Abstract
Recent advances in the speed and sensitivity of mass spectrometers and in analytical methods, the exponential acceleration of computer processing speeds, and the availability of genomic databases from an array of species and protein information databases have led to a deluge of proteomic data. The development of a lab-based automated proteomic software platform for the automated collection, processing, storage, and visualization of expansive proteomic datasets is critically important. The high-throughput autonomous proteomic pipeline (HTAPP) described here is designed from the ground up to provide critically important flexibility for diverse proteomic workflows and to streamline the total analysis of a complex proteomic sample. This tool is comprised of software that controls the acquisition of mass spectral data along with automation of post-acquisition tasks such as peptide quantification, clustered MS/MS spectral database searching, statistical validation, and data exploration within a user-configurable lab-based relational database. The software design of HTAPP focuses on accommodating diverse workflows and providing missing software functionality to a wide range of proteomic researchers to accelerate the extraction of biological meaning from immense proteomic data sets. Although individual software modules in our integrated technology platform may have some similarities to existing tools, the true novelty of the approach described here is in the synergistic and flexible combination of these tools to provide an integrated and efficient analysis of proteomic samples.
Keywords: Automation, LIMS, MS/MS database search, Peptide analysis, Relational database
1 Introduction
Dramatic progress has recently been made in expanding the sensitivities, resolving power, mass accuracy, and scan rates of mass spectrometers that can fragment and identify peptides through tandem mass spectrometry (MS/MS) [1–4]. Unfortunately, this enhanced ability to acquire proteomic data has not been accompanied by increased availability of tools able to assimilate, explore, and analyze these data efficiently. The typical proteomics experiment can generate tens of thousands of spectra per hour, and the use of multidimensional LC/MS, as with the MUDPIT technique [5], can generate even larger datasets.
Computational tools for the collection and analysis of proteomic data lag far behind analytical methods for proteomic data creation [6]. In a typical experiment, collection and analysis of data is a fully manual process requiring repetitive and laborious sample- and data-processing steps with much unnecessary user intervention [6]. Proteomic datasets are expansive; adequate systems for the initial storage of proteomic data and its relationships to data from other external protein knowledge sources are inflexible and not integrated with the software used in data acquisition.
There are two options for handling the massive and diverse workflows in the modern proteomics lab: either provide a completely integrated software platform that is malleable to the users’ needs, or provide independent software tools that require extensive user intervention to complete a total analysis of the data. Great progress has been made in providing independent software tools such as software focused on a single aspect of the proteomic pipeline. However, proteomic end users are left to fend for themselves in passing data amongst the various software tools and in modifying the individual software tools to provide the processing and analysis needed for interpretation of their specific data. For example, one software tool is used for data acquisition, (such as Xcalibur or Analyst). A second tool interprets tandem mass spectra (such as X!Tandem [7, 8], Mascot [9], SEQUEST [10], OMSSA [11]) or statistical validation of database search results (such as Peptide/Protein Prophet [12], or Ascore [13]). A third tool provides quantitation of proteomic data (such as Xcalibur XDK, or MSQuant [14]), and a fourth provides a relational database for data warehousing (such as PRIME [15] or PeptideAtlas [16]) or a database graphical user interface for visual analysis of proteomic database search results (CPAS [17]). An assortment of web-based protein knowledge resources such as Swiss-Prot [18], HPRD [19], Genbank [20], OMIM [21], BLAST [22], IPI [23], and STRING [24] provide rich annotation of the proteins revealed in high-throughput proteomic experiments. These web-based metadata tools do not permit users to organize these external information sources relationally within the expansive proteomic datasets or to archive user observations. Although each of these tools provides essential functionality, they have not necessarily been engineered to adapt to diverse proteomic workflows or to work together efficiently.
Recent progress has been made in developing integrated systems for post-acquisition processing of data from high-throughput proteomic analysis. Notably, the Trans-Proteomic Pipeline [25] (TPP) integrates many critical aspects of post-acquisition proteomic analysis, including user initiated MS/MS sequence assignment, validation, quantitation and interpretation. To further expand the concepts driving the creation of workflow automation systems for proteomics such as TPP, we have now integrated sample management, data acquisition, post acquisition analysis, and data visualization as integral components of a fully autonomous analysis pipeline called HTAPP.
2 Materials and Methods
2.1 Overall Scheme
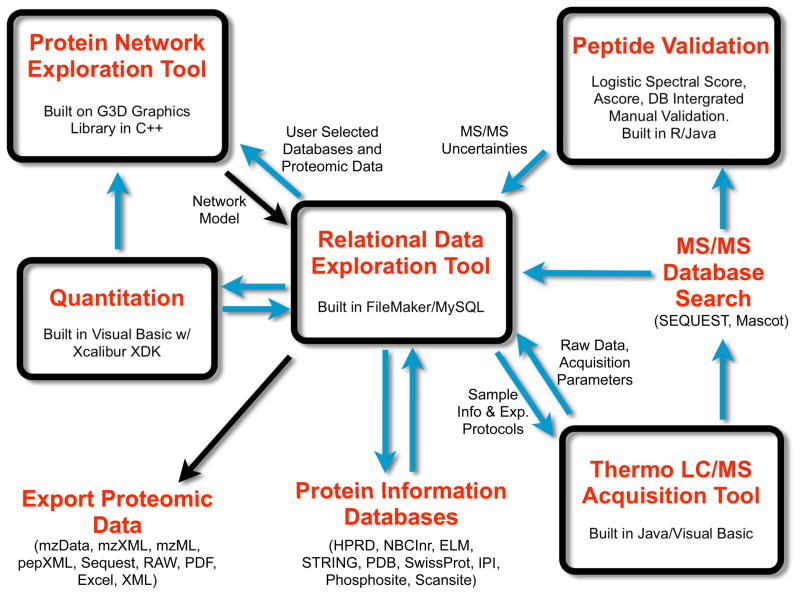
The overall scheme of HTAPP is illustrated in Figure 1. While each individual component of the integrated system can provide critical functionality independently, it is the interoperability of the components that provides a complete technology platform integrating data collection, storage, and visualization. In parallel with the development of HTAPP we have also developed a new relational database for proteomic analysis called PeptideDepot [26]. HTAPP automatically directs the incoming data stream into PeptideDepot where a user may then interact with the processed proteomic data.
Figure 1.
Overview of the HTAPP proteomic pipeline. HTAPP streamlines the collection and autonomous analysis of proteomic data. This system performs automated LC/MS data generation, identification, validation, quantitation, and integration with external protein information databases and enables protein network exploration. Blue arrows indicate actions that are performed automatically and black arrows describe tasks that require users’ intervention.
2.2 Parallel Processing
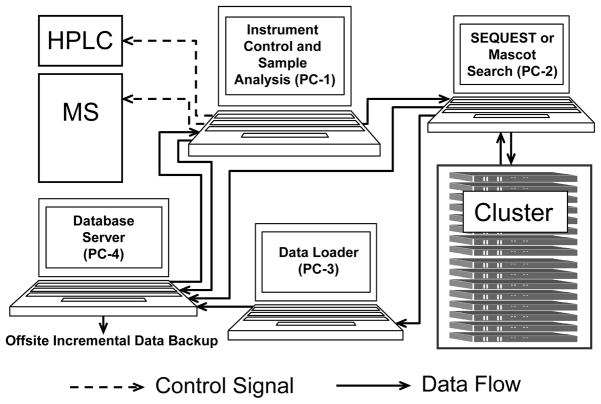
To accelerate data processing and enhance system performance through parallel processing, the system components of HTAPP reside separately on several computers running Windows Server 2003 or Windows XP (Figure 2). Proteomic data are exchanged among those computers using HTAPP programs with a build-in TCP-based file transfer server/client. Inter-component communications are also achieved using HTAPP programs with TCP/IP protocol. Most of user-specific parameters such as server IP, port number and file directories are externalized to a user-configurable parameter file.
Figure 2.
Diagram showing intercomponent data flow and communication scheme for HTAPP. HPLC: high performance liquid chromatography system; MS: mass spectrometer; PC-1: Data acquisition component; PC-2: transfer raw data files and perform SEQUEST or Mascot database search; Cluster: perform clustered SEQUEST or Mascot search; PC-3: autonomous post-acquisition analysis such as spectral validation, peptide quantitation, and upload proteomics data into PeptideDepot database; PC-4: database server (PeptideDepot) for visualization of proteomic data. Once data is deposited into PeptideDepot, it is incrementally backed-up offsite daily.
The modular design of HTAPP allows increased throughput as each component of the analysis workflow is performed simultaneously on separate computers. Through use of a distributed system, parallel processing enables the complete analysis of a proteomic data set within the acquisition time of the next proteomic sample. For example, an experiment containing 10,000 total MS/MS spectra in which ~1,000 spectra are high-quality (as defined by user determined thresholds) requires 1.5 hours to acquire the raw data on the mass spectrometer coupled to PC-1, 1 hour to perform a clustered SEQUEST search on PC-2 and the database search cluster, and 1.5 hours to complete the post-processing tasks including loading of data into the PeptideDepot relational database. Since SEQUEST search and post-processing can be quite CPU-intensive, sequential processing of the data on a single computer requires approximately 4 hours per sample. However, with the distributed system the overall time is reduced to a total of 1.5 hours per sample.
2.3 Data Acquisition Software Module
An automated data acquisition tool developed in Microsoft Visual Basic 6.0 (VB6) runs on PC-1 to organize the predefined sample queue for analysis and to control a set of instrument manufacture software using Visual Basic (Figure 2, and 3D). The extensibility of this tool is derived from flexible instrument control using Visual Basic SendKeys commands allowing the autonomous operation of any instrument control software. This central component of the automated acquisition of LC/MS data controls the unmonitored separation of peptides in, at most, three dimensions of chromatography and a simplified version has been described previously [27].
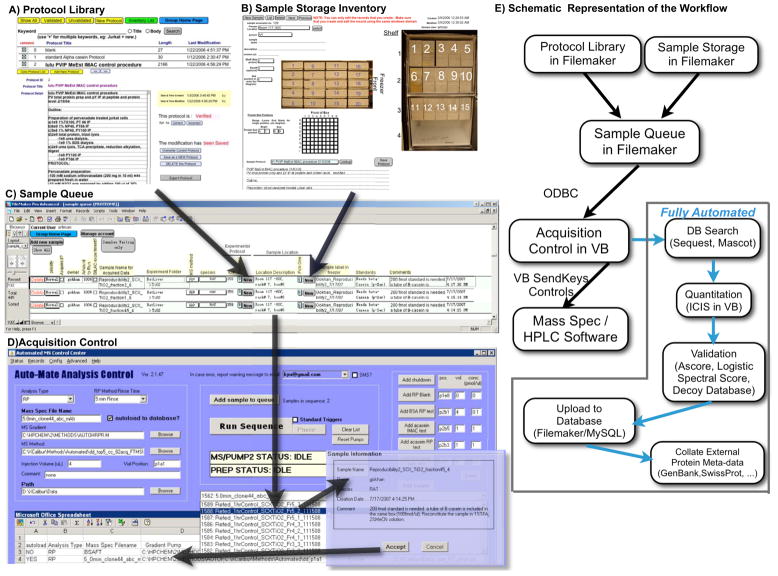
Figure 3.
Integration among relational database component, data acquisition control, and fully autonomous post-acquisition analysis of HTAPP. A) A sample-generating user enters the protocol used in sample creation, B) the location of the sample, and any critical post-acquisition parameters into a C) sample queue located within our FileMaker database. D) The mass spectrometer operator then selects the sample for analysis with the acquisition control software component of our integrated system, which resides on the mass spectrometer control computer. All preferences for post-acquisition analysis, such as database search parameters and quantitation choices, are passed automatically to the acquisition control software from the sample queue and may be optionally modified by the instrument operator. When ‘run sequence’ is clicked, the acquisition control software communicates directly with data acquisition software provided by the instrument manufacturers via flexible VB SendKeys controls. E) Immediately after data acquisition is complete, the acquisition control software initiates automated data analysis, including MS/MS database searching, quantitation of relative peptide abundance, validation of peptide sequence assignments, loading of resulting data into FileMaker/MySQL, and caching of relationships between newly collected proteomic data and existing protein knowledge imported from external protein information databases and located internally within FileMaker/MySQL.
Here, we expand this data acquisition tool to integrate it within a data analysis pipeline that includes a relational database organized sample queue, MS/MS database searching, validation, and quantitation pipeline that automatically deposits the proteomic data and associated analysis within a relational database called PeptideDepot. An ODBC connection (Figure 3E) between the sample queue in PeptideDepot (Figure 3C) and the Visual Basic data acquisition software (Figure 3D) allows retrieval of selected sample information from the PeptideDepot database (FileMaker, version 9.0v3, FileMaker Inc., Santa Clara, CA). During the run, real-time instrument status information such as HPLC pressure profiles (Figure 4D), automated evaluation of selected ion chromatogram peak areas of peptides from standard mixes (Figure 4F), and screen captures (Figure 4E) are archived in the MySQL (version 5.1.40; MySQL Inc., Cupertino, CA) component of PeptideDepot using a VB6 program. This data is available remotely through a website (Figure 4D–F) driven by Apache 2.2.4 (The Apache Software Foundation, Los Angeles, CA) and PHP 5.2.1 (http://www.php.net/).
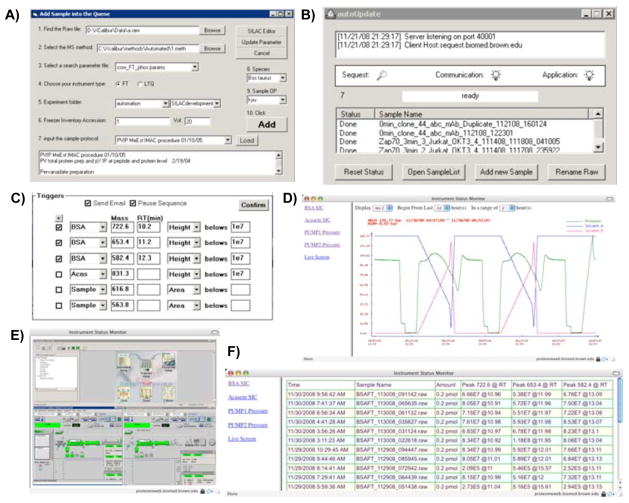
Figure 4.
Automated analysis and troubleshooting within the HTAPP LC/MS data acquisition module. A) Designation of automated SEQUEST search and database deposition parameters for proteomic samples; B) Post-acquisition data pusher for initiation of autonomous post-acquisition analysis; C) Thresholds set for automated real-time monitoring of selected ion chromatogram peak heights or areas of bovine serum albumin (BSA), alpha casein peptides and user-selected masses in users’ proteomic samples that triggers email alerts and/or halts the automated acquisition queue; D) Historical archive of HPLC gradient and pressure profile monitoring for multiple pumps displayed in a web browser; E) A webpage to monitor the live running status of LC/MS; F) Historical archive of three selected BSA peptide ion chromatogram peak areas from automated standard runs.
2.4 Automated Post-Processing Software Modules
Once a sample tagged as ‘Autoload’ is acquired on PC-1, a VB6 program running on PC-2 is notified and raw data files are downloaded from PC-1 via TCP/IP communication over a user-configurable port (Figure 2). MS/MS spectra are extracted from Thermo RAW files using extract_msn.exe (version 4.0; Thermo Scientific, Waltham, MA) or extracted from mzData, mzXML and mzML format data files using ExtractMSMS.jar (in-house developed in Java 1.6.0; Sun Microsystems, Santa Clara, CA) to generate DTA files. A SEQUEST cluster (version 27; Thermo Scientific) or Mascot cluster (version 2.2.1; Matrix Science) MS/MS database search is initiated via a networked computer cluster.
After completion of SEQUEST or Mascot searching, using HTAPP’s build-in file transfer service, proteomic data is pushed to a third computer, PC-3 over a user-configurable port. (Figure 2) On PC-3, a variety of independent post-acquisition calculations are performed on the proteomic data. A VB6 program called “AutoLoad” orchestrates the initiation and transfer of data amongst these separate software tools. A peptide quantitation tool and SILAC [28, 29] calculation tool are available to quantify each identified peptide hits. A phosphosite localization tool that calculates Ascore as described previously [13] is written in Java 1.6.0, and MS/MS validation tool implementing a new user-trainable logistic regression algorithm that more than doubles peptide identifications at a user selected false discovery rate compared to XCorr [30] is implemented in R 2.4.1 (The R Foundation, http://www.r-project.org/). Once the calculations are finished, proteomic data are immediately uploaded to a FileMaker/MySQL relational database called PeptideDepot hosted on the remote server PC-4 using FileMaker script. The proteomic data is then accessible from a graphical FileMaker client (version 9.0v3) running both on Mac and Windows. The database files are synchronized daily without user intervention to an offsite server for the incremental backup using either the commercial software tool Retrospect 7.5 (EMC Insignia; Pleasanton, CA) or the Carbonite backup service (http://carbonite.com; Boston, MA).
3 Results
3.1 Automated Acquisition Control
To create a robust infrastructure capable of high-throughput analysis of proteomic samples, we sought tight integration between the bioinformatic tools used in analyzing proteomic data and the software involved in acquiring mass spectral data. This system can flexibly automate projects ranging from simple LC/MS of in-gel digested proteins to more complex proteomic analyses, such as 2D nano-LC/MS experiments or protein post-translational modification analyses. To maximize the capability of controlling various venders’ instruments, software utilizing a VB SendKeys API was built to automatically run the underlying native HPLC and mass spectrometry software. Such a design transcends any limitations that are artificially imposed by API limitations of the instrument manufacturer’s data acquisition software. SendKeys controls are utilized solely for communication between HTAPP and data acquisition software such as Xcalibur and Chemstation and not used for post-acquisition analysis.
3.2 Sample Queue Management and Automated Workflows
A sample queue capability within the FileMaker component of PeptideDepot relational database integrates sample creation, and metadata annotation with data acquisition control and automated post-acquisition analysis (Figure 3A–D; Figure 4A, B). This system provides unparalleled flexibility to the user by 1) letting any user tailor the sample queue in FileMaker for automation of any lab-specific post-acquisition analysis task or association of any experimental meta-data with the nascent proteomic data, and 2) providing an array of choices for post-acquisition analysis for the automated or manual interpretation of proteomic data.
The laboratory information management system (LIMS) components of the PeptideDepot database are created in the user-friendly FileMaker environment, allowing proteomic end-users to tailor the associated fields and layouts to their specific needs (Figure 3A–C). For example, users wanting to store a new piece of information within the system to be automatically associated with the analyzed proteomic data may quickly add a field for this data in FileMaker and position it precisely within user-defined layouts with FileMaker’s WYSIWYG layout tools (such as illustrated for the protocol library and sample storage inventory in Figure 3A–B). With this flexibility, the end-user need not wait for a programmer or database engineer to add the desired functionality; it may be implemented directly.
3.3 Automation of Post-Acquisition Data Analysis
Although sample metadata may vary dramatically from lab to lab, the processing of proteomic data after acquisition most commonly involves some combination of database searching, quantitation, validation of database search results, and storage of proteomic data within a relational database. A variety of software tools are used in each step of this standard analysis pipeline (summarized in Figure 1). For database searching, our automated system currently supports SEQUEST, Mascot, or any other algorithm that exports to pepXML. For quantitation, our automated system currently uses the ICIS algorithm available in the Xcalibur XDK to calculate peak areas for label free or isotopic labeling methods such as SILAC from any Thermo Scientific Xcalibur (RAW) file and uses an existing software tool ProteinQuant [31] for the label-free quantitation from standard proteomic data formats mzXML and mzData. For validation, our system currently automates the analysis of reversed database searches [32], performs peptide validation using a recently developed logistic spectral score that more than doubles peptide yield at a fixed FDR [30], and phosphorylation site localization using the Ascore algorithm [13]. Our relational database PeptideDepot also provides unique tools, namely SpecNote for database-integrated manual validation and annotation of spectra [30]. Our current system provides for unmonitored import of proteomic data and proteomic analyses into our flexible PeptideDepot relational database that utilizes a FileMaker generated user interface.
3.4 Sample Tracking Database and Protocol Library
We have also created a sample tracking database and protocol library (Figure 3A, B) that organize information about sample preparation and storage and associate this information tightly with the nascent proteomic data. This tool enhances the ability to find correlations between proteomic results and the conditions used to prepare and store samples while facilitating post-acquisition analysis by specification of data processing parameters prior to data acquisition. These tools are dynamically integrated within our data acquisition and automation tools to facilitate the automation and documentation of samples awaiting proteomic analysis. By requiring the entry of sample protocols before data acquisition, critical experimental conditions and metadata are captured, organized, and associated with complex proteomic datasets. Also, the protocol library allows assimilation of all protocols used in the lab within a lab-based relational database and provides a mechanism by which protocols can be reviewed and optionally approved by other researchers.
3.5 Automated Monitoring and System Troubleshooting
To promote efficient troubleshooting of fluctuations in system performance, the automated data acquisition includes the capability to store and analyze metadata captured during spectral acquisition in a fully automated fashion. Information such as the pressure profiles and chromatography gradients are all automatically archived in the MySQL component of the PeptideDepot relational database that is linked to the raw data and SEQUEST results and accessible through a web-based PHP interface (Figure 4D). Selected ion chromatogram (SIC) peak areas of either Bovine Serum Albumin (BSA) or α-casein peptides from automated standard runs, or of user-selected standard peptides incorporated into user samples, are monitored automatically. If any selected peptide falls below a user-defined threshold, the operator is optionally alerted via email or instant SMS and the acquisition queue can be set by the user to pause until the problem is solved (Figure 4C). A user may also explore the historical BSA and α-casein SIC data acquired on the instrument in an interactive web browser layout driven by PHP (Figure 4F) or in a VB6 program to track and troubleshoot instrument sensitivity over time. Remote access capabilities allow any operator to monitor the status of the system in real time (Figure 4E) and to control the system through an encrypted network connection.
3.6 Relational Database for Proteomic Data Exploration
Proteomic results are automatically imported to a networked relational database called PeptideDepot which is described in detail elsewhere [26]. Tight integration of external protein information sources is a critical aspect of this system. Once newly acquired data are deposited into the PeptideDepot database, many data-mining calculations are triggered automatically by querying externally available protein information databases such as PDB [33], IPI [23], HPRD [19], Swiss-Prot [34], STRING [24], Phosphosite [35] and Scansite [36] by peptide sequence across locally cached databases. All possible protein names associated with a given peptide sequence are collated from the locally cached external protein information databases. This capability overcomes the limitation of alternative protein naming by allowing for users to “deep search” the data sets across an index of all possible protein names in every database.
After automated analysis and deposition of the data within PeptideDepot, users may explore the data with flexible FileMaker WSIWYG layouts. PeptideDepot features an extensive collection of predefined data filters that enable users to limit false-discovery rates estimated by reversed database search while focusing on specific peptide qualities such as tyrosine phosphorylation, etc. Comparative analysis views, useful in comparing peptides observed in different cellular states such as disease versus healthy tissue, are provided to facilitate quantitative comparison among samples using either label-free or stable-isotope incorporation quantitation strategies such as SILAC.
4 Discussion
One of the largest impediments to truly high-throughput proteomic methods is the lack of automation after the acquisition of spectra and lack of capture of critical acquisition-specific metadata. In addition, there is a fundamental need not only to acquire data more quickly but also to increase the quality of data acquired. An ideal high-throughput proteomic pipeline would provide for the thorough documentation of a sample’s provenance: the protocol used in sample preparation, sample storage information, environmental conditions such as temperature and humidity during the analysis, and HPLC gradients and pressure profiles.
4.1 Integrative Approach to Proteomic Analysis
One of the fundamental goals of the work described here is to provide truly high-throughput multidimensional acquisition of spectra coupled to automated database searching, data archiving, data filtering, visualization, analysis, quantification, and statistical validation of spectra. The software described here uses an integrative approach in which all information concerning a proteomic experiment is archived automatically along with the raw data and database assignments. All components of analysis are integrated within a lab-centric relational database. Capturing a myriad of experimental metadata in addition to spectral acquisition enables the organization and documentation of complex experiments and facilitates troubleshooting. Unlike other currently available proteomic software, our integrated platform utilizes a sample queue in which post-processing parameters and user-provided proteomic sample annotation are passed directly to data acquisition control software and are associated automatically with proteomic data as it is collected and processed within a lab’s relational database. This tight integration greatly increases efficiency by automating labor-intensive post-processing tasks and reduces the chances that critical connections between newly collected proteomic data and experimental metadata will be lost.
This work provides an integrated yet extensible technology platform for the automated processing, storage, and visual analysis of expansive proteomic datasets. Instead of trying to patch together a variety of preexisting software tools that fit together awkwardly, match analytic needs only marginally, and lack critically important functionality, we have created from the ground up an optimized set of integrated tools that provides automated acquisition, processing and visual analysis of proteomic data. Although many aspects of our software implementations are both unique and essential for a thorough analysis of these types of data, the main novelty of our approach is the direct software integration of the collection, quantitative processing, and visual analysis of proteomic data. No publicly available software tool currently available provides this level of integration. Current proteomic end-users must either develop their own proteomic pipeline software systems in each lab or else perform tedious data manipulation steps manually to extract biological meaning from the immense datasets.
4.2 Flexible Workflow Support
The HTAPP software is designed to provide critical flexibility and functional extensibility for users to implement alternative proteomic workflows as needed. Although the software tool that performs automated data acquisition currently incorporates a Thermo Scientific hybrid linear ion trap – Fourier Transform mass spectrometer (LTQ-FTICR) and Agilent 1100/1200 HPLC pumps, our control software is adaptable to any mass spectrometer and chromatography system through the use of flexible Visual Basic SendKeys controls [37]. In its current implementation, SendKeys works through Xcalibur and Chemstation software to control the automated acquisition of data. Using SendKeys controls, our software sends keyboard commands to any currently running software. By using SendKeys, control of additional mass spectrometer data acquisition software systems can be rapidly implemented to provide critically important extensibility to our automated platform.
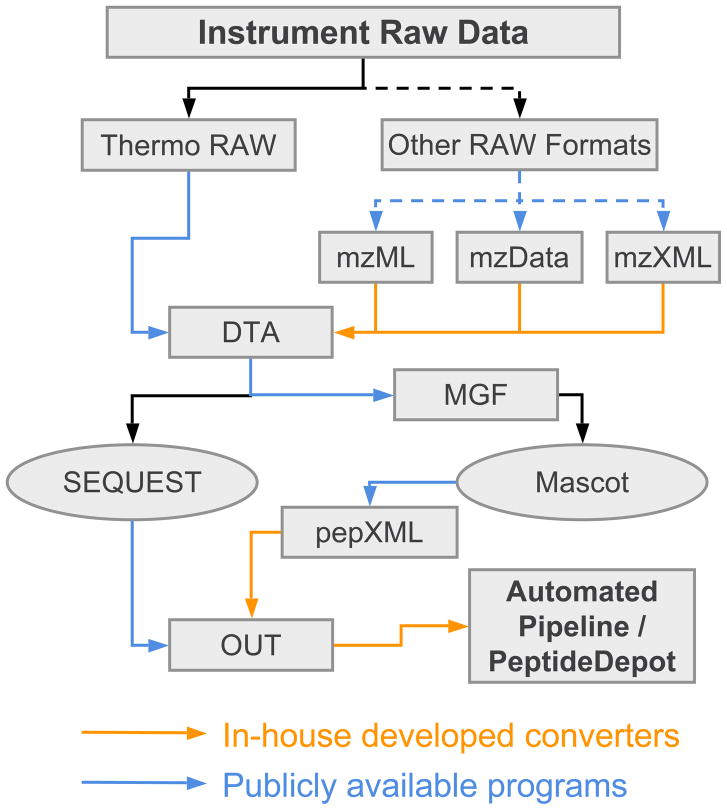
HTAPP also supports the analysis of any additional mass spectrometer MS/MS data that may be converted to the standard proteomic data formats, i.e. mzData [38], mzXML [39] and mzML [40] (Figure 5). Tools to convert manufacturer-specific raw data to standard formats are publicly available (http://tools.proteomecenter.org/wiki/index.php?title=Formats:mzXML). For Thermo Scientific RAW files, the analysis pipeline is fully automated. If a user desires to analyze data from other types of mass spectrometers, the user first converts the data to either mzData, mzXML or mzML format using publicly available software prior to autonomous analysis through HTAPP. We have implemented a Java program in HTAPP to convert MS/MS spectra from standard formats and initiate autonomous data analysis. This software was successfully tested with publicly available proteomic datasets acquired on Agilent, LCQ-Deca, LTQ and QSTAR mass spectrometers [41].
Figure 5.
Flexible workflows through support of standard proteomic data exchange formats. In the figure, mzML, mzXML and mzData are standard XML formats for MS/MS data. DTA is the generic format for SEQUEST input. MGF is the Mascot Generic Format. pepXML is the standard XML format for database search output. OUT is the generic format of SEQUEST search output. Conversions indicated with solid arrows are accomplished autonomously within HTAPP while the dashed lines indicate the tasks that require user intervention. Post-processing of additional mass spectrometer specific raw data formats is provided through support of mzML, mzXML and mzData formats. Database search engines beyond SEQUEST and Mascot can be implemented by converting DTA to a particular format for that search engine and exporting search output in pepXML format, as is illustrated for Mascot here.
After data acquisition, the peptide sequences are assigned through a SEQUEST or Mascot cluster, peptides quantitated, uncertainties of peptide and phosphorylation site placement are accessed, and proteomic data are deposited into a networked relational database (Figure 3E). If a user’s workflow includes additional analysis tasks beyond the core functionality already available within HTAPP, these additional calculations may be automated through FileMaker scripts which export the proteomic data in standard formats, trigger external analysis software, and import the analysis results back into the PeptideDepot database into user-defined fields that are displayed on user-configured layouts.
4.3 Extensibility
To support expansion for future software to interact with the automated pipeline, samples awaiting analysis reside in two independent flat-file formatted sample queues. The first sample queue resides on the data acquisition component (PC-1; Figure 2) while the second queue resides downstream of the database search component on the data loader (PC-3; Figure 2). By adding, removing, or altering the text formatted sample queues, a user can integrate their own software within the HTAPP pipeline (See Supplemental Data 1 for formatting details of the sample flat-file).
To incorporate a new database search engine such as X!Tandem for MS/MS interpretation, the proteomic researcher only need to configure the database search program to export the results in the standard pepXML [25] format and trigger existing pepXML import scripts that are already available in HTAPP (Figure 5). Once imported to FileMaker, the parsed database search results would be integrated into user-defined flexible layouts.
To accomplish any additional post-acquisition data analysis task, the sample queue table within FileMaker has a unique counter field that is transferred throughout the data analysis pipeline and stored with the analyzed proteomic data. Using this counter field, proteomic end users may add any post acquisition preferences to the sample queue, and optionally trigger the execution of external software tools using FileMaker scripts that export the proteomic data from PeptideDepot, trigger the external program and import the results of the external analysis back into FileMaker for display on user-defined custom layouts. For fully automated post-acquisition analysis, the existing FileMaker data import script can then optionally trigger these external calculations.
4.4 Overall Benefit
The laboriousness of current proteomics software manual implementations distracts the proteomics investigator from the biological meaning of the data, leading to the all-too-frequent deposition of data into the scientific literature with minimal biological or clinical interpretation. Instead of treating individual steps in the proteomic pipeline as separate events whose integration depends on end-user intervention, we let the user focus on the interpretation of the data through automation of routine data manipulations and caching of comparisons between newly collected proteomic data and external bioinformatic resources within a lab-based relational database. The software described here are available for nonprofit use free of charge from http://peptidedepot.com after the completion of a license agreement.
Acknowledgments
We thank Samuel P. Ulin of the Brown’s Department of Molecular Biology, Cell Biology, and Biochemistry for help in the preparation of this manuscript. This work was supported by National Institutes of Health Grant 2P20RR015578 and by a Beckman Young Investigator Award.
Footnotes
The authors have declared no conflict of interests.
Supplemental Material
For quick evaluation of the utility of this software, we have provided a phosphoproteomic dataset from a mast cell stimulation timecourse [42] and a simple BSA protein digest. These datasets are available, along with the software described here, at http://peptidedepot.com/.
References
- 1.Chernushevich IV, Loboda AV, Thomson BA. An introduction to quadrupole-time-of-flight mass spectrometry. J Mass Spectrom. 2001;36:849–865. doi: 10.1002/jms.207. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz JC, Senko MW, Syka JE. A two-dimensional quadrupole ion trap mass spectrometer. J Am Soc Mass Spectrom. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 3.Syka JE, Marto JA, Bai DL, Horning S, et al. Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J Proteome Res. 2004;3:621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 4.Yates JR, Cociorva D, Liao L, Zabrouskov V. Performance of a linear ion trap-Orbitrap hybrid for peptide analysis. Anal Chem. 2006;78:493–500. doi: 10.1021/ac0514624. [DOI] [PubMed] [Google Scholar]
- 5.Washburn MP, Wolters D, Yates JR. 3rd, Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 6.Topaloglou T. Informatics solutions for high-throughput proteomics. Drug Discov Today. 2006;11:509–516. doi: 10.1016/j.drudis.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom. 2003;17:2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 8.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 9.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Eng JK, McCormack AL, Yates JR. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 11.Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 13.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature biotechnology. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 14.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 15.Ulintz PJ, Bly MJ, Hurley MC, Haynes HA, Andrews PC. 4th Siena 2D Electrophoresis Meeting; Siena, Italy. 2000. [Google Scholar]
- 16.Desiere F, Deutsch EW, Nesvizhskii AI, Mallick P, et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cottingham K. CPAS: a proteomics data management system for the masses. J Proteome Res. 2006;5:14. doi: 10.1021/pr0626839. [DOI] [PubMed] [Google Scholar]
- 18.Wu CH, Apweiler R, Bairoch A, Natale DA, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 24.von Mering C, Jensen LJ, Snel B, Hooper SD, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Molecular systems biology. 2005;1:2005 0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu K, Salomon AR. PeptideDepot: Flexible relational database for visual analysis of quantitative proteomic data and integration of existing protein information. Proteomics. 2009;9:5350–5358. doi: 10.1002/pmic.200900119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ficarro SB, Salomon AR, Brill LM, Mason DE, et al. Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- 28.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 29.Amanchy R, Kalume DE, Pandey A. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci STKE. 2005;2005:pl2. doi: 10.1126/stke.2672005pl2. [DOI] [PubMed] [Google Scholar]
- 30.Yu K, Sabelli A, DeKeukelaere L, Park R, et al. Integrated platform for manual and high-throughput statistical validation of tandem mass spectra. Proteomics. 2009;9:3115–3125. doi: 10.1002/pmic.200800899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann B, Madera M, Sheng Q, Tang H, et al. ProteinQuant Suite: a bundle of automated software tools for label-free quantitative proteomics. Rapid Commun Mass Spectrom. 2008;22:3823–3834. doi: 10.1002/rcm.3781. [DOI] [PubMed] [Google Scholar]
- 32.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 33.Berman HM, Westbrook J, Feng Z, Gilliland G, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 36.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2. 0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao L, Yu K, Salomon AR. Phosphoproteomic analysis of lymphocyte signaling. Adv Exp Med Biol. 2006;584:277–288. doi: 10.1007/0-387-34132-3_19. [DOI] [PubMed] [Google Scholar]
- 38.Orchard S, Taylor C, Hermjakob H, Zhu W, et al. Current status of proteomic standards development. Expert review of proteomics. 2004;1:179–183. doi: 10.1586/14789450.1.2.179. [DOI] [PubMed] [Google Scholar]
- 39.Pedrioli PG, Eng JK, Hubley R, Vogelzang M, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nature biotechnology. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 40.Deutsch E. mzML: a single, unifying data format for mass spectrometer output. Proteomics. 2008;8:2776–2777. doi: 10.1002/pmic.200890049. [DOI] [PubMed] [Google Scholar]
- 41.Klimek J, Eddes JS, Hohmann L, Jackson J, et al. The standard protein mix database: a diverse data set to assist in the production of improved Peptide and protein identification software tools. J Proteome Res. 2008;7:96–103. doi: 10.1021/pr070244j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao L, Yu K, Banh C, Nguyen V, et al. Quantitative time-resolved phosphoproteomic analysis of mast cell signaling. J Immunol. 2007;179:5864–5876. doi: 10.4049/jimmunol.179.9.5864. [DOI] [PMC free article] [PubMed] [Google Scholar]