Abstract
Photorhabdus temperata K122 is an entomopathogenic bacterium symbiotically associated with nematodes of the family Heterorhabditidae. Surface fimbriae are important for the colonization of many pathogenic bacteria, and here we report the nucleotide sequence and analysis of the expression of a 12-kbp fragment encoding the mannose-resistant fimbriae of P. temperata (mrf). The mrf gene cluster contains 11 genes with an organization similar to that of the mrp locus from Proteus mirabilis. mrfI (encoding a putative recombinase) and mrfA (encoding pilin), the first gene in an apparent operon of nine other genes, are expressed from divergent promoters. The mrfI-mrfA intergenic region contains inverted repeats flanking the mrfA promoter. This region was shown to be capable of inversion, consistent with an ON/OFF regulation of the operon. In in vitro liquid cultures, both orientations were detected. Nevertheless, when we analyzed the expression of all of the genes in the mrf locus by semiquantitative reverse transcription-PCR during infection of Galleria mellonella (greater wax moth) larvae, expression of mrfA was not detected until 25 h postinfection, preceding the death of the larvae at 32 h. In contrast, mrfJ (a putative inhibitor of flagellar synthesis) was expressed throughout infection. Expression of mrfI was also detected only late in infection (25 to 30 h), indicating a possible increase in inversion frequency at this stage. In both in vitro liquid cultures and in vivo larval infections, the distal genes of the operon were expressed at substantially lower levels than mrfA. These results indicate the complex regulation of the mrf cluster during infection.
Photorhabdus luminescens K122 is a gram-negative insect pathogen belonging to the gamma subdivision of the Enterobacteriaceae (10, 16, 38, 39). In a recent reclassification, P. luminescens K122 was renamed Photorhabdus temperata (19). Despite the fact that it is a pathogen to insects, P. temperata also forms a natural symbiotic association in the intestines of nematodes of the family Heterorhabditidae (20). P. temperata presents numerous advantages as a model system for the study of both symbiosis and virulence in the same organism, since the bacteria and the nematode together constitute a highly efficient pathogenic interaction with a broad host range of insects.
For initiation of the pathogenic process, the nematode transports the bacteria into the interior of insect larvae, where they are released into the hemolymph. By releasing several toxins (6, 12, 18), the bacteria rapidly kill the larvae and multiply to large numbers (14). This virulence phase is followed, as the bacteria reach stationary phase, by the secretion of many hydrolytic enzymes (proteases [13, 15, 40], lipase [42], phospholipase, and DNase) and other enzymes, which degrade the macromolecular structures of the larva. This phase, which can be called the bioconversion phase, is essential for the development and reproduction of the hermaphrodite nematode (11, 15). In addition, during the bioconversion phase, the bacteria secrete a number of antibiotics which ensure that the insect cadaver remains monoxenic (2, 24). With the completion of nematode development and reproduction, the bacteria and infective juvenile nematodes reestablish a symbiotic interaction. Finally, the infective juvenile nematodes leave the cadaver and eventually enter a new host, thus repeating the life cycle.
From the bacterial perspective, this system appears to represent a true symbiosis, since the bacteria have never been isolated in nature independently of the nematode (20). Nevertheless, the organism can be cultivated in the laboratory in a variety of media, and when P. temperata is grown in rich medium, it is frequently possible to identify two phenotypic forms, termed variants I and II. Phenotypic variant I cells are the only form found in nature to date. Variant II cells are distinguishable by colony morphology, the qualitative and quantitative profile of proteins secreted to the medium (33, 42), a substantial reduction in the secretion of antibiotics, and the loss of bioluminescence, which is normally displayed by variant I cells as they enter stationary phase (22). In addition, variant II cells have lost the capacity to enter into symbiosis with nematode hosts (7) but nevertheless remain virulent and kill larvae with great efficiency when injected directly into the hemocoel (41).
Pathogenesis in animals and humans is usually associated with several phases. These include the important early phase of attachment and colonization of surfaces through the action of a wide variety of fimbriae or pili and a variety of adhesins on the surfaces of the bacteria (36). The paradigms for fimbrial expression and regulation are pyelonephritis-associated pili (type P pili encoded by pap) and type 1 fimbriae (encoded by fim) (for a review, see reference 8). These structures and their associated adhesin proteins permit the colonization of epithelial cells through attachment of bacteria to specific surface glycoproteins (3, 17, 28, 30). Another class of fimbriae, the mannose-resistant fimbriae, have also been shown to be essential for the virulence of several bacterial species. Mannose-resistant pili are structures 6 to 7 nm in diameter. They are composed of large numbers of a single pilin molecule of about 17 kDa, and they carry specific adhesins and a few copies of other proteins localized at their tips (36). The genes encoding the different polypeptides of the mannose-resistant pilus are organized in an operon; the best-studied example is present in Proteus mirabilis, which promotes urogenital infections in humans (4, 29, 44).
In this study, we have identified and characterized the composition and expression of the mrf operon of P. temperata K122, encoding a mannose-resistant pilus, in both variant I and variant II cells in in vitro liquid culture and during in vivo infection of an insect host, Galleria mellonella (greater wax moth).
MATERIALS AND METHODS
Bacterial strains.
P. temperata K122 was obtained from D. Clarke (21). Escherichia coli DH5α [supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used as a host strain for all transformations (except for the library construction) and as a negative control for all experiments.
The P. temperata K122 plasmid library was transformed into XL1-Blue MRF′ Supercompetent cells (Stratagene).
Plasmids.
Plasmid pPTF 100 contains the entire mannose-resistant fimbrial mrf operon. It was isolated from a genomic library of P. temperata K122 by hybridization against mrfA and mrfG genes.
Media and chemicals.
All bacteria were grown in Luria broth (LB) at 29°C. When DH5α/pPTF 100 was grown, ampicillin was added to a final concentration of 100 μg/ml. All enzymes were supplied by Promega, unless otherwise stated.
DNA probes were labeled with [α-32P]dCTP (Amersham) by using the Rediprime II kit (Amersham). After labeling, they were purified on Microspin columns of Sephacryl S-200 HR (Amersham).
Insect infection.
An overnight culture of P. temperata K122 was diluted in LB to 5 × 105 cells · ml−1. Ten microliters of this suspension was injected into the hemolymph of G. mellonella 5th-instar larvae. Injected larvae were incubated at 28°C, and at various times after infection, larvae were crushed, following freezing in liquid nitrogen, with a mortar and pestle. Larvae were obtained from G. Dumond, Earl La Teigne Dorée, St. Germain les Vergnes, France.
Chromosomal DNA extraction from P. temperata.
A 30-ml overnight culture of P. temperata was centrifuged at 10,000 × g for 15 min. The cell pellet was resuspended in 3 ml of lysis buffer (10 mM NaCl, 20 mM Tris-HCl [pH 8], 1 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 100 mg of proteinase K · ml−1), and the mixture was incubated at 50°C overnight. The lysate was treated with 3 ml of phenol-chloroform (21:1, vol/vol) and centrifuged as described above, and the DNA was precipitated from the aqueous phase in the presence of 0.3 M sodium acetate and 2 volumes of ethanol.
Recombinant DNA techniques.
Plasmid DNA was isolated by using minicolumns (Macherey-Nagel) as specified by the manufacturer. Electroporation, transformation, and restriction endonuclease digestion were performed according to the methods of Sambrook et al. (35).
Genomic library of P. temperata K122.
Genomic DNA was partially digested with Sau3AI and separated on a sucrose density gradient. Fragments isolated in the range of 5 to 15 kbp were cloned into plasmid pUC18, which had previously been linearized with BamHI, and dephosphorylated with bacterial alkaline phosphatase (Pharmacia Amersham Biotech).
RNA preparation. (i) Bacterial RNA (E. coli and P. temperata).
Total RNA was prepared from bacterial cultures by using the SV Total RNA Isolation kit as specified by the manufacturer (Promega). Each RNA sample was used as a template in a 35-cycle PCR as a control to ensure the absence of DNA contamination.
(ii) RNA from infected larvae.
Total RNA was obtained from whole infected larvae (1 larva per time point) by using the TRI reagent (Sigma) and was treated with 5 U of DNase RQ1 (Promega). Then the RNA was precipitated in the presence of 0.3 M sodium acetate and 2 volumes of ethanol. Fifty nanograms of each total-RNA sample was used as a template in a 35-cycle PCR, as a control to ensure the absence of DNA contamination.
Hemagglutination assays:.
P. temperata variant I and variant II bacteria were grown to either exponential or stationary phase in LB liquid culture with or without agitation or were resuspended in LB from colonies on LB agar plates. Cells were washed in 1× phosphate-buffered saline (PBS) and resuspended to a final concentration of 106 bacteria per ml. Serial dilutions of resuspended cells were made by factors of 2, and 50 μl was added to 50 μl of horse erythrocytes that had been washed and resuspended in 144 mM NaCl in the wells of a microtiter plate. Mannose resistance was determined by using erythrocytes resuspended in 144 mM NaCl-50 mM mannose. Plates were incubated for 30 min at 25°C, and hemagglutination was determined by the accumulation of precipitated erythrocyte pellets at the bottoms of the wells.
DNA sequencing and analysis.
DNA templates were sequenced by using the Big Dye Terminator RR Mix (Applied Biosystems). Reactions were run on an ABI 373 DNA sequencer (Perkin-Elmer Applied Biosystems). Data bank searches and analysis were performed with BLASTX and BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/).
PCR.
DNA fragments spanning the invertible mrfA promoter element were amplified by 30 cycles of PCR. The PCR was performed on a GeneAmp PCR system 2700 (Applied Biosystems) using Taq DNA polymerase. The oligonucleotides used for amplification of mrfA were A5-5 (5′ TCACCCCTGAAACAGTTG 3′) and A5-3 (5′ CCTGAAGATAAGCAGCGA 3′), and those used for amplification of mrfG were mrfG-5 (5′ CGTGTTGCGCGTTGGTC 3′) and mrfG-3 (5′ CTACCCGCACTTAATGC 3′).
Primer extension.
Primer extension experiments were performed using two oligonucleotides: Con2Bis (5′ GAAGAAGAACGACCG 3′), to identify the transcriptional start site of the operon upstream of mrfA, and Seq 2700 (5′ CAGCATGACGACCTTGACG 3′), for the analysis of mrfI. Two micrograms of RNA extracted from P. temperata variant I cells in the exponential-growth phase was denatured at 100°C for 5 min in the presence of the corresponding oligonucleotide. The sample was cooled at 50°C for 5 min. A mixture containing the Moloney murine leukemia virus (M-MLV) buffer (5×), 1 U of RNasin (Promega), 0.5 μl of 100 mM dGTP, 0.5 μl of 100 mM dTTP, 0.5 μl of 100 mM dATP, 0.5 μl of [α-32P]dCTP, and 1 μl of M-MLV reverse transcriptase (Promega) was prepared. The mixture was incubated for 10 min at 37°C. Following the addition of 0.5 μl of 100 mM dCTP, the sample was incubated at 37°C for 50 min and purified on Microspin columns of Sephacryl S-200 HR (Amersham). The samples were denatured at 100°C in the presence of 6 μl of stop buffer (Promega) and loaded onto a 6% polyacrylamide DNA sequencing gel (Ultrapure Sequagel-6; National Diagnostics). A DNA sequencing ladder provided the markers for accurate determination of the transcript initiation site.
Western blotting.
Cells (0.25 OD450 [optical density at 450 nm] unit) were centrifuged and resuspended in PBS. Proteins were precipitated with 10% trichloroacetic acid for 30 min on ice, washed with 80% ice-cold acetone, denatured in loading buffer containing β-mercaptoethanol as described by Sambrook et al. (35), and then incubated at 100°C for 10 min. Samples were subjected to SDS-polyacrylamide gel electrophoresis on an 11% acrylamide gel (BDH Laboratory Supplies), and proteins were transferred to a nitrocellulose membrane (Trans-Blot; Bio-Rad Laboratories) in 25 mM Tris-192 mM glycine buffer. Western blots were incubated for 1 h with a rabbit polyclonal antiserum against MrpA of P. mirabilis (25) at a dilution of 1:20,000. Following three 15-min washes in an excess of PBS, blots were incubated with a 1:10,000 dilution of the secondary antibody, an alkaline phosphatase-conjugated goat anti-rabbit antibody (Promega). Blots were developed with the ECL detection system (Amersham) according to the manufacturer's instructions.
Northern blotting.
Northern blotting was performed according to the method of Sambrook et al. (35). Four to 30 μg of total RNA was denatured at 50°C for 1 h in 0.5 M dimethyl sulfoxide-0.1 M phosphate buffer-144 mM glyoxal. Denatured RNA was separated on a 1% agarose gel in 10 mM phosphate buffer. Following migration, RNA was transferred in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to a nylon membrane (Hybond N+; Amersham). Probes were labeled with [α-32P]dCTP by using the Rediprime II kit and were purified on Sephacryl S-200 HR Microspin columns (Amersham). Purified probes were added to the prehybridization solution, and the mixture was incubated at 65°C overnight. Membranes were washed twice in 2× SSC-0.5% SDS at 65°C, air dried, and exposed to X-ray film (Kodak).
Semiquantitative RT-PCR (Q-RT-PCR).
Single-step reverse transcription-PCRs (RT-PCRs) were performed using the Lightcycler RNA amplification kit with SYBR Green I (Roche Molecular Biochemicals). Reverse transcription was performed at 55°C for 10 min, followed by PCR (55 cycles of 0 s at 94°C, 5 s at 55°C, and 20 s at 72°C). Single-point fluorescence acquisition from the incorporation of SYBR Green into the double-stranded PCR product was measured at the end of each cycle. Melting analysis was performed at the end of each PCR to determine the homogeneity of PCR products, and results were confirmed by the migration of PCR bands on 1.5% agarose gels. The efficiency (E) of each pair of PCR primers for each of the mrf genes, and that for the control P. temperata K122 16S gene, was determined by PCR of serial dilutions of P. temperata K122 genomic DNA (100 ng to 10 pg); E was consistently 1.93 ± 0.05. The pair of oligonucleotide primers for the 16S RNA was specific for P. temperata and was obtained from R. ffrench-Constant (15). All semiquantitative RT-PCRs were performed with four replicate reactions.
Immunogold electron microscopy.
P. temperata K122 variants I and II were grown either on LB agar or in aerated or static liquid LB. For labeling experiments, bacterial suspensions were prepared in PBS. Immunogold labeling was performed as described by Zhao et al. (44). Diluted cultures were placed on an N Formvar grid, from which the excess liquid was removed after a 5-min incubation. The grid was air dried and blocked for 30 min in the presence of PBS containing 1% bovine serum albumin (BSA). The grid was incubated with a primary antiserum solution (1/100) against MrpA for 1 h (25) and then washed twice for 5 min in PBS-1% BSA. Gold-labeled goat anti-rabbit immunoglobulin G (AuroProbe; Amersham Pharmacia Biotech) was used as a secondary antibody for 2 h. Grids were washed twice with PBS-BSA, and bacteria coupled with antisera were fixed with 3% glutaraldehyde for 5 min. Following a wash in sterile water, grids were stained with 5% uranyl acetate for 10 s, and observations were performed by transmission electron microscopy using a Philips EM 208 at 80 kV.
Nucleotide sequence accession number.
The nucleotide sequence of the P. temperata K122 mrf operon has been deposited in GenBank under accession no. AF396083.
RESULTS
Identification of a gene, mrfA, encoding a mannose-resistant pilus in P. temperata K122.
In the course of an investigation of differential gene expression in P. temperata K122 variant I and II cells, using RNA fingerprinting by arbitrarily primed PCR (43) and BLASTX and BLASTN analysis, we identified, for the first time in this organism, a gene fragment with 90% similarity to mrpA, which encodes a mannose-resistant pilin in P. mirabilis. Although the P. temperata gene, which we designated mrfA, appeared to be expressed in both variant I and variant II cells in liquid culture, we considered it worthy of further investigation, in anticipation that it may play an important role in pathogenicity toward insects. In addition, in these experiments we also identified a fragment of a second gene with strong homology to another gene in the P. mirabilis mrp operon, mrpG, encoding a minor component of the pilus.
Screening of a P. temperata library for the intact mrf operon.
A plasmid bank of P. temperata grown on LB agar medium plus ampicillin was screened by hybridization with a probe for the mrfA gene as described in Materials and Methods. Several positive clones were obtained, and one was selected for further study. The plasmid was extracted, and the DNA was amplified by PCR using primers derived from mrfA and mrfG. The plasmid was analyzed by various restriction enzymes, and the results indicated an insert of approximately 12 kbp possessing both mrfA and mrfG, as shown by Southern blotting. Moreover, the extremities of the insert did not correspond to genes in the mrp operon from P. mirabilis, indicating that the entire mrf operon was present on this fragment.
DNA sequence of the P. temperata K122 mrf operon.
DNA sequencing of the insert was achieved by walking along the fragment, and the sequence was aligned and compared with the P. mirabilis sequence as described in Materials and Methods, by using FredFrap and Autoassembler.
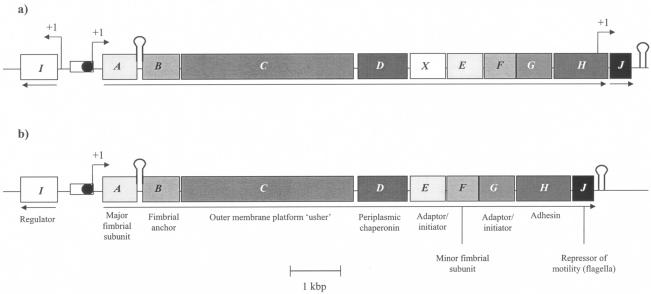
As shown in Fig. 1, the resulting sequence (GenBank accession no. AF396083) contains 11 open reading frames (ORFs), with a predicted stem-loop structure between mrfA and mrfB, together with a Rho independent terminator following the last gene, mrfJ. The mrf genes are organized in an apparent operon (Fig. 1a) like the mrp operon from P. mirabilis (Fig. 1b). Similarly, the mrf locus has an additional gene, mrfI, on the opposing strand and upstream of mrfA, as in P. mirabilis. However, we also noted some differences between the mrf and mrp operons. Most clearly, the mrf operon contains an additional gene, mrfX, upstream of mrfE. Although this gene displays 44.8% identity with mrfE at the DNA level (Table 1), there is only 20.7% identity at the protein level. Similarly, mrfX and mrpE exhibit 43.1% DNA identity and 20.5 and 32.7% amino acid identity and similarity, respectively. The origin of mrfX and the function of MrfX remain unknown, although a database BLASTP search indicates the presence of a conserved fimbrial motif (motif pfam00419.8), and the closest database homologue is the long polar fimbrial protein A precursor from Salmonella enterica serovar Typhimurium LT2 (GenBank accession no. AAL22500).
FIG. 1.
Genetic organization of the P. temperata K122 mrf (GenBank accession no. AF396083) (a) and P. mirabilis mrp (GenBank accession no. Z32686) (b) operons. “+1” indicates transcription initiation sites. The stem-loop following mrfA is predicted on the basis of sequence data. The predicted function of each gene, based on the data for P. mirabilis, is given. Arrows indicate direction of transcription.
TABLE 1.
Global alignmenta comparison between individual mrp and mrf genes and encoded protein sequences
| Mrf vs Mrp comparison | % DNA similarity | % Amino acid identity | % Amino acid similarity |
|---|---|---|---|
| I | 67.1 | 73.3 | 84.8 |
| A | 63.7 | 66.7 | 77.2 |
| B | 51.9 | 41.9 | 55.5 |
| C | 61.3 | 60.1 | 76.8 |
| D | 58.9 | 61.7 | 82.6 |
| E | 53.4 | 44.3 | 63.5 |
| F | 62.3 | 57.1 | 71.8 |
| G | 56.0 | 48.2 | 64.9 |
| H | 56.6 | 52.5 | 71.2 |
| J | 55.1 | 52.3 | 65.8 |
| MrfX vs MrpE | 43.1 | 20.5 | 32.7 |
| MrfX vs MrfE | 44.8 | 20.7 | 32.3 |
Alignments were performed with the algorithm from EMBOSS (34) available at http://www.ebi.ac.uk/emboss/align/index.html.
Table 1 further details the DNA and amino acid comparisons between the conserved P. temperata mrf and P. mirabilis mrp ORFs. Overall, genes encoding the major fimbrial subunit (mrfA and mrpA), minor subunit (mrfF and mrpF), regulator protein (mrfI and mrpI), outer membrane usher (mrfC and mrpC), and periplasmic chaperone (mrfD and mrpD) are all highly conserved at both the DNA and amino acid sequence levels. The exceptions are mrfD and mrpD, where DNA conservation is relatively low (58.9%) while amino acid sequence similarity is high (82.6%). Genes encoding the putative fimbrial anchor (mrfB), adaptor/initiator proteins (mrfE and mrfG), adhesin (mrfH), and repressor of flagellar synthesis (mrfJ) exhibit lower levels of DNA and amino acid sequence conservation.
The intergenic distance between the divergent genes mrfA and mrfI in P. temperata is slightly longer (684 bp) than that between mrpI and mrpA in P. mirabilis (678 bp), and as described below, the most distal gene, mrfJ, unlike mrpJ, can apparently be transcribed independently of the rest of the operon, at least under some conditions.
P. temperata-mediated horse erythrocyte hemagglutination.
In view of the differences between the mrf and mrp operons, we tested if mrf encodes fimbriae functionally similar to those encoded by mrp by examining mannose-resistant erythrocyte hemagglutination. P. temperata variant I and II cells from exponential- and stationary-phase liquid LB cultures grown with or without agitation and from resuspended colonies from LB plates were tested for their abilities to hemagglutinate horse erythrocytes (see Materials and Methods). In all cases, addition of bacteria to erythrocyte suspensions in PBS resulted in bacterial-dose-dependent hemagglutination. Addition of 50 mM mannose to hemagglutination tests had no significant effect. Therefore, we concluded that the mrf fimbriae, like the mrp fimbriae in P. mirabilis, are indeed mannose resistant.
Primer extension analysis to identify transcription start sites.
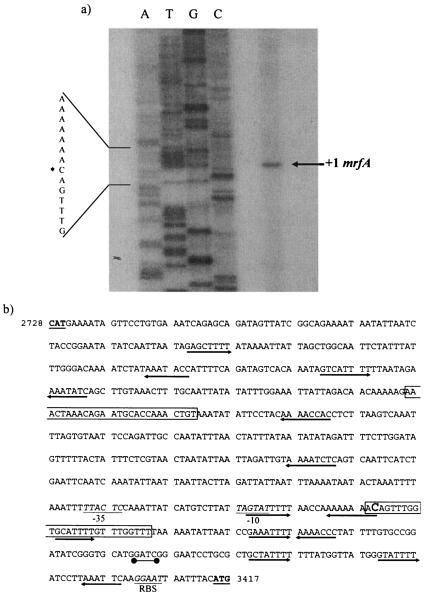
Primer extension analysis was performed as described in Materials and Methods following total-RNA extraction from variant I cells growing exponentially in LB medium. As shown in Fig. 2a, the results of the first extension analysis demonstrated that a transcriptional start site was detected at nucleotide C3260, upstream of, and on the coding strand for, mrfA.
FIG. 2.
(a) +1 position of mrfA. Asterisk indicates the +1 nucleotide in the sequence, corresponding to the mrfA transcriptional initiation site. (b) Organization of the mrfA promoter and invertible element. Inverted repeats are boxed, and the mrfA transcriptional start site, C3260, is enlarged in the second repeat. mrfI and mrfA initiation codons are boldfaced and underlined. The mrfA ribosome binding site (RBS) and −10 and −35 sequences are italicized and underlined. Putative Lrp binding sites are indicated by arrows, and the putative dam methylation site (positions 3343 to 3346) is indicated by a dumbbell. Numbers correspond to sequence positions in GenBank accession no. AF396083.
In addition, by using primer Seq2700 (bp 2655 to 2673; located within the 3′ end of mrfI), we identified a transcriptional start site upstream of mrfI (data not shown). However, we cannot indicate the nucleotide with precision in this case, since the sequencing ladder was illegible. We attributed this to a possible mixed population of DNA molecules being in the ON and OFF states (see below) with respect to the mrfI-mrfA intergenic region, thus resulting in a double DNA sequencing ladder.
Subsequent studies of the expression of mrf genes during infection of larvae (see below) also indicated that mrfJ, whose equivalent in P. mirabilis encodes a repressor of flagellar synthesis (27), might in fact be transcribed independently of the other mrf genes. However, our attempts at locating a transcription start site upstream of mrfJ were not conclusive.
Sequence analysis of the phase inversion region in the mrf operon.
Regulation of phase variation expression of the fim type 1 fimbrial operon, similarly to P. mirabilis mrp regulation, involves inversion of a short DNA element, fimS, between fimE and fimA (1). Regulation of fim expression involves other recombinases, FimB and FimE, together with the regulatory proteins Lrp (leucine-responsive regulatory protein) and IHF (integration host factor), which stimulate recombinase activity (8). In contrast, recombinase activity is repressed by H-NS (37). Expression of the pap operon does not involve an invertible element but is regulated by Lrp, together with PapB and PapI, H-NS, the cyclic AMP receptor protein (CRP), and Dam methylation of two GATC sites within the papI-papB intergenic region (see reference 8). In both the fim and pap operons, Lrp, together with regulators specific to each system, is involved in the formation of a nucleoprotein complex in the regulatory region, binding to a consensus motif, 5′-GN2-3TTT(T)-3′ (32). In the case of the pap system, this also implicates the Dam methylation sites.
Inspection of the DNA sequence of the mrfA locus identified a 319-bp sequence containing the two 25-bp inverted repeats flanking a 269-bp core. In contrast to those in P. mirabilis, the 25-bp inverted repeats are not identical but have two mismatches (indicated by the asterisks): 2966-AAACT*AAACAG*AATGCACCAAACTGT-2991 and 3259-ACAGTTTGGTGCATTT*TGTTTG*GTTT-3284. The positions of the relevant control elements of the mrfI-mrfA intergenic region in the ON position are shown in Fig. 2b. Sequence analysis of this region not only reveals the inverted-repeat motifs but also identifies 14 consensus Lrp binding sites (seven 5′ to 3′ and seven 3′ to 5′), a GATC Dam methylation site consensus (positions 3342 to 3345), putative −35 (TTACTC; bp 3214 to 3220) and −10 (TAGTA; bp 3238 to 3242; 18 bp upstream of the transcription initiation site, C3260) sequences, and a ribosome binding site (GGAAT; bp 3402 to 3406) 8 bp upstream of the mrfA initiation codon.
In vitro inversion of the mrfI-mrfA inversion element.
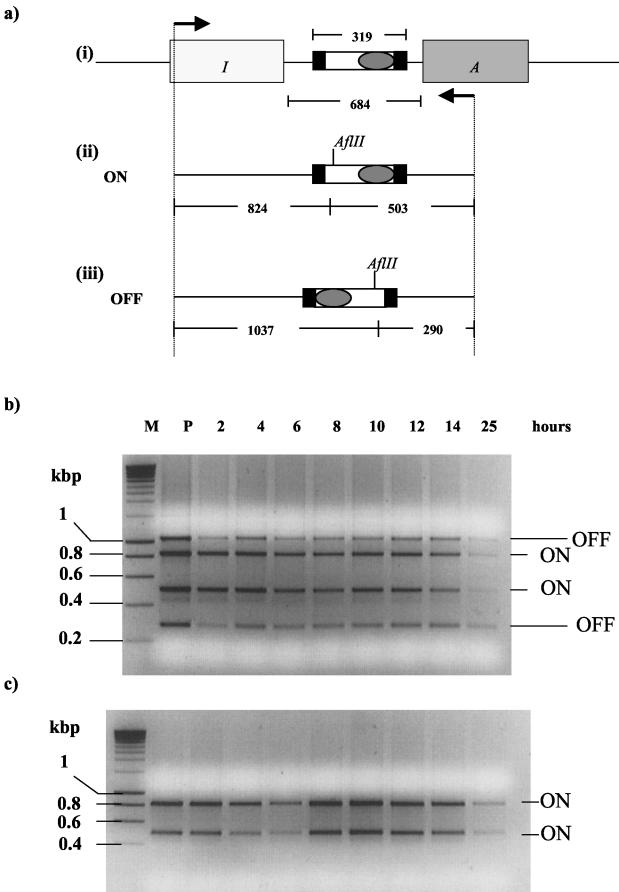
Figure 3a indicates the two alternative positions of the inverted element for transcription of mrfA, the ON and the OFF positions. The two alternative positions should be detectable by restriction enzyme digestion of the intergenic and flanking regions, yielding two AflII fragments of different lengths.
FIG. 3.
(a) (i) Organization of the mrfA-mrfI intergenic region, showing the invertible element bearing the mrfA promoter (shaded oval) flanked by two inverted repeats (solid boxes). (ii and iii) Schematic representations of the PCR products obtained after amplification of the intergenic region. The relative positions of the AflII site and the sizes of the fragments obtained after restriction digestion are indicated, allowing for discrimination between the ON (ii) and OFF (iii) states of the invertible element. (b and c) Analysis of the inversion states of the intergenic element during growth of P. temperata variants I and II. Growth times (in hours) are given at the top of the upper gel. P, overnight culture. (b) PCR products of the variant I intergenic region digested with AflII and analyzed on a 2% agarose gel. (c) A similar experiment with variant II cells.
Accordingly, a 1,327-bp fragment from a site in mrfI or mrfA (Fig. 3a) was first amplified by PCR from genomic DNA isolated at different times throughout the growth of a culture of P. temperata variant I and was then digested with AflII. The results (Fig. 3b) identified two intense bands corresponding to the intergenic region in the ON position. However, two additional bands of lower intensity, with sizes corresponding to the OFF position, were also detected. This indicated that a substantial proportion (approximately 40%) of variant I cells under these conditions have an inactive mrfA promoter. Furthermore, the relative proportions of ON and OFF bands appear to remain constant throughout growth.
When an identical experiment was performed with variant II cells, the results (Fig. 3c) were quite clear-cut, with the whole population displaying the ON position.
Expression of mrfA in liquid cultures.
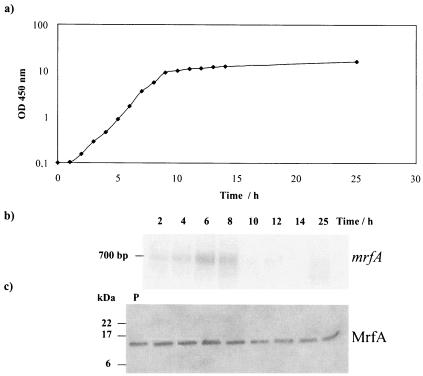
Cultures of both variant I and variant II bacteria were grown in LB medium, and samples were removed for mRNA and protein analysis throughout growth (Fig. 4a). Total RNA was extracted, denatured, and analyzed by Northern blotting followed by probing with a denatured [α-32P]dCTP-labeled mrfA probe. Figure 4b shows that the mrfA transcript could be detected throughout most of the exponential-growth phase in variant I cells but then disappeared in stationary phase. Similar results were obtained for variant II cells (data not shown). Furthermore, the estimated size of the mrfA transcript (700 bp) is consistent with the size of the mrfA gene (540 bp) plus the 155-bp untranslated leader, indicating that transcription termination occurs at the stem-loop structure downstream of mrfA. We were unable to detect a larger operon transcript by Northern blotting. This may be attributed to the limit of detection in total-RNA samples by this technique.
FIG. 4.
Production of mrfA transcripts and the MrfA protein during the growth of variant I cells in LB culture. (a) An overnight culture of P. temperata K122 variant I was diluted in LB medium to an OD450 of 0.1. Growth at 29°C was monitored by measurement of OD450 at each hour. (b) Northern blot of total RNA during growth. mrfA was used as a probe. (c) Western blot analysis of MrfA during growth, using an antiserum against MrpA.
In contrast to the expression of mrfA, Northern blot analysis of mrfG expression failed to detect any transcript, suggesting that expression of the distal genes is regulated in some way, presumably involving the mrfA distal stem-loop structure. Expression of mrfA in both variant I and variant II cells was also examined by semiquantitative RT-PCR. As shown in Fig. 5, the results confirmed expression in exponentially growing cells (taken at 5 h of growth from the culture for which results are shown in Fig. 4a) at significantly higher levels, as might be expected from the results shown in Fig. 3c for the variant II population. Importantly, semiquantitative RT-PCR now also detected transcripts from mrfG (data not shown), although at levels 50- to 70-fold lower than those of mrfA in variant I and II populations, respectively. This supports the idea that most mrfA transcripts may terminate at the predicted stem-loop between mrfA and mrfB (see Fig. 1a).
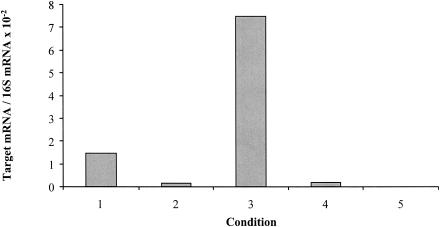
FIG. 5.
Relative abundance and expression of mrfA transcripts (with 16S RNA used as a standard) at different growth points in liquid cultures of P. temperata variants I and II and E. coli (used as a negative control). Bar 1, P. temperata K122 variant I cells growing exponentially; bar 2, variant I cells in stationary phase; bar 3, variant II cells in exponential phase; bar 4, variant II cells in stationary phase; bar 5, E. coli cells in exponential phase.
Samples were also analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting with an antibody to the P. mirabilis MrpA protein (see Materials and Methods). The results for variant I cells showed that the MrfA protein was present throughout the growth phase and remained quite stable during stationary phase (Fig. 4c). Essentially identical results were obtained for variant II cells (data not shown). Subsequent studies with immunogold electron microscopy confirmed that Mrf fimbriae are indeed present on the P. temperata K122 cell surface.
Immunogold detection of Mrf fimbriae.
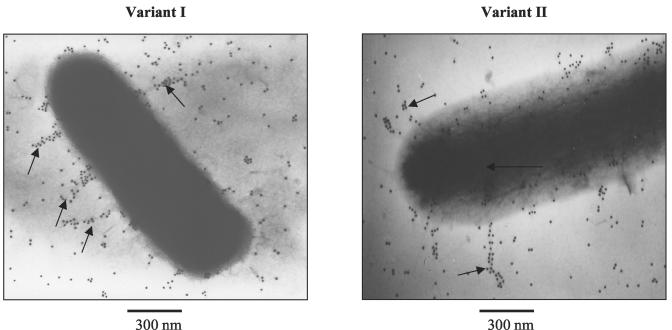
The experiments described above clearly demonstrated the expression of mrfA, encoding the pilin, in both variant I and variant II cells growing exponentially. The results also indicated a lower-level expression of the downstream genes necessary for production of complete pili. As shown in Fig. 6, by using an antibody to the homologous MrpA pilin from P. mirabilis (a gift from H. Mobley), we were able to confirm by immunogold electron microscopy the production of a small number of approximately 300-nm-long Mrf fimbriae in both variant I and variant II cells. Analysis of cells from colonies grown on solid LB agar medium or from liquid LB culture throughout the growth phase demonstrated that Mrf fimbriae are always present on the surface of P. temperata K122 and in approximately similar numbers. Notably, however, in Fig. 6, very few intact fimbriae can be detected, indicating their apparent fragility under these conditions.
FIG. 6.
Immunogold detection of P. temperata K122 variant I and variant II Mrf fimbriae. Cells were grown on LB agar. Arrows indicate the localization of mannose-resistant fimbriae on cells.
RT-PCR analysis of the expression of the mrf operon during infection.
Previous studies have indicated that following injection of G. mellonella larvae with approximately 5,000 cells of P. temperata, death of larvae occurs after approximately 30 h. The bacteria accumulate to 1010 cells after 48 h, and the specific secreted metalloprotease PrtA, associated with the bioconversion phase (13), is first detected 5 to 10 h after the death of the larvae (15). In this study, we analyzed the expression of the mrf operon during the infection of G. mellonella larvae by using semiquantitative RT-PCR. Since it is generally accepted that prokaryotic 16S rRNA genes, like eukaryotic actin, are constitutively expressed at a constant level under most growth conditions, the bacterial 16S rRNA gene was used as an internal standard for expression studies.
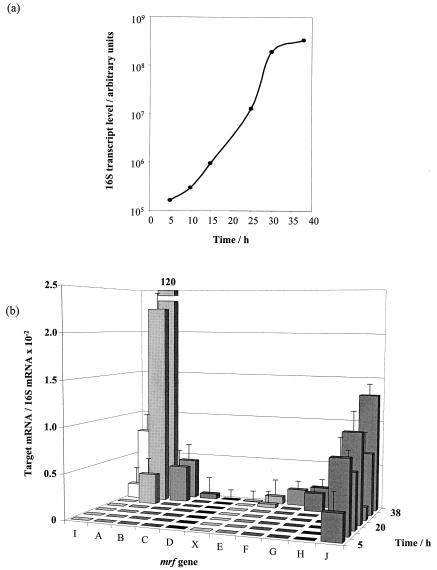
G. mellonella larvae were infected with 5,000 variant I bacteria as described in Materials and Methods. Total RNA was extracted from individual larvae at intervals up to 38 h. First, as shown in Fig. 7a, the accumulation of 16S RNA itself during infection was measured by semiquantitative RT-PCR. This reflects the growth of the bacteria, with a short, approximately 5-h lag, an exponential phase, and entry into stationary phase at approximately 30 h, with an exponential generation time of approximately 2 h 26 min. By use of primers specific for each gene in the mrf locus, the relative levels of transcription of these genes were determined by comparison with the level of 16S RNA (see Materials and Methods). Total-RNA samples were tested by PCR with all oligonucleotide pairs, and all samples used in Q-RT-PCR were negative for DNA contamination.
FIG. 7.
(a) Growth curve of P. temperata K122 variant I bacteria in infected G. mellonella larvae as determined from the Q-RT-PCR crossing-point data obtained by amplification of 16S mRNA. (b) Expression patterns of different genes of the mannose-resistant fimbrial locus during infection of G. mellonella by P. temperata K122 variant I.
The results of this analysis are presented in Fig. 7b. The first gene whose expression was detected in infected larvae was mrfJ at 5 h, and its levels remained relatively high to the end of the infection. This result clearly supports the independent expression of mrfJ, encoding a putative inhibitor of flagellar biosynthesis, from its own promoter. In contrast, mrfA expression was detected only at 25 h, about 5 to 6 h before the death of the larvae in nonsacrificed controls. Expression of several downstream genes of the mrf operon was detected between 30 and 38 h. However, the levels of the latter, for example, mrfB, mrfC, mrfF, and mrfG, were on average 2 orders of magnitude lower than the level of mrfA, again suggesting a role for the predicted stem-loop following the mrfA gene, in vivo as in vitro (see Fig. 1a), in downregulating expression of the distal genes. Barely detectable levels of expression were obtained for the mrfD and mrfX genes, and no expression was observed for mrfH. We should stress that all oligonucleotide primer pairs, including those for mrfH, were verified for PCR efficiency (see Materials and Methods) and that E values were 1.93 ± 0.05 in all cases (theoretical maximum = 2). Moreover, expression of mrfH could be demonstrated by Q-RT-PCR from total RNA isolated from exponentially growing P. temperata K122 cells grown in vitro in LB liquid culture. Therefore, the failure to detect mrfH expression or the low-level expression of mrfD and mrfX in in vivo infection samples was not due to a physical detection limit but rather to some factor related to conditions of extraction from larvae.
The analysis shown in Fig. 7b indicates that mrfA transcription is blocked in the early stages of growth of the bacteria, despite the fact that the liquid-grown cells used for the inoculum normally contain a majority (60%) with the invertible region in the ON position. The regulation of transcription under these infection conditions may therefore be complex. In this context, it is interesting that, as also shown in Fig. 7b, the expression of mrfI is detected at approximately 30 h, consistent with a possibly increased frequency of switching of the invertible mrfA promoter region at this time.
DISCUSSION
Surface appendages such as pili or fimbriae are now well established as important pathogenicity factors in several bacteria (30). In P. mirabilis the mannose-resistant MR/P fimbriae are essential for uropathogenic infections leading to bladder colonization in mice (26). Such MR/P fimbriae are also implicated in pellicle formation (26) and swarming in this organism (23). As in other cases, MR/P fimbriae are therefore likely to be involved both in interbacterial interactions and in adhesion to host cell surface receptors. Previous studies with Xenorhabdus nematophilus, a related insect pathogen also capable of a highly specific symbiotic relationship with nematodes (31), have indicated that adhesion is important for colonization of the nematode gut, although the molecular basis of this phenomenon was not investigated (5). In this study, we have identified and characterized the genetic locus, mrf, in P. temperata. The mrf operon is highly similar to the mrp operon encoding the mannose-resistant pili in P. mirabilis. These pili are related to the type P and type 1 pili in E. coli and are secreted by the “chaperone-usher” pathway (36).
The mrf operon, like its mrp counterpart, contains the highly conserved structural genes for the outer membrane usher, the PapD-like chaperone, and different components of the pilus itself. The latter include the major pilin, encoded by the first gene, mrfA, and the putative adhesin, encoded by mrfH. While the structural components of pili are well conserved, the global regulation of expression of different fimbrial operons in different bacteria varies considerably, although the different regulators are usually encoded at either end of the corresponding operon. These regulatory mechanisms include modulation of the methylation of GATC sites within the promoter region in E. coli (9), trans-acting regulatory factors (Lrp, H-NS, IHF), and site-specific recombination switches (inversion) of the promoter. In fact, as in the mrp locus from P. mirabilis (44), we also identified a putative recombinase gene, mrfI, transcribed in the opposite direction from mrfA, in the mrf locus. The presence of inverted repeats upstream of mrfA, and direct evidence from PCR analysis for inversions in this region, clearly shows that the mrf operon is also regulated by an inversion mechanism, controlling an ON/OFF switch. Similarly, both the mrp and mrf operons contain a predicted stem-loop structure between the first and the second genes in the operon which could participate in the modulation of transcription to ensure that the pilin subunit, MrfA, is produced in excess of the other Mrf proteins. Indeed, both in in vitro liquid cultures and in vivo, in the insect, mrfA messenger was detected in excess (up to 2 orders of magnitude) of the transcripts for the downstream genes. By using primer extension analysis, transcriptional start sites for the divergent mrfI and mrfA genes were confirmed, as found in P. mirabilis for the mrp operon.
Several studies have previously indicated that the production of pili or flagella, required for relatively sedentary versus motile behavior, respectively, might be mutually exclusive (23). Indeed, Li et al. (27) have shown that the MrpJ protein in P. mirabilis is a repressor of flagellar synthesis, although the molecular basis of this effect was not elucidated. The MrfJ protein is 52.3% identical to MrpJ and is highly likely to fulfill a similar function in regulating motility in P. temperata.
Photorhabdus species display a characteristic phenotypic variation, in some cases in response to stress conditions, whereby stationary-phase cultures of variant II cells lack the characteristic bioluminescence, strong yellow pigmentation, and many proteins normally secreted to the medium (22). Our results clearly showed that while laboratory cultures of variant I populations displayed both ON and OFF variations for the mrfI-mrfA intergenic region, variant II cells were all apparently locked in the ON position. Unfortunately, neither the physiological significance of this phase change nor its molecular basis is known; therefore, it is not clear how the conversion to the ON state is effected in variant II cells. Furthermore, the intergenic region between mrfI and mrfA, containing the inversion element, also contains 14 consensus Lrp binding sites and one potential methylation GATC sequence (positions 3342 to 3345) between the mrfA ribosome binding site and the −10 promoter sequence on the untranslated 5′ leader. We cannot, therefore, exclude the formal possibility that these sequences are also involved in regulation of mrf expression.
From previous studies with P. luminescens W14 (15) and from the present study with the related species P. temperata K122, the time course and program of events during infection of G. mellonella larvae may be summarized as follows. The injected bacteria grow exponentially during most of the 10- to 30-h postinfection period; the larvae then appear to die between 30 and 40 h (particularly in the case of K122), while the bioconversion phase, signaled by the production of a secreted protease (13) and the production of the orange pigment secreted by the K122 strain, commences around 40 to 48 h (data not shown). In liquid cultures of Photorhabdus, protease and pigment production occurs at the onset of stationary phase. In the case of P. luminescens W14 (15), the presence of at least one secreted toxin can be detected in infected larvae after about 18 h, consistent with a role in the killing of host cells. In the present study, which used Q-RT-PCR, the major pilin gene, mrfA, apparently was transcribed only between 20 and 25 h postinfection. This time point apparently corresponds to the mid-exponential phase of growth of strain K122 in the larvae, according to the accumulation of 16S mRNA (Fig. 7a), which is still many hours before the death of the larvae. Interestingly, in liquid culture (Fig. 4b), early-exponential-phase cells also appear to express very low levels of mrfA messenger compared to those expressed by late-exponential-phase cells. This temporal control of mrfA expression indicates that the mrf operon may be regulated in other ways in addition to the ON/OFF promoter inversion. Moreover, since the inoculum used in these infection experiments usually contains a majority of cells (60%) with the mrf operon in the ON position (Fig. 3b), these results imply that early in infection the ON switch is inverted or, alternatively, that transcription from cells with mrfA in the ON position is repressed by some other mechanism. In either case, the results suggest that Mrf pili may be most important for some late stages of infection. Expression of other downstream genes in the operon was also detected between 25 and 30 h postinfection, but, as found in liquid cultures, their levels were substantially lower than that of mrfA. Every effort was made to ensure the efficacy of the oligonucleotide probes, including that for mrfH, which, while detecting the product in liquid cultures, failed to detect anything in larvae. It is also unlikely that the difficulties with some distal genes or the absence of mrfA transcripts early in infection is due to an inherent limit in transcript detection, since mrfJ messenger was detectable from the earliest times.
As a further illustration of the possible complexity of mrf expression in vivo, transcription of the presumed recombinase gene, mrfI, was also detected in these experiments only after about 25 to 30 h. As in the case of P. mirabilis, we have identified a transcriptional start for this gene, which is quite distinct from that of the inverting pA promoter and outside the inversion element. However, these results indicate that in the different cells of the bacterial population in the infected larvae, both mrfI and mrfA can be activated in the later stages of infection, with the implication that some cells may have the mrf operon in the ON state and others may have it in the OFF state. This remains to be tested. The presence of two opposed promoters, pI and pA, in the mrfI-mrfA intergenic region, together with a putative GATC Dam methylation site and Lrp binding motifs, is similar to the situation in the papI-papB intergenic region. The presence of an inversion element and Lrp binding sites is similar to that in the fimE-fimA intergenic region. Regulation of expression of the P. temperata mrf and P. mirabilis mrp fimbrial operons may, therefore, possess elements from both of the well-characterized paradigms of fimbrial expression, fim and pap. The in vivo Q-RT-PCR experiments (Fig. 7b) analyzing mrfJ expression demonstrate a profile clearly different from that of the other mrf genes. We attempted to identify an independent mrfJ transcriptional start site and promoter element upstream of mrfJ by primer extension, but results were inconclusive. This intriguing profile for mrfJ expression will be studied further.
Overall, the in vivo experiments and analysis of mrfJ expression suggest that P. temperata K122 flagellar synthesis is regulated even early during infection. In addition, however, pilin synthesis is also blocked and is renewed only after 25 h or more. This perhaps may coincide with a new bacterial colonization phase, not yet identified, preceding larval death, which requires these surface structures. However, it is not possible to determine from these experiments whether the production of Mrf pili is specifically required for the eventual killing of the host or for some other function.
Although much remains to be determined concerning the nature of the molecular switches which control the different stages of Mrf pilus production in vivo, the results presented here further illuminate the differential regulation of gene expression in the pathogenicity of P. temperata and in particular strongly suggest that the mannose-resistant pili play an important role in this process.
Acknowledgments
We thank D. Jaillard for technical assistance with electron microscopy and Y. Zivanovic for helping with the DNA sequence assembly and alignments. We are very grateful to H. L. Mobley for antibodies against MrpA and to G. Dumond for kindly providing G. mellonella larvae.
This work was supported by the CNRS and Université Paris-Sud. L.M.M-C. acknowledges the receipt of a Fondation pour la Recherche Médicale (FRM) fellowship.
REFERENCES
- 1.Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:5724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhurst, R. J. 1982. Antibiotic activity of Xenorhabdus spp., bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J. Gen. Microbiol. 128:3061-3065. [DOI] [PubMed] [Google Scholar]
- 3.Bahrani, F. K., G. Massad, C. V. Lockatell, D. E. Johnson, R. G. Russell, J. W. Warren, and H. L. Mobley. 1994. Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 62:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahrani, F. K., and H. L. Mobley. 1994. Proteus mirabilis MR/P fimbrial operon: genetic organization, nucleotide sequence, and conditions for expression. J. Bacteriol. 176:3412-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binnington, K. C., and L. Brooks. 1993. Fimbrial attachment of Xenorhabdus nematophilus to intestine of Steinernema carpocapsae, p. 147-155. In R. Bedding, R. Akhurst, and H. Kaya (ed.), Nematodes and the biological control of insect pests. CSIRO, Melbourne, Australia.
- 6.Blackburn, M., E. Golubeva, D. Bowen, and R. H. ffrench-Constant. 1998. A novel insecticidal toxin from Photorhabdus luminescens, toxin complex a (Tca), and its histopathological effects on the midgut of Manduca sexta. Appl. Environ. Microbiol. 64:3036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleakley, B., and K. H. Nealson. 1988. Characterization of primary and secondary forms of Xenorhabdus luminescens strain Hm. FEMS Microbiol. Ecol. 53:241-250. [Google Scholar]
- 8.Blomfield, I. C. 2001. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv. Microb. Physiol. 45:1-49. [DOI] [PubMed] [Google Scholar]
- 9.Blyn, L. B., B. A. Braaten, and D. A. Low. 1990. Regulation of pap pilin phase variation by a mechanism involving differential Dam methylation states. EMBO J. 9:4045-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boemare, N., R. Akhurst, and R. G. Mourant. 1993. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int. J. Syst. Bacteriol. 43:249-255. [Google Scholar]
- 11.Bowen, D., M. Blackburn, T. Rocheleau, C. Grutzmacher, and R. H. ffrench-Constant. 2000. Secreted proteases from Photorhabdus luminescens: separation of the extracellular proteases from the insecticidal Tc toxin complexes. Insect Biochem. Mol. Biol. 30:69-74. [DOI] [PubMed] [Google Scholar]
- 12.Bowen, D., T. A. Rocheleau, M. Blackburn, O. Andreev, E. Golubeva, R. Bhartia, and R. H. ffrench-Constant. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280:2129-2132. [DOI] [PubMed] [Google Scholar]
- 13.Bowen, D. J., T. A. Rocheleau, C. K. Grutzmacher, L. Meslet, M. Valens, D. Marble, A. Dowling, R. ffrench-Constant, and M. A. Blight. 2003. Genetic and biochemical characterization of PrtA, an RTX-like metalloprotease from Photorhabdus. Microbiology 149:1581-1591. [DOI] [PubMed] [Google Scholar]
- 14.Clarke, D., and B. Dowds. 1995. Virulence mechanisms of Photorhabdus sp. strain K122 toward wax moth larvae. J. Invertebr. Pathol. 66:149-155. [Google Scholar]
- 15.Daborn, P. J., N. Waterfield, M. A. Blight, and R. H. ffrench-Constant. 2001. Measuring virulence factor expression by the pathogenic bacterium Photorhabdus luminescens in culture and during insect infection. J. Bacteriol. 183:5834-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehlers, R. U., S. Stoessel, and U. Wyss. 1990. The influence of phase variants of Xenorhabdus spp. and Escherichia coli (Enterobacteriaceae) on the propagation of entomopathogenic nematodes of the genera Steinernema and Heterhabditis. Rev. Nématol. 13:417-424. [Google Scholar]
- 17.Emmerth, M., W. Goebel, S. I. Miller, and C. J. Hueck. 1999. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J. Bacteriol. 181:5652-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ffrench-Constant, R. H., and D. J. Bowen. 2000. Novel insecticidal toxins from nematode-symbiotic bacteria. Cell Mol. Life Sci. 57:828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer-Le Saux, M., V. Viallard, B. Brunel, P. Normand, and N. E. Boemare. 1999. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov., and P. asymbiotica sp. nov. Int. J. Syst. Bacteriol. 49:1645-1656. [DOI] [PubMed] [Google Scholar]
- 20.Forst, S., B. Dowds, N. Boemare, and E. Stackebrandt. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47-72. [DOI] [PubMed] [Google Scholar]
- 21.Griffin, C. T., J. F. Moore, and M. J. Downes. 1991. Occurrence of insect pathogenic nematodes (Steinernematidae, Heterorhabditidae) in the Republic of Ireland. Nematologica 37:92-100. [Google Scholar]
- 22.Joyce, S. A., and D. J. Clarke. 2003. A hexA homologue from Photorhabdus regulates pathogenicity, symbiosis and phenotypic variation. Mol. Microbiol. 47:1445-1457. [DOI] [PubMed] [Google Scholar]
- 23.Latta, R. K., A. Grondin, H. C. Jarrell, and G. R. Nicholls. 1999. Differential expression of nonagglutinating fimbriae and MR/P pili in swarming colonies of Proteus mirabilis. J. Bacteriol. 181:3220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, J., G. Chen, H. Wu, and J. M. Webster. 1995. Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl. Environ. Microbiol. 61:4329-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, X., D. E. Johnson, and H. L. Mobley. 1999. Requirement of MrpH for mannose-resistant Proteus-like fimbria-mediated hemagglutination by Proteus mirabilis. Infect. Immun. 67:2822-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, X., C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 2002. Identification of MrpI as the sole recombinase that regulates the phase variation of MR/P fimbria, a bladder colonization factor of uropathogenic Proteus mirabilis. Mol. Microbiol. 45:865-874. [DOI] [PubMed] [Google Scholar]
- 27.Li, X., D. A. Rasko, C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 2001. Repression of bacterial motility by a novel fimbrial gene product. EMBO J. 20:4854-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, X., H. Zhao, L. Geymonat, F. Bahrani, D. E. Johnson, and H. L. Mobley. 1997. Proteus mirabilis mannose-resistant, Proteus-like fimbriae: MrpG is located at the fimbrial tip and is required for fimbrial assembly. Infect. Immun. 65:1327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massad, G., and H. L. Mobley. 1994. Genetic organization and complete sequence of the Proteus mirabilis pmf fimbrial operon. Gene 150:101-104. [DOI] [PubMed] [Google Scholar]
- 30.Morrow, B. J., J. E. Graham, and R. Curtiss III. 1999. Genomic subtractive hybridization and selective capture of transcribed sequences identify a novel Salmonella typhimurium fimbrial operon and putative transcriptional regulator that are absent from the Salmonella typhi genome. Infect. Immun 67:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moureaux, N., T. Karjalainen, A. Givaudan, P. Bourlioux, and N. Boemare. 1995. Biochemical characterization and agglutinating properties of Xenorhabdus nematophilus F1 fimbriae. Appl. Environ. Microbiol. 61:2707-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nou, X., B. Braaten, L. Kaltenbach, and D. A. Low. 1995. Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 14:5785-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popowska, M., K. Pancer, and Z. Markiewicz. 1997. Structure of cell envelope components of the primary and secondary forms of Xenorhabdus luminescens. Acta Microbiol. Pol. 46:19-25. [PubMed] [Google Scholar]
- 34.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spears, P. A., D. Schauer, and P. E. Orndorff. 1986. Metastable regulation of type 1 piliation in Escherichia coli and isolation and characterization of a phenotypically stable mutant. J. Bacteriol. 168:179-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szallas, E., C. Koch, A. Fodor, J. Burghardt, O. Buss, A. Szentirmai, K. H. Nealson, and E. Stackebrandt. 1997. Phylogenetic evidence for the taxonomic heterogeneity of Photorhabdus luminescens. Int. J. Syst. Bacteriol. 47:402-407. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, G. M., and G. O. J. Poinar. 1979. Xenorhabdus gen. nov., a genus of entomopathogenic, nematophilic bacteria of the family Enterobacteriaceae. Int. J. Syst. Bacteriol. 29:352-360. [Google Scholar]
- 40.Valens, M., A. C. Broutelle, M. Lefebvre, and M. A. Blight. 2002. A zinc metalloprotease inhibitor, Inh, from the insect pathogen Photorhabdus luminescens. Microbiology 148:2427-2437. [DOI] [PubMed] [Google Scholar]
- 41.Volgyi, A., A. Fodor, A. Szentirmai, and S. Forst. 1998. Phase variation in Xenorhabdus nematophilus. Appl. Environ. Microbiol. 64:1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, H., and B. C. Dowds. 1993. Phase variation in Xenorhabdus luminescens: cloning and sequencing of the lipase gene and analysis of its expression in primary and secondary phases of the bacterium. J. Bacteriol. 175:1665-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welsh, J., K. Chada, S. S. Dalal, R. Cheng, D. Ralph, and M. McClelland. 1992. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 20:4965-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhoa, H., X. Li, D. E. Johnson, I. Blomfield, and H. L. Mobley. 1997. In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Mol. Microbiol. 23:1009-1019. [DOI] [PubMed] [Google Scholar]