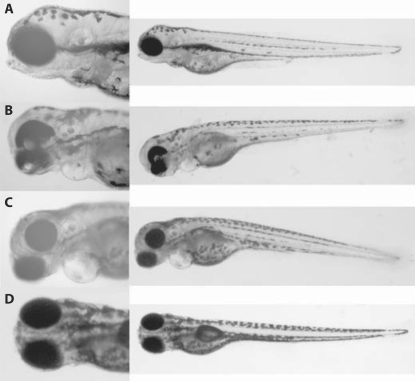
To investigate the potential role of foxd1l further, its expression was knocked down by injection of 0.4–0.8–1.6 ng/nl solution of anti-foxd1l morpholino. Even at the highest doses, the embryos developed normally and no specific morphological changes were observed. The branchial arches appeared to develop normally (fig. 3D). As a control, a tfap2a morpholino was injected at 1–2.5–5 ng/nl. In all embryos, severe branchial arch malformations were observed, which were dose dependent. This phenotype was previously described [O'Brien et al., 2004] (fig. 3C).
Fig. 3.
Phenotype of the zebrafish embryos. A Wild-type embryo. B Embryos injected with anti-foxd1l and anti-tfap2a morpholinos. Note the lack of pigmentation of the ventral part of the eye and ventral displacement of the eye globe axis. C Embryos injected with anti-tfap2a only. The branchial arches are severely affected, as described previously [O'Brien et al., 2004; Bassett et al., 2010]. D Embryos injected with anti-foxd1l only. The embryos appear to have normal morphology.
To determine whether TFAP2A and FOXD1 might interact or act in a common pathway, anti-foxd1l and tfap2a morpholinos were injected together in order to simultaneously block the translation of both mRNAs. We used a combination of 2.5 ng/nl and 0.4 ng/nl tfap2a and anti-foxd1l, respectively. All embryos injected with the combination of both morpholinos lacked pigmentation of the ventral part of the eye and showed displacement of the lens more ventrally as compared to the wild-type embryos. This ocular phenotype was not present in the embryos injected with the anti-tfap2a or anti-foxd1l only (fig. 3B).
Discussion
In a family with BOFS-like features, an additional copy of a 1.1-Mb region on chromosome 5q13 was found segregating with the phenotype in both affected individuals. Similarly to the features described in BOFS, the affected child presented with branchial cysts at the cervical area and his mother had a white forelock. However, they did not show other features characteristic of BOFS, such as cleft, bifid nose, nasolacrimal duct obstruction, eye abnormalities and skin defects. In contrast to patients with BOFS, both members of the affected family had delayed mental development (table 1). The duplication was not detected as a benign polymorphism in over 4,000 individuals from a local database and it is not reported in the Database of Genomic Variants. Patients with large, microscopically visible 5q13 duplications have been described. Two presented with preauricular pits and tags [Rojas-Martinez et al., 1990; Breslau-Siderius et al., 1993]. Other patients with a duplication of the same region did not show similar characteristics [Zou et al., 2010]. This suggests variable expressivity and penetrance of these features. The overlap of the clinical phenotype between the patients presented here and the families described in the literature as well as the absence of benign variants in this region strongly suggest that this duplication is causal for the BOFS-like phenotype.
Table 1.
Comparison between the BOFS patients with detected TFAP2A mutation from Milunsky et al. [2008] and the present patients
| Clinical features | Patients with BOFS [Milunsky et al, 2008] | Present patients |
|---|---|---|
| Genetic defect | mut or del TFAP2A | del5ql3 |
| Family history | 2/6 | + |
| Intellectual disability | 0/6 | + |
| Branchial anomalies | 6/6 | + |
| Ocular anomalies | 5/6 | − |
| Facial anomalies | ||
| Auricular malformation | 6/6 | − |
| Characteristic face | 5/6 | − |
| Chin dimple or cleft | 3/6 | _ |
| Cleft lip and palate | 4/6 | _ |
| Facial nerve palsy | 1/6 | _ |
| Preauricular pits | 3/6 | − |
| Prematurely gray hair | 2/6 | + |
| Hearing loss | 5/6 | − |
The duplicated region encompasses 7 annotated genes: LOC100289045, RGNEF, UTP15, ANKRA2, FUNDC2P, BTF3 and FOXD1. RGNEF (rho guanine nucleotide exchange factor) was found to promote cytoskeleton contraction and to inhibit neurite outgrowth in a mouse neuroblastoma cell line [Gebbink et al., 1997]. ANKRA2 (ankyrin repeat containing protein, family A, member 2) encodes a protein interacting with the cytoplasmic domain of megalin and might facilitate the endocytosis in the proximal tubules of the kidney [Rader et al., 2000]. FUNDC2P is a pseudogene. BTF3 (basic transcription factor 3) has been shown to be important for the processes of transcription and apoptosis [Wiedmann et al., 1994]. These genes haveubiquitous expression. FOXD1 (OMIM 601091), on the contrary, has specific expressionin the brain, ear, eye, skin and branchial arches during the embryonic development. Therefore, we considered FOXD1 as the most likely candidate gene to explain the phenotype. FOXD1 is a member of the forkhead transcription factor family. Members of this family are known to underlie disorders with craniofacial and eye involvement through dosage sensitive mechanisms. FOXC1 is important for the formation of the structures of the ocular anterior chamber and heterozygous mutations in this gene cause Axenfeld-Rieger or other eye anomalies [Nishimura et al., 1998]. Another member of this gene family, FOXL2, leads to Blepharophimosis, Ptosis, Epicantus inversus syndrome upon mutation or deletion [Crisponi et al., 2001]. In the developing zebrafish, foxd1l is expressed in the diencephalon, neural crest, branchial arches and eye [Thisse, 2004]. Its role for the specification of the naso-temporal axis of the retina and the formation of the optic chiasma has been reported [Takahashi et al., 2003; Herrera et al., 2004]. The increased dosage of FOXD1 possibly leads to the developmental abnormalities observed in our patients. The expression of the gene in the branchial arches can explain the cervical cysts in the child, while the white forelock could be due to abnormal neural crest cell migration. Both patients have delayed mental development and FOXD1 is expressed in the central neural system during the embryonic life.
Classical BOFS patients have alterations in the TFAP2A gene. However, there is evidence for clinical heterogeneity [Stoetzel et al., 2009; Tekin et al., 2009]. The phenotype varies from severe to very mild between family members carrying the same genetic defect [Milunsky et al., 2008; Stoetzel et al., 2009]. The clinical variability suggests that probably additional factors modify the effect of TFAP2A alterations. As a transcription activator, TFAP2A interacts with other developmental genes. This has been demonstrated by the binding of TFAP2A with the IRF6 gene enhancer, where disruption of the TFAP2A binding site leads to nonsyndromic cleft lip [Rahimov et al., 2008]. Reduced tfap2a function sensitizes the developing eye for the effect of mutations in bmp4 and otx2 in zebrafish, leading to a more pronounced phenotype [Gestri et al., 2009].
We hypothesized an interaction between FOXD1 and TFAP2A, based on the phenotypic overlap of the observed family with BOFS. To investigate this, we performed experiments in zebrafish demonstrating the occurrence of a novel phenotype when both tfap2a and foxd1l translation is blocked. The embryos injected with a combination of both morpholinos showed lack of pigmentation of the ventral part of the eye and ventral displacement of the optic globe axis. Most likely, an abnormal retinal patterning was occurring. When injected separately, the anti-foxd1l did not cause any ocular phenotype even at higher concentrations. Anti-tfap2a morpholino leads to ocular abnormalities, microphthalmia and coloboma, at higher doses. However, these phenotypes differ significantly from the ones due to the combined inhibition of both genes. As shown by others, FOXD1 is important for the formation of the optic axis [Takahashi et al., 2009] and possibly the additional inhibition of tfap2a leads to a more severe perturbation of this process. Moreover, Tfap2a knockout mice were recently shown to exhibit optic cup patterning defects.
Establishing genetic interactions contributes to our understanding of the different developmental pathways and the pathogenesis of genetic disorders. Our data provides evidence for an interaction between TFAP2A and FOXD1, however, the exact mechanisms underlying the observed phenotypes remain unknown and require further studies.
Supplementary Material
Acknowledgements
We would like to thank the family with BOFS-like phenotype, the staff of the Zebrafish Facility of the K.U. Leuven for their collaboration and competent advices and Bernard Thienpont for the fruitful discussions.
I.B. is supported by grant OE/08/24 of the K.U. Leuven. This work was made possible in part by grants from the IWT (SBO-60848) and GOA/2006/12, and the SymBioSys Center of Excellence (Research Council, K.U. Leuven, EF/05/007) to J.R.V. and K.D.
References
- Bassett EA, Williams T, Zacharias AL, Gage PJ, Fuhrmann S, West-Mays JA. AP-2alpha knockout mice exhibit optic cup patterning defects and failure of optic stalk morphogenesis. Hum Mol Genet. 2010;19:1791–1804. doi: 10.1093/hmg/ddq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslau-Siderius EJ, Wijnen JT, Dauwerse JG, de Pater JM, Beemer FA, Khan PM. Paternal duplication of chromosome 5q11.2–5q14 in a male born with craniostenosis, ear tags, kidney dysplasia and several other anomalies. Hum Genet. 1993;92:481–485. doi: 10.1007/BF00216455. [DOI] [PubMed] [Google Scholar]
- Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet. 2001;27:159–166. doi: 10.1038/84781. [DOI] [PubMed] [Google Scholar]
- Gebbink MF, Kranenburg O, Poland M, van Horck FP, Houssa B, Moolenaar WH. Identification of a novel, putative Rho-specific GDP/GTP exchange factor and a RhoA-binding protein: control of neuronal morphology. J Cell Biol. 1997;137:1603–1613. doi: 10.1083/jcb.137.7.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestri G, Osborne RJ, Wyatt AW, Gerrelli D, Gribble S, et al. Reduced TFAP2A function causes variable optic fissure closure and retinal defects and sensitizes eye development to mutations in other morphogenetic regulators. Hum Genet. 2009;126:791–803. doi: 10.1007/s00439-009-0730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E, Marcus R, Li S, Williams SE, Erskine L, et al. Foxd1 is required for proper formation of the optic chiasm. Development. 2004;131:5727–5739. doi: 10.1242/dev.01431. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Luo T, Lee YH, Saint-Jeannet JP, Sargent TD. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proc Natl Acad Sci USA. 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menten B, Maas N, Thienpont B, Buysse K, Vandesompele J, et al. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J Med Genet. 2006;43:625–633. doi: 10.1136/jmg.2005.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milunsky JM, Maher TA, Zhao G, Roberts AE, Stalker HJ, et al. TFAP2A mutations result in branchio-oculo-facial syndrome. Am J Hum Genet. 2008;82:1171–1177. doi: 10.1016/j.ajhg.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, et al. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19:140–147. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- Nottoli T, Hagopian-Donaldson S, Zhang J, Perkins A, Williams T. AP-2-null cells disrupt morphogenesis of the eye, face, and limbs in chimeric mice. Proc Natl Acad Sci USA. 1998;95:13714–13719. doi: 10.1073/pnas.95.23.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard CD. A practical approach. Oxford: Oxford University Press; 2002. Zebrafish. [Google Scholar]
- O'Brien EK, d'Alencon C, Bonde G, Li W, Schoenebeck J, et al. Transcription factor Ap-2alpha is necessary for development of embryonic melanophores, autonomic neurons and pharyngeal skeleton in zebrafish. Dev Biol. 2004;265:246–261. doi: 10.1016/j.ydbio.2003.09.029. [DOI] [PubMed] [Google Scholar]
- Rader K, Orlando RA, Lou X, Farquhar MG. Characterization of, ANKRA, a novel ankyrin repeat protein that interacts with the cytoplasmic domain of megalin. J Am Soc Nephrol. 2000;11:2167–2178. doi: 10.1681/ASN.V11122167. [DOI] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh E, Papsin BC, Forte V. Branchio-oculo-facial syndrome. Int J Pediatr Otorhinolaryngol. 2000;53:149–156. doi: 10.1016/s0165-5876(00)00310-4. [DOI] [PubMed] [Google Scholar]
- Rojas-Martinez A, Garcia-Cruz D, Medina C, Moller M, Restrepo CM, Rivera H. Tandem duplication of proximal 5q. Ann Genet. 1990;33:228–230. [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Riehm S, Bennouna Greene V, Pelletier V, Vigneron J, et al. Confirmation of TFAP2A gene involvement in branchio-oculo-facial syndrome (BOFS) and report of temporal bone anomalies. Am J Med Genet A. 2009;149A:2141–2146. doi: 10.1002/ajmg.a.33015. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Shintani T, Sakuta H, Noda M. CBF1 controls the retinotectal topographical map along the anteroposterior axis through multiple mechanisms. Development. 2003;130:5203–5215. doi: 10.1242/dev.00724. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Sakuta H, Shintani T, Noda M. Functional mode of FoxD1/CBF2 for the establishment of temporal retinal specificity in the developing chick retina. Dev Biol. 2009;331:300–310. doi: 10.1016/j.ydbio.2009.05.549. [DOI] [PubMed] [Google Scholar]
- Tekin M, Sirmaci A, Yuksel-Konuk B, Fitoz S, Sennaroglu L. A complex TFAP2A allele is associated with branchio-oculo-facial syndrome and inner ear malformation in a deaf child. Am J Med Genet A. 2009;149A:427–430. doi: 10.1002/ajmg.a.32619. [DOI] [PubMed] [Google Scholar]
- Thisse BT: Fast release clones: A high throughput expression analysis. ZFIN Direct Data Submission (http://zfin org) (2004).
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al: Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034 http://genomebiology.com/2002/3/7/research/0034 (2002). [DOI] [PMC free article] [PubMed]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, et al. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- Zou YS, Newton S, Milunsky JM. A complex maternal rearrangement results in a pure 10.8 Mb duplication of the 5q13.1–q14.1 region in an affected son. Am J Med Genet A. 2010;152A:498–503. doi: 10.1002/ajmg.a.33266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.