Abstract
Rationale: Basophils contribute to anaphylaxis and allergies. We examined the utility of assessing basophil-associated surface antigens (CD11b/CD63/CD123/CD203c/CD294) in characterizing and monitoring subjects with nut allergy. Methods: We used flow cytometry to analyze basophils at baseline (without any activation) and after ex vivo stimulation of whole blood by addition of nut or other allergens for 2, 10, and 30 min. We also evaluated whether basophil expression of CD11b/CD63/CD123/CD203c/CD294 was altered in subjects treated with anti-IgE monoclonal antibody (omalizumab) to reduce plasma levels of IgE. Results: We demonstrate that basophil CD203c levels are increased at baseline in subjects with nut allergy compared to healthy controls (13 subjects in each group, p < 0.0001). Furthermore, we confirm that significantly increased expression of CD203c occurs on subject basophils when stimulated with the allergen to which the subject is sensitive and can be detected rapidly (10 min of stimulation, n = 11, p < 0.0008). In 5 subjects with severe peanut allergy, basophil CD203c expression following stimulation with peanut allergen was significantly decreased (p < 0.05) after 4 and 8 weeks of omalizumab treatment but returned toward pretreatment levels after treatment cessation. Conclusions: Subjects with nut allergy show an increase of basophil CD203c levels at baseline and following rapid ex vivo stimulation with nut allergen. Both can be reduced by omalizumab therapy. These results highlight the potential of using basophil CD203c levels for baseline diagnosis and therapeutic monitoring in subjects with nut allergy.
Key Words: Anaphylaxis, Atopic, CD203c, Cockroach allergen, Flow cytometry, Human basophils, Omalizumab, Peanut allergy, Tree nut allergy
Introduction
Nut allergy is one of the most severe clinical presentations of food allergy [1]. Although not every subject with peanut or other food allergies develops near-fatal symptoms when exposed to the offending allergen, most of them, particularly young children, experience restrictions in their quality of life and nutrition [2]. For this reason, objective endpoints reflecting allergic predisposition towards a given allergen are important for the proper identification of at-risk subjects. Currently, such endpoints are limited to skin tests and double-blind placebo-controlled food challenges. Both are cumbersome in vivo procedures with significant associated risks, including anaphylaxis [3]. Hence, reliable ex vivo outcome measures are needed for the diagnosis of allergic subjects.
Furthermore, although some allergy reduction therapies show promise [4,5], definitive demonstration of their efficacy has yet to be accomplished. Omalizumab, a humanized monoclonal antibody against IgE, has been used to treat subjects with peanut allergy in a phase II trial [6], and there are currently several clinical trials in progress testing omalizumab for treatment of other food allergies. A combination of efficacy endpoints [7] and reliable ex vivo outcome measures would be beneficial for evaluating treatment responses in these trials [8]. However, no ex vivo outcome measures have as yet proven reliable to track the effectiveness of omalizumab therapy.
Basophils are involved in mediating anaphylaxis and several studies have presented evidence that basophils can be useful in diagnosing allergies [9,10,11]. Basophil activation, in addition to activation of mast cells and other immune cells, and subsequent production of chemical mediators (such as histamine) can contribute to the initiation of anaphylaxis [12]. Basophils express the surface activation markers CD11b, CD63, CD123, CD203c and CD294, some of which previously have been shown to reflect allergen-dependent activation of basophils ex vivo [9,13,14,15,16,17,18,19].
Our studies presented here demonstrate that CD203c expression on blood basophils is a useful ex vivo outcome measure for subjects with nut allergy. First, we show that basophil CD203c levels at baseline (without any ex vivo stimulation) were significantly higher in subjects with nut allergy than in healthy controls. Second, ex vivo allergen stimulation induced an increase in basophil CD203c levels that appears to be specific to the offending allergen for a given donor. Third, we show that basophil CD203c levels at baseline and upon ex vivo allergen stimulation are reduced by omalizumab therapy in subjects with nut allergy and therefore track treatment responsesin vivo.
Methods
Human Subjects
The study was approved by the Stanford Administrative Panel of Human Subjects in Medical Research. All 16 food-allergic subjects (or parents, for minors) and all healthy controls (n = 13) signed informed consent forms before the subjects underwent study procedures. Details regarding the inclusion of subjects in the various parts of the study are available in the online supplementary material (www.karger.com/doi/10.1159/000321824). Clinical nut or apple allergy (specific to peanut: 12; to cashew: 2; to walnut: 1; to apple: 1; tables 1, 2) was diagnosed by clinical history of food allergy reaction, nut-specific IgE ≥15 kUA/l (ImmunoCAP; Phadia, Uppsala, Sweden) and/or positive skin prick test to nut allergen (tables 1, 2), and severity was graded based on published scores of anaphylaxis symptoms, as noted by Nowak- Wegrzyn et al. [20]. Five of the subjects with a history of anaphylactic reaction to peanuts were enrolled in a phase I open-label study of omalizumab (investigator-initiated study, Stanford IND 103, 711). Omalizumab (Genentech/Novartis, South San Francisco, Calif., USA) was dosed as per product insert guidelines based on total IgE levels and the subject's body weight (tables 1, 2).
Table 1.
Characteristics of the subjects
| Patients | Age years | Gender | Duration years | Peanut allergy symptoms | Severity |
|---|---|---|---|---|---|
| 1 | 8 | female | 7 | wheezing, throat swelling | severe |
| 2 | 3 | male | 2 | asthma, throat swelling, loss of voice | severe |
| 3 | 5 | female | 4 | abdominal pain, systemic urticaria, throat itching | moderate |
| 4 | 9 | female | 8 | urticaria, angioedema, wheezing | severe |
| 5 | 4 | female | 5 | wheezing | mild |
| 6 | 7 | female | 7 | angioedema | severe |
| 7 | 9 | male | 9 | wheezing, angioedema | severe |
| 8 | 18 | female | 19 | wheezing, angioedema | severe |
| 9 | 6 | male | 5 | wheezing, angioedema | severe |
| 10 | 21 | male | 20 | wheezing, angioedema | severe |
| 11* | 5 | female | 4 | urticaria, angioedema, wheezing | severe |
| 12* | 24 | male | 23 | urticaria, angioedema, abdominal pain, vomiting, diarrhea, difficulty wheezing | severe |
| 13* | 8 | male | 7 | wheezing, hypotension | severe |
| 14* | 7 | male | 6 | lip swelling, throat swelling | severe |
| 15* | 6 | male | 5 | throat swelling | severe |
| 16 | 28 | female | 27 | oral food challenge | moderate |
These subjects were part of a phase I open-label study of omalizumab.
Table 2.
Biological characteristics of the study subjects
| Subject | Total IgE kU/l | Cockroach-specific IgE, kU/l | Allergen-specific IgE, kU/l | Allergen-specific skin test | Omalizumab dose, mg/kg per IU/ml IgE |
|---|---|---|---|---|---|
| 1 | 591 | 2.98 | >100 (P) | 4+ (P) | |
| 2 | 257 | <0.35 | 43.6 (C) | 3+ (C) | |
| 3 | 1,480 | ND, SK- | >100 (P) | 4+ (P) | |
| 4 | 180 | ND, SK- | 12.4 (P) | 3+ (P) | |
| 5 | 126 | <0.35 | 0.35 (P) | 3+ (P) | |
| 6 | 3,206 | 3.81 | 83.5 (P) | 3+ (P) | |
| 7 | 1,975 | 5.07 | 67 (P) | 4+ (P) | |
| 8 | 3,050 | 0.85 | 13.5 (P) | 2+ (P) | |
| 9 | 1,752 | 0.32 | 79 (W) | 3+ (W) | |
| 10 | 434 | ND, SK- | 83 (C) | 4+ (C) | |
| 11* | 1,850 | 17.3 | 100 (P) | 4+ (P) | 0.008 |
| 12* | 284 | <0.35 | 15.1 (P) | 3+ (P) | 0.017 |
| 13* | 321 | 3.53 | 36.4 (P) | 3+ (P) | 0.02 |
| 14* | 784 | 2.24 | 69.7 (P) | 3+ (P) | 0.022 |
| 15* | 167 | 6 | 88.67 (P) | 4+ (P) | 0.06 |
| 16 | 432 | <0.35 | 41(A) | 2+ (A) |
Specific allergens are indicated between parentheses. Skin test results were obtained as per standard methods and were judged positive as compared to a histamine control. A = Apple; C = cashew; P = peanut; W = walnut; ND, SK- = not done because of negative skin test.
These subjects were part of a phase I open-label study of omalizumab.
Sample Collection and Processing
Blood was collected in ethylene diamine tetraacetic acid (EDTA) tubes by venipuncture, centrifuged (400 g, 10 min, 4°C), and the leukocyte pellet retained while the plasma was further centrifuged (3,000 g for 10 min, 4°C) to remove platelets. The blood sample was then reconstituted to its original volume by adding the platelet-free plasma to the pelleted leukocytes. We removed platelets from the plasma to avoid unwanted aggregation and clotting in the course of our 30-min assay at 37°C (see below).
Basophil Stimulation Assay
Three microliters of phosphate-buffered saline (PBS) or of an allergen extract (peanut, cockroach, cashew, walnut, apple) used clinically for skin testing (Greer, Lenoir, N.C., USA) were added to 200 μl of blood (processed as above) and the mixture was incubated for varying times (2, 10 or 30 min) at 37°C. The peanut extract contained shelled, uncooked/unsalted Virginia peanuts, which were ground and then treated with acetone to remove fat. The defatted ground peanuts were dried to remove the acetone and sent to the extract laboratory, where they were extracted in Coca's solution at 1:10 w/v, pre-filtered. The incubation was stopped by adding ice-cold PBS-EDTA (EDTA is a calcium chelator that blocks cell activation) [21], and the cells were pelleted by centrifugation (490 g, 5 min, 4°C). The supernatant was removed and stored at −80°C.
Immunoglobulins
Total blood IgE levels were measured in all subjects by the Clinical Laboratories at Stanford Hospital and Clinics using a standard Immunocap (Phadia) assay [22]. Free IgE (non-omalizumab bound) to peanut was measured in the subset of 5 subjects with peanut allergy under ongoing omalizumab therapy, according to the method described by Hamilton et al. [23].
Surface Marker Profiling
For surface staining, 50 to 200 μl of blood were stained with the Live/Dead near-infrared viability probe (Invitrogen, Carlsbad, Calif., USA) and several antibodies against surface determinants for 20 min on ice, in the dark. These antibodies (clone numbers indicated in parentheses) included CD3 (UCHT1), CD11b (VIM12) and CD16 (3G8) from Invitrogen; CD20 (2H7), CD41a (96.2C1), CD56 (B157), CD63 (H5C6), CD66b (G10F5), CD123 (7G3), CD294 (BM16) and HLA-DR (L243) from BD Biosciences (San Jose, Calif., USA), and CD203c (NP4D6) from Biolegend (San Diego, Calif., USA). The gating strategy to identify basophils is illustrated in figure 1. After staining, cells were washed with excess PBS-EDTA, centrifuged (490 g, 5 min, 4°C) and the supernatant was removed. Upon resuspension in 100 μl of PBS-EDTA (2.5 mM final), cells were fixed with 2 ml of 1× Lyze/Fix PhosFlow (BD Biosciences) for 30 min on ice, in the dark. Then, the cells were washed once by centrifugation (490 g, 5 min, 4°C) before acquisition on the flow cytometer.
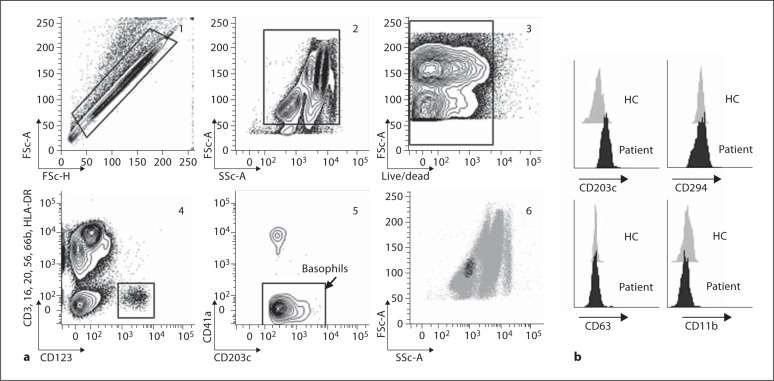
Fig. 1.
Flow cytometry enables basophil phenotyping from one drop of blood. a (1) Doublets were excluded based on forward scatter area versus height; (2) leukocytes were selected based on forward and side scatter; (3) dead cells were excluded using the viability marker Live/Dead Near InfraRed; (4) basophils were selected as the CD3–/CD16–/CD20–/CD56–/CD66b–/HLA-DR– and CD123+ population; (5) platelets were excluded using CD41a; (6) confirmation of gated basophil subset as intermediate for forward scatter and low for side scatter. b Basophil CD11b, CD63, CD203c and CD294 levels at baseline from 1 healthy control (HC) and 1 subject with peanut allergy.
Flow Cytometry Data Acquisition and Analysis
Data for 150,000–200,000 cells per sample were acquired on an LSRII digital flow cytometer equipped with 4 lasers (535, 488, 633, 405 nm), 2 light scatter detectors (yielding forward and side scatter data) and 18 fluorescent detectors (BD Biosciences). Acquisition was controlled using the DiVa software (BD Biosciences). Compensation was done using single-stained beads or cells during postacquisition analysis in the FlowJo software (Treestar, Ashland, Oreg., USA), as previously described [24,25].
Statistical Analysis
Between- and within-group comparisons used the nonparametric Wilcoxon rank sum and signed rank tests, respectively. Pairwise correlations used the Spearman test. Predictive values of tests (such as blood basophil CD203c) for subject identification (subjects with nut allergy vs. healthy controls) were calculated by means of nominal logistic regression, yielding p values for negative log-likelihood χ2 tests. Predictive abilities were expressed as the area under the receiver operating characteristics (ROC) curve [26], which plots the frequency of true positive (sensitivity) against the frequency of false positive (1 – specificity) results. Area values were considered excellent at more than 0.9 (1.0 is the maximal). Differences or correlations were considered significant at a p value of less than 0.05.
Results
Basophil CD203c Levels Are Increased at Baseline in Subjects with Nut Allergy
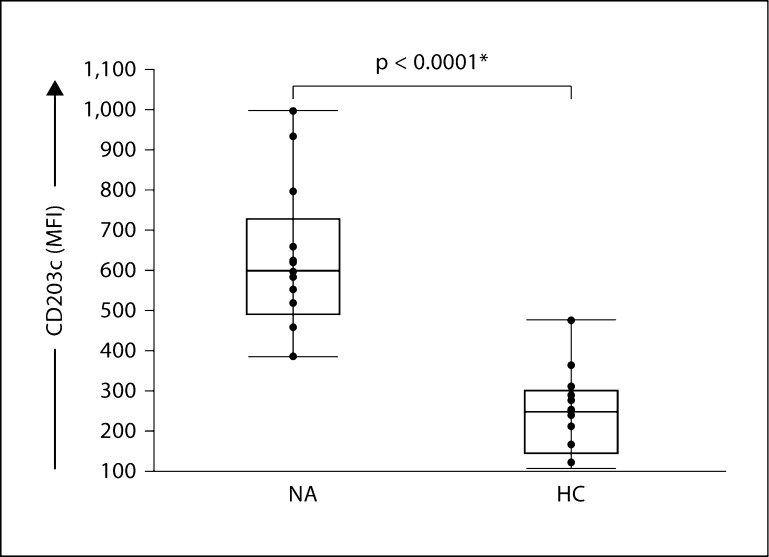
We used flow cytometry and sequential gating methods to discriminate live basophils from cell clumps (cells that are bound to each other and cannot be characterized individually) [27], dead cells, platelets and platelet-leukocyte aggregates and nonbasophil leukocytes in peripheral blood samples (fig. 1). Basophils represent approximately 0.5% of the leukocyte population in peripheral blood. We found that basophil CD203c levels at baseline (measured as median fluorescence intensity, MFI) were significantly increased in subjects with nut allergy compared to healthy controls (fig. 2; 13 in each group, p < 0.0001). Baseline levels for the other activation markers tested, that is, CD11b, CD63, CD123 and CD294, were not different between these two groups (online suppl. fig. 1). Consistent with the limited overlap between the relative distributions of values in subjects with nut allergy compared to healthy controls (fig. 2), basophil CD203c levels at baseline yielded a statistically predictive value between the two groups (area under the ROC curve = 0.96, 13 in each group, p < 10–4).
Fig. 2.
Predictive value of basophil CD203c levels at baseline for the clinical classification of subjects with nut allergy. Basophil CD203c levels at baseline were significantly increased in subjects with nut allergy (NA) compared to healthy controls (HC), n = 13 in each group, p < 10–4. Statistical significance was assessed by Wilcoxon rank sum nonparametric test. Each point represents a single sample.
Ex vivo Nut Allergen Stimulation Further Increases Basophil CD203c Levels in Subjects with Nut Allergy
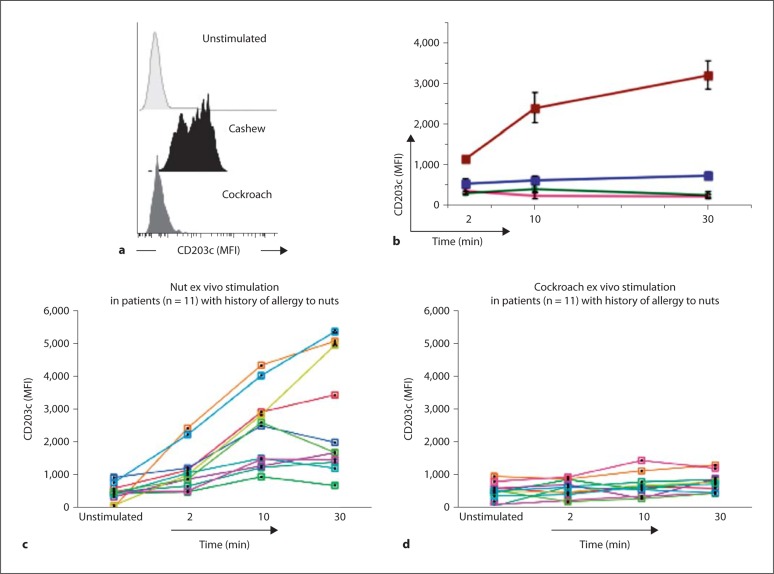
We confirmed the work of others in observing an upregulation of blood basophil CD203c levels after stimulation with the offending allergen (peanut, cashew or walnut) [9,13,14,15,16,17,18,19]. Basophils in an unstimulated blood sample were mostly CD203clo; by contrast, a sizeable fraction of basophils in ex vivo stimulated samples showed increased CD203c expression and became CD203chi (fig. 3a). Quantitatively, this shift was reflected by an increase in CD203c MFI (fig. 3) which was >8 fold higher in basophils from subjects with nut allergy than in basophils from healthy controls stimulated with the same nut allergen (fig. 3; p < 0.003). Basophils from subjects with nut allergy exhibited significantly higher responses to the specific offending nut allergen than to an aeroallergen (cockroach) to which they had no history of allergy (fig. 3; n = 11; nut vs. cockroach allergen stimulation: 2-, 4- and 5-fold differences, in CD203c MFI increases after 2-, 10-and 30-minute stimulations, respectively; p < 0.002, p < 0.0008 and p < 0.003). Additionally, we confirmed that there was no increase in basophil CD203c levels when the stimulation occurred with a nonoffending, nut allergen (for example, cashew for a subject with peanut allergy, or vice versa; online suppl. fig. 2). Altogether, our results establish this method as a rapid, specific, ex vivo assay for detecting allergic predisposition to specific nut allergens in human subjects.
Fig. 3.
Ex vivo stimulation with nut allergen further increases basophil CD203c levels in subjects with nut allergy. a Basophils in the sample from a single subject stimulated with the offending allergen (cashew) showed increased CD203c expression, some clustering in a CD203chi peak (bimodal distribution); b time course analysis (2-, 10- and 30-min stimulations) showing group means and standard errors for basophil CD203c levels (MFI) from healthy controls (n = 8) stimulated with peanut (pink line) or cockroach (green line) allergens, compared to subjects with nut allergy (n = 11) stimulated with nut (10 with peanut, 1 with cashew) (red line) or cockroach (blue line) allergens. In c and d, we represent basophil CD203c levels (MFI) as measured from the same individual subjects, each indicated by a line of a different color, upon stimulation with nut (c) or cockroach (d) allergens.
CD203c Provides a More Efficient and Reliable Index of Nut Allergy than Other Basophil Surface Markers
CD63 levels have been previously reported to be increased on basophils from allergic subjects in response to stimulation with offending allergens [13,19]. However, in our study, CD63 expression was not a reliable endpoint in that its levels increased on basophils in 11 of the 13 subjects tested, whereas increased levels of CD203c were detected in all 13. Moreover, in the 13 subjects tested, following ex vivo stimulation with nut allergen, the percentage of increase in MFI for CD63 was significantly lower than that for CD203 (online suppl. fig. 3b). Basophils from subjects with allergy to a specific nut allergen exhibited higher responses to this allergen than to an aeroallergen (cockroach) to which they had no relevant history of clinical allergy (online suppl. fig. 3a; n = 11; nut vs. cockroach allergen challenge: 2- and 3.5-fold increases, respectively, in CD63 expression after 10- or/and 30-min stimulation with each of the allergens; p < 0.028 or p < 0.032, respectively). However, no increase in basophil CD63 levels was detected in our assay at 2 min. Other basophil markers tested (CD11b, CD123, CD294) were not significantly increased by nut allergen exposure at any of the 3 time points analyzed (data not shown).
Blood Basophil CD203c Levels at Baseline and upon ex vivo Stimulation Are Largely Independent of Plasma IgE and Other Plasma Factors
Total IgE levels in plasma are a common indicator of allergic predisposition. Here, we found that total IgE levels in plasma from subjects with nut allergy at baseline did not correlate with basophil CD203c levels at baseline, nor did they predict CD203c levels upon 2 or 30 min of ex vivo stimulation with the offending nut allergen. However, a significant correlation existed between total IgE levels in plasma at baseline and basophil CD203c levels upon 10 min of ex vivo stimulation with the offending nut allergen (n = 15, Spearman rho = 0.554, p < 0.0396). It is known that IgE is strongly bound to basophil Fc∊RI and will remain bound even if plasma is removed from the cells and cells are subsequently placed in PBS. When we replaced subject plasma by PBS during the procedure, we found that the increase in basophil CD203c levels upon allergen stimulation still occurred (online suppl. fig. 4).
Basophil CD203c Expression at Baseline and following Stimulation with Nut Allergen Is Significantly Decreased during Omalizumab Treatment but Returns to Pretreatment Levels after Treatment Cessation
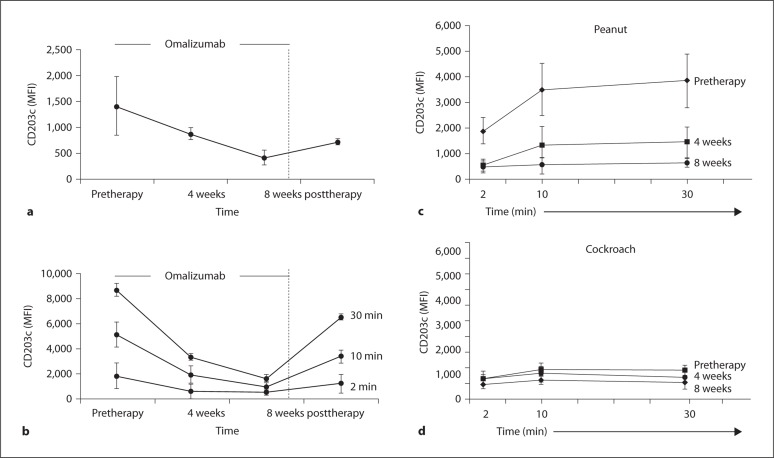
Longitudinal studies of basophil CD203c levels at baseline in our cohort of omalizumab-treated subjects with peanut allergy (tables 1, 2) demonstrated a significant decrease at 4 and 8 weeks of treatment (p < 0.05 for pretreatment vs. 4 weeks; for 4 weeks vs. 8 weeks and for pretreatment vs. 8 weeks after 30 min of ex vivo stimulation) (fig. 4a). Importantly, baseline basophil CD203c levels tended to increase 1 month after cessation of treatment. Furthermore, we show that the increase in basophil CD203c levels upon ex vivo stimulation with peanut allergen was significantly lowered during omalizumab treatment (fig. 4c; p < 0.033 at either 4 or 8 weeks of omalizumab treatment vs. pretreatment, n = 5). The abrogation of the allergen-induced increase in basophil CD203c levels seems to be temporary (p < 0.07 for comparison of pre-therapy vs. post-therapy levels of CD203c). One month after cessation of omalizumab treatment, we observed an increase in basophil CD203c levels upon ex vivo stimulation with peanut allergen after 2, 10 and 30 min ofex vivo stimulation (fig. 4b). Reduced levels of CD63 were observed at (1) the 10-min time points, between pretreatment and both 4 or 8 weeks of omalizumab treatment (online suppl. fig. 5b; p < 0.039 at both 4 and 8 weeks of omalizumab treatment vs. pre-treatment, n = 5) and (2) at the 30-min time points, between only pre-treatment and 8 weeks of omalizumab treatment (online suppl. fig. 5b; p < 0.041 at 8 weeks of omalizumab treatment vs. pre-treatment, n = 5). By one month after cessation of omalizumab treatment, the recovery of baseline basophil CD63 levels was marginal (online suppl. fig. 5a, b). We also measured the levels of free IgE (non-omalizumab bound) in the cohort of omalizumab-treated subjects (pretreatment vs. on treatment). The level of free IgE was decreased in 4 of the 5 subjects tested (online suppl. table 1).
Fig. 4.
Levels of basophil CD203c expression at baseline and following stimulation with peanut allergen are significantly decreased during omalizumab treatment but return to pretreatment levels after treatment cessation. a Basophil CD203c levels at baseline measured before omalizumab treatment, during treatment (4 and 8 weeks) and 4 weeks after treatment cessation. b Basophil CD203c levels following ex vivo stimulation (2-, 10- and 30-min stimulations) with peanut measured before omalizumab treatment, during treatment (4 and 8 weeks) and 4 weeks after treatment cessation. The figure represents the group means and standard errors for basophil CD203c levels (MFI) from peanut allergic subjects (n = 5) stimulated with peanut allergen. c, d Comparative data for basophil CD203c levels following ex vivo stimulation of subjects with peanut allergy with peanut allergen (c) or cockroach allergen (d) before omalizumab treatment and during treatment (4 and 8 weeks).
Discussion
Several groups have analyzed the expression of various basophil surface structures in an effort to develop new diagnostic and prognostic indicators of allergy [9,13,14,15,16,17,18,19]. Here, we report that basophil CD203c levels are increased at baseline in a cohort of subjects with nut allergy compared to healthy controls, suggesting that basophils may be in a ‘primed’ state in such allergic subjects. In addition to demonstrating this baseline difference, our studies confirmed previous reports demonstrating that basophil CD203c levels increase upon ex vivo stimulation with the allergen to which the subject is sensitive and that this response can be detected rapidly [9,17]. Allergen-induced increases in basophil CD203c levels proceeded equally well when the basophils were tested either in absence or presence of the subject's plasma. Finally, in 5 subjects with peanut allergy, basophil CD203c levels at baseline were decreased and their increase upon ex vivo stimulation with peanut allergen was reduced after 4 and 8 weeks of omalizumab treatment but returned toward pretreatment levels after cessation of treatment.
Our study shows that measuring baseline levels of basophil CD203c directly (that is, without any cell separation or activation) in whole blood can effectively discriminate between subjects with nut allergy and healthy controls. CD203c is a transmembrane molecule that belongs to the family of ectonucleotide pyrophosphatases/phosphodiesterases [28]; it has been reported that this is the only surface antigen which, among hematopoietic cells, is specifically expressed at baseline by basophils and mast cells in healthy subjects (and CD34+ progenitor cells) [9]. While further work will be required to establish the mechanisms which account for the increased levels of surface CD203c on basophils in nut-allergic subjects, it is interesting to note that the promoter region of the CD203c gene contains binding sites for three critical transcription factors which may be activated in basophils from allergic subjects. For example, basophils from allergic subjects can be chronically exposed to IL-3 [27], which mediates its function via the transcription factor cyclic AMP-response element binding protein 1 [29]. Alternatively, basophil exposure to IgE [30,31,32] leads to activation of the Syk tyrosine kinase, downstream of Fc∊RI ligation. Syk then activates the transcription factors Ets like gene1 [33] and activation transcription factor 2 [34]. A better understanding of these signaling cascades might lead to the discovery of additional biomarkers and/or targets for therapy in allergic subjects.
Further work will be needed to extend this analysis and examine baseline levels of basophil CD203c in whole blood from subjects with aeroallergen allergies. Based on the current literature, it seems unlikely that a similar elevation of basophil CD203c will be found in subjects with aeroallergen allergy, since such allergens are generally known to be less likely to cause basophil priming and anaphylaxis than nut allergens. That said, to our knowledge, most of the research in this area has focused on changes in levels of expression of basophil CD203c following ex vivo stimulation with the offending allergen [10,15,16,35]. In patients who received rush immunotherapy for Japanese cedar pollinosis, the authors observed a reduction in CD203c expression after the therapy, suggesting the utility of the basophil CD203c level for monitoring immunotherapy [15,16]. Interestingly, baseline basophil CD203c levels have also been found to be upregulated in subjects with chronic urticaria [36].
The ex vivo activation of basophils and upregulation of their CD203c occurred in whole blood reconstituted with autologous platelet-free plasma (from the original EDTA-containing tube). We found that CD203c upregulation also occurred when the autologous platelet-poor plasma (less than 10% of platelets were left) was replaced by regular PBS (without either EDTA or calcium). Thus, in our hands, increases in basophil CD203c (and CD63) upon ex vivo basophil activation with specific antigen did not require exogenous calcium, indicating that intracellular calcium stores were sufficient to promote basophil activation upon antigen binding in this setting, as has also been observed in other studies [15,16,35,37,38,39,40]. However, it is possible that the relatively weak CD63 basophil response following ex vivo stimulation with an offending allergen reflected, at least in part, the use of experimental conditions that were not optimal for induction of basophil degranulation (that is, there was no extracellular calcium in the medium used).
Other studies have shown increased levels of CD203c on blood basophils upon exposure to pollen, wheat or cat allergens ex vivo [9,13,14,15,16,17,18,19], or increased levels of CD63 on basophils exposed to peanut, wheat, milk, egg, cedar pollen or amoxicillin [13,16,17,19,40]. When we compared CD203c to CD63, our results were most consistent with those of Ocmant et al. [38], in that CD203c represented a more reliable marker of basophil activation. However, in a recent study of subjects with milk allergy, changes in basophil CD63 levels upon ex vivo stimulation were more pronounced than those of CD203c [19]. Taken together, results from our group and others suggest that it may be useful to assess levels of both CD63 and CD203c on basophils; while both markers can be increased upon basophil stimulation, these two markers may be modulated with different kinetics, perhaps reflecting different mechanisms for their mobilization, or may be differentially activated based on the composition of the medium used to perform the test.
In our study, basophil CD203c levels were decreased at baseline, and their increase upon ex vivo stimulation with peanut allergen was reduced after 4 and 8 weeks of omalizumab treatment but returned toward pretreatment levels after treatment cessation. These findings suggest a potential clinical relevance for the method described here (that is, obtaining blood basophil CD203c measurements at baseline followed by ex vivo stimulation with an offending allergen). Omalizumab is a humanized IgG1κ that binds to the C3 domain of the IgE molecule. This binding prevents IgE ligation to its receptors, including Fc∊RIα, on effector cells. Omalizumab has been shown to downregulate surface expression of Fc∊RIα on basophils after 2 weeks of therapy, presumably because of its ability to reduce levels of free IgE [31,41]. Basophils from allergic subjects treated with omalizumab can exhibit reduced activation for degranulation and mediator release in vitro when exposed to antigens to which the subject is sensitive, again presumably reflecting the reduced expression of Fc∊RI in these cells compared to baseline values [42]. In light of those findings, we are not surprised that basophils from omalizumab-treated subjects also exhibit reduced IgE- and antigen-dependent increases in surface CD203c.
In summary, basophils from subjects with nut allergy exhibit increased surface levels of CD203c at baseline and following rapid ex vivo stimulation with nut allergen. Both can be reduced by omalizumab therapy. These results suggest that basophil CD203c levels can be used for baseline diagnosis and therapeutic monitoring in subjects with nut allergy. The assay described here might also be useful for the clinical monitoring of subjects who undergo omalizumab therapy for other food allergies. Finally, it is possible that the rapid measurement of basophil CD203c levels in whole blood, as performed here, can be used as an in vitro screening method to identify agents which inhibit basophil activation and as an ex vivo outcome measure to test the efficacy of new anti-allergy treatments.
Supplementary Material
Basophil CD11b (A), CD63 (B), CD294 (C), and CD123 (D) levels at baseline were similar in subjects with nut allergy compared to healthy controls. Statistical signi?cance was evaluated by the Wilcoxon rank sum non-parametric test. Each point represents a single sample. Two outliers were observed for CD11b, and one outlier was observed for CD63, their MFI were outside the confidence interval (2 standard deviations). NA: subjects with nut allergy; HC: healthy controls; NS: not significant.
Basophil CD203c levels were specifically increased after stimulation with the offending allergen. Data for 4 representative subjects are shown (subjects 7, 8, 10, 16 as listed in Tables I and II), each with specific allergy to given food, i.e., (1) peanut; (2) walnut; (3) cashew; and (4) apple. Offending allergens are indicated in bold.
Changes in basophil CD63 levels in subjects with nut allergy or healthy controls upon ex vivo stimulation with nut or cockroach allergen. (A) time course analysis (2–, 10– and 30-minute stimulation) showing group means and standard errors for basophil CD63 levels (MFI) from healthy controls (N = 8) stimulated with nut (pink line) or cockroach (green line) allergens, compared to subjects with nut allergy (N = 11) stimulated with nut (red line) or cockroach (blue line) allergens. (B) Percentage increase for CD63 and CD203c following ex vivo stimulation with nut allergen in 13 subjects with nut allergy. The percentage increase for CD63 was significantly lower than the percentage increase for CD203c following ex vivo stimulation with nut allergen in patients with nut allergy (P < 0.035, P < 0.020 and P < 0.021 at, respectively, 2, 10, and 30 minute-time points). % for percentage and * for P < 0.05. In panels (C) and (D), we represent basophil CD63 levels (MFI) as measured from individual subjects upon stimulation with (C) nut or (D) cockroach allergens.
Basophil CD203c levels following ex vivo stimulation with an offending allergen were similar in the presence or absence of plasma factors. In blood samples depleted of plasma from subjects with nut allergy, CD203c levels were significantly increased following 10 minutes of ex vivo stimulation with nut allergen. Data shown are from 3 representative subjects.
Modulation of basophil CD63 levels in subjects with peanut allergy treated with omalizumab. (A) basophil CD63 levels at baseline measured before omalizumab treatment, during treatment (4 and 8 weeks) and 4 weeks after treatment cessation (post therapy). (B) comparative data for basophil CD63 levels following ex vivo stimulation of nut allergy subjects with peanut allergen before omalizumab treatment and during treatment (4 and 8 weeks). W: week.
Level of free plasma IgE (non-omalizumab bound) and basophil CD203c levels upon 10-minute ex vivo stimulation with peanut allergen measured in subjects with peanut allergy before and during omalizumab treatment (4 weeks for subjects 12-15 and 12 weeks for subject 15).
Acknowledgements
We thank our subjects for their participation. All free IgE measurements were performed at Johns Hopkins University (Hamilton Laboratory). We also thank Drs. R. Moss and J. Faix for their review of the manuscript, C. Crumpton and J. Van Dyke at the Stanford FACS Facility, Drs. M. Makam, E. Engleman, K. Atkuri and P. Ngatchou-Sadate for critical advice, and E. Hoyte, D. Robinson and M. Becard for technical assistance.
We would like to thank: Stanford School of Medicine's Dean Fellowship (Y.G.) and the Stanford Institute of Immunity, Transplantation and Infectious Diseases Seed Grant (K.C.N.), Orsak Family Fund (K.C.N.), Lucile Packard Children's Hospital Children's Health Research Program (Y.G. and K.C.N.), ARARD (Association Régionale d'Assistance Respiratoire à Domicile) (Y.G.), Skippy Frank Foundation (R.T. and Y.G.), and United States Public Health Service grants AI23990, AI070813 and CA72074 (M.T. and S.J.G.).
References
- 1.Burks AW. Peanut allergy. Lancet. 2008;371:1538–1546. doi: 10.1016/S0140-6736(08)60659-5. [DOI] [PubMed] [Google Scholar]
- 2.Sicherer SH, Sampson HA. 9. Food allergy. J Allergy Clin Immunol. 2006;117:S470–S475. doi: 10.1016/j.jaci.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 3.Cox L, Williams B, Sicherer S, Oppenheimer J, Sher L, Hamilton R, Golden D. Pearls and pitfalls of allergy diagnostic testing: report from the American College of Allergy, Asthma and Immunology/American Academy of Allergy, Asthma and Immunology Specific IgE Test Task Force. Ann Allergy Asthma Immunol. 2008;101:580–592. [PubMed] [Google Scholar]
- 4.Clark AT, Islam S, King Y, Deighton J, Anagnostou K, Ewan PW. Successful oral tolerance induction in severe peanut allergy. Allergy. 2009;64:1218–1220. doi: 10.1111/j.1398-9995.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 5.Jones SM, Pons L, Roberts JL, Scurlock AM, Perry TT, Kulis M, Shreffler WG, Steele P, Henry KA, Adair M, Francis JM, Durham S, Vickery BP, Zhong X, Burks AW. Clinical efficacy and immune regulation with peanut oral immunotherapy. J Allergy Clin Immunol. 2009;124:292–300. doi: 10.1016/j.jaci.2009.05.022. 300, e291–e297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung DY, Sampson HA, Yunginger JW, Burks AW, Jr, Schneider LC, Wortel CH, Davis FM, Hyun JD, Shanahan WR., Jr Effect of anti-IgE therapy in patients with peanut allergy. N Engl J Med. 2003;348:986–993. doi: 10.1056/NEJMoa022613. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton RG, Marcotte GV, Saini SS. Immunological methods for quantifying free and total serum IgE levels in allergy patients receiving omalizumab (xolair) therapy. J Immunol Methods. 2005;303:81–91. doi: 10.1016/j.jim.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 8.MacGlashan D., Jr Therapeutic efficacy of omalizumab. J Allergy Clin Immunol. 2009;123:114–115. doi: 10.1016/j.jaci.2008.10.053. [DOI] [PubMed] [Google Scholar]
- 9.Buhring HJ, Streble A, Valent P. The basophil-specific ectoenzyme e-npp3 (cd203c) as a marker for cell activation and allergy diagnosis. Int Arch Allergy Immunol. 2004;133:317–329. doi: 10.1159/000077351. [DOI] [PubMed] [Google Scholar]
- 10.Aerts NE, Dombrecht EJ, Bridts CH, Hagendorens MM, de Clerck LS, Stevens WJ, Ebo DG. Simultaneous flow cytometric detection of basophil activation marker cd63 and intracellular phosphorylated p38 mitogen-activated protein kinase in birch pollen allergy. Cytometry B Clin Cytom. 2008;76B:8–17. doi: 10.1002/cyto.b.20437. [DOI] [PubMed] [Google Scholar]
- 11.Kleine-Tebbe J, Erdmann S, Knol EF, MacGlashan DW, Jr, Poulsen LK, Gibbs BF. Diagnostic tests based on human basophils: potentials, pitfalls and perspectives. Int Arch Allergy Immunol. 2006;141:79–90. doi: 10.1159/000094495. [DOI] [PubMed] [Google Scholar]
- 12.Simons FE, Frew AJ, Ansotegui IJ, Bochner BS, Golden DB, Finkelman FD, Leung DY, Lotvall J, Marone G, Metcalfe DD, Muller U, Rosenwasser LJ, Sampson HA, Schwartz LB, van Hage M, Walls AF. Practical Allergy (PRACTALL) report: risk assessment in anaphylaxis. Allergy. 2008;63:35–37. doi: 10.1111/j.1398-9995.2007.01605.x. [DOI] [PubMed] [Google Scholar]
- 13.Abuaf N, Rostane H, Rajoely B, Gaouar H, Autegarden JE, Leynadier F, Girot R. Comparison of two basophil activation markers CD63 and CD203C in the diagnosis of amoxicillin allergy. Clin Exp Allergy. 2008;38:921–928. doi: 10.1111/j.1365-2222.2008.02960.x. [DOI] [PubMed] [Google Scholar]
- 14.de Weck AL, Sanz ML, Gamboa PM, Aberer W, Bienvenu J, Blanca M, Demoly P, Ebo DG, Mayorga L, Monneret G, Sainte-Laudy J. Diagnostic tests based on human basophils: more potentials and perspectives than pitfalls. Int Arch Allergy Immunol. 2008;146:177–189. doi: 10.1159/000115885. [DOI] [PubMed] [Google Scholar]
- 15.Fujisawa T, Nagao M, Hiraguchi Y, Hosoki K, Tokuda R, Usui S, Masuda S, Shinoda M, Hashiguchi A, Yamaguchi M. Biomarkers for allergen immunotherapy in cedar pollinosis. Allergol Int. 2009;58:163–170. doi: 10.2332/allergolint.09-RAI-0097. [DOI] [PubMed] [Google Scholar]
- 16.Nagao M, Hiraguchi Y, Hosoki K, Tokuda R, Usui T, Masuda S, Yamaguchi M, Fujisawa T. Allergen-induced basophil CD203c expression as a biomarker for rush immunotherapy in patients with japanese cedar pollinosis. Int Arch Allergy Immunol. 2008;146(suppl 1):47–53. doi: 10.1159/000126061. [DOI] [PubMed] [Google Scholar]
- 17.Ocmant A, Mulier S, Hanssens L, Goldman M, Casimir G, Mascart F, Schandene L. Basophil activation tests for the diagnosis of food allergy in children. Clin Exp Allergy. 2009;39:1234–1245. doi: 10.1111/j.1365-2222.2009.03292.x. [DOI] [PubMed] [Google Scholar]
- 18.Tokuda R, Nagao M, Hiraguchi Y, Hosoki K, Matsuda T, Kouno K, Morita E, Fujisawa T. Antigen-induced expression of CD203c on basophils predicts IgE-mediated wheat allergy. Allergol Int. 2009;58:193–199. doi: 10.2332/allergolint.08-OA-0023. [DOI] [PubMed] [Google Scholar]
- 19.Wanich N, Nowak-Wegrzyn A, Sampson HA, Shreffler WG. Allergen-specific basophil suppression associated with clinical tolerance in patients with milk allergy. J Allergy Clin Immunol. 2009;123:789–794. doi: 10.1016/j.jaci.2008.12.1128. e720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowak-Wegrzyn A, Assa'ad AH, Bahna SL, Bock SA, Sicherer SH, Teuber SS. Work group report: oral food challenge testing. J Allergy Clin Immunol. 2009;123:S365–S383. doi: 10.1016/j.jaci.2009.03.042. [DOI] [PubMed] [Google Scholar]
- 21.Gibbs BF, Plath KE, Wolff HH, Grabbe J. Regulation of mediator secretion in human basophils by p38 mitogen-activated protein kinase: phosphorylation is sensitive to the effects of phosphatidylinositol 3-kinase inhibitors and calcium mobilization. J Leukoc Biol. 2002;72:391–400. [PubMed] [Google Scholar]
- 22.Miller RL, Eppinger TM, McConnell D, Cunningham-Rundles C, Rothman P. Analysis of cytokine signaling in patients with extrinsic asthma and hyperimmunoglobulin E. J Allergy Clin Immunol. 1998;102:503–511. doi: 10.1016/s0091-6749(98)70141-1. [DOI] [PubMed] [Google Scholar]
- 23.Hamilton RG, Franklin Adkinson N., Jr In vitro assays for the diagnosis of IgE-mediated disorders. J Allergy Clin Immunol. 2004;114:213–225. doi: 10.1016/j.jaci.2004.06.046. quiz 226. [DOI] [PubMed] [Google Scholar]
- 24.Herzenberg LA. FACS innovation: a view from Stanford. Clin Invest Med. 2004;27:240–252. [PubMed] [Google Scholar]
- 25.Tung JW, Heydari K, Tirouvanziam R, Sahaf B, Parks DR, Herzenberg LA. Modern flow cytometry: a practical approach. Clin Lab Med. 2007;27:453–468. doi: 10.1016/j.cll.2007.05.001. v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 27.Ghosn EE, Yang Y, Tung J, Herzenberg LA. CD11b expression distinguishes sequential stages of peritoneal b-1 development. Proc Natl Acad Sci USA. 2008;105:5195–5200. doi: 10.1073/pnas.0712350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buhring HJ, Seiffert M, Giesert C, Marxer A, Kanz L, Valent P, Sano K. The basophil activation marker defined by antibody 97a6 is identical to the ectonucleotide pyrophosphatase/phosphodiesterase 3. Blood. 2001;97:3303–3305. doi: 10.1182/blood.v97.10.3303. [DOI] [PubMed] [Google Scholar]
- 29.Gubina E, Luo X, Kwon E, Sakamoto K, Shi YF, Mufson RA. Betac cytokine receptor-induced stimulation of camp response element binding protein phosphorylation requires protein kinase c in myeloid cells: a novel cytokine signal transduction cascade. J Immunol. 2001;167:4303–4310. doi: 10.4049/jimmunol.167.8.4303. [DOI] [PubMed] [Google Scholar]
- 30.Didichenko SA, Spiegl N, Brunner T, Dahinden CA. IL-3 induces a PIM1-dependent antiapoptotic pathway in primary human basophils. Blood. 2008;112:3949–3958. doi: 10.1182/blood-2008-04-149419. [DOI] [PubMed] [Google Scholar]
- 31.MacGlashan D, Jr, Lichtenstein LM, McKenzie-White J, Chichester K, Henry AJ, Sutton BJ, Gould HJ. Upregulation of FC∊RI on human basophils by IgE antibody is mediated by interaction of IgE with FC∊RI. J Allergy Clin Immunol. 1999;104:492–498. doi: 10.1016/s0091-6749(99)70399-4. [DOI] [PubMed] [Google Scholar]
- 32.Park BL, Kim LH, Choi YH, Lee JH, Rhim T, Lee YM, Uh ST, Park HS, Choi BW, Hong SJ, Park CS, Shin HD. Interleukin 3 (IL3) polymorphisms associated with decreased risk of asthma and atopy. J Hum Genet. 2004;49:517–527. doi: 10.1007/s10038-004-0184-x. [DOI] [PubMed] [Google Scholar]
- 33.Bhoumik A, Ronai Z. ATF2: a transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle. 2008;7:2341–2345. doi: 10.4161/cc.6388. [DOI] [PubMed] [Google Scholar]
- 34.Shore P, Bisset L, Lakey J, Waltho JP, Virden R, Sharrocks AD. Characterization of the ELK-1 ETS DNA-binding domain. J Biol Chem. 1995;270:5805–5811. doi: 10.1074/jbc.270.11.5805. [DOI] [PubMed] [Google Scholar]
- 35.Ebo DG, Bridts CH, Hagendorens MM, Aerts NE, De Clerck LS, Stevens WJ. Basophil activation test by flow cytometry: present and future applications in allergology. Cytometry B Clin Cytom. 2008;74:201–210. doi: 10.1002/cyto.b.20419. [DOI] [PubMed] [Google Scholar]
- 36.Yasnowsky KM, Dreskin SC, Efaw B, Schoen D, Vedanthan PK, Alam R, Harbeck RJ. Chronic urticaria sera increase basophil CD203c expression. J Allergy Clin Immunol. 2006;117:1430–1434. doi: 10.1016/j.jaci.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 37.Chirumbolo S, Conforti A, Ortolani R, Vella A, Marzotto M, Bellavite P. Stimulus-specific regulation of CD63 and CD203c membrane expression in human basophils by the flavonoid quercetin. Int Immunopharmacol. 2010;10:183–192. doi: 10.1016/j.intimp.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 38.Ocmant A, Peignois Y, Mulier S, Hanssens L, Michils A, Schandene L. Flow cytometry for basophil activation markers: the measurement of CD203c up-regulation is as reliable as CD63 expression in the diagnosis of cat allergy. J Immunol Methods. 2007;320:40–48. doi: 10.1016/j.jim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Ono E, Taniguchi M, Higashi N, Mita H, Kajiwara K, Yamaguchi H, Tatsuno S, Fukutomi Y, Tanimoto H, Sekiya K, Oshikata C, Tsuburai T, Tsurikisawa N, Otomo M, Maeda Y, Hasegawa M, Miyazaki E, Kumamoto T, Akiyama K. CD203c expression on human basophils is associated with asthma exacerbation. J Allergy Clin Immunol. 2010;125:483–489. doi: 10.1016/j.jaci.2009.10.074. e483. [DOI] [PubMed] [Google Scholar]
- 40.Tokuda R, Nagao M, Hiraguchi Y, Hosoki K, Matsuda T, Kouno K, Morita E, Fujisawa T. Antigen-induced expression of CD203c on basophils predicts IgE-mediated wheat allergy. Allergol Int. 2009;58:193–199. doi: 10.2332/allergolint.08-OA-0023. [DOI] [PubMed] [Google Scholar]
- 41.Asai K, Kitaura J, Kawakami Y, Yamagata N, Tsai M, Carbone DP, Liu FT, Galli SJ, Kawakami T. Regulation of mast cell survival by IgE. Immunity. 2001;14:791–800. doi: 10.1016/s1074-7613(01)00157-1. [DOI] [PubMed] [Google Scholar]
- 42.MacGlashan DW, Jr, Bochner BS, Adelman DC, Jardieu PM, Togias A, McKenzie-White J, Sterbinsky SA, Hamilton RG, Lichtenstein LM. Down-regulation of FC∊RI expression on human basophils during in vivo treatment of atopic patients with anti-IgE antibody. J Immunol. 1997;158:1438–1445. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Basophil CD11b (A), CD63 (B), CD294 (C), and CD123 (D) levels at baseline were similar in subjects with nut allergy compared to healthy controls. Statistical signi?cance was evaluated by the Wilcoxon rank sum non-parametric test. Each point represents a single sample. Two outliers were observed for CD11b, and one outlier was observed for CD63, their MFI were outside the confidence interval (2 standard deviations). NA: subjects with nut allergy; HC: healthy controls; NS: not significant.
Basophil CD203c levels were specifically increased after stimulation with the offending allergen. Data for 4 representative subjects are shown (subjects 7, 8, 10, 16 as listed in Tables I and II), each with specific allergy to given food, i.e., (1) peanut; (2) walnut; (3) cashew; and (4) apple. Offending allergens are indicated in bold.
Changes in basophil CD63 levels in subjects with nut allergy or healthy controls upon ex vivo stimulation with nut or cockroach allergen. (A) time course analysis (2–, 10– and 30-minute stimulation) showing group means and standard errors for basophil CD63 levels (MFI) from healthy controls (N = 8) stimulated with nut (pink line) or cockroach (green line) allergens, compared to subjects with nut allergy (N = 11) stimulated with nut (red line) or cockroach (blue line) allergens. (B) Percentage increase for CD63 and CD203c following ex vivo stimulation with nut allergen in 13 subjects with nut allergy. The percentage increase for CD63 was significantly lower than the percentage increase for CD203c following ex vivo stimulation with nut allergen in patients with nut allergy (P < 0.035, P < 0.020 and P < 0.021 at, respectively, 2, 10, and 30 minute-time points). % for percentage and * for P < 0.05. In panels (C) and (D), we represent basophil CD63 levels (MFI) as measured from individual subjects upon stimulation with (C) nut or (D) cockroach allergens.
Basophil CD203c levels following ex vivo stimulation with an offending allergen were similar in the presence or absence of plasma factors. In blood samples depleted of plasma from subjects with nut allergy, CD203c levels were significantly increased following 10 minutes of ex vivo stimulation with nut allergen. Data shown are from 3 representative subjects.
Modulation of basophil CD63 levels in subjects with peanut allergy treated with omalizumab. (A) basophil CD63 levels at baseline measured before omalizumab treatment, during treatment (4 and 8 weeks) and 4 weeks after treatment cessation (post therapy). (B) comparative data for basophil CD63 levels following ex vivo stimulation of nut allergy subjects with peanut allergen before omalizumab treatment and during treatment (4 and 8 weeks). W: week.
Level of free plasma IgE (non-omalizumab bound) and basophil CD203c levels upon 10-minute ex vivo stimulation with peanut allergen measured in subjects with peanut allergy before and during omalizumab treatment (4 weeks for subjects 12-15 and 12 weeks for subject 15).