Summary
Prostasin is expressed at the apical surface of normal epithelial cells and suppresses in vitro invasion of cancer cells. Prostasin re-expression in the PC-3 prostate carcinoma cells down-regulated the epidermal growth factor receptor (EGFR) protein expression and EGF-induced phosphorylation of the extracellular signal-regulated kinases (Erk1/2). We report here that prostasin and its activating enzyme matriptase are capable of inducing proteolytic cleavages in the EGFR extracellular domain (ECD) when co-expressed in the FT-293 cells, generating two amino-terminally truncated fragments EGFR135 and EGFR110, at 135 and 110 kDa. Prostasin’s role in EGFR cleavage is dependent on the serine active site but not the GPI-anchor. The modifications of EGFR were confirmed to be on the primary structure by deglycosylation. EGFR135 and EGFR110 are not responsive to EGF stimulation, indicating loss of the ligand-binding domains. EGFR110 is constitutively phosphorylated and in its presence Erk1/2 phosphorylation is increased in the absence of EGF. The protease-induced EGFR cleavages are not dependent on EGFR phosphorylation. The EGFR ECD proteolytic modification by matriptase-prostasin is also observed in the BEAS-2B normal lung epithelial cells, the BPH-1 benign prostate hyperplasia and the MDA-MB-231 breast cancer cell lines; and represents a novel mechanism for epithelial cells to modulate EGF-EGFR signaling.
Keywords: ErbB Receptor Tyrosine Kinases, GPI-anchor, Transmembrane Glycoprotein, Extracellular Signal-regulated Kinases, MT-SP1, PRSS8
Introduction
Prostasin (PRSS8), a glycosylphosphatidylinositol (GPI)-anchored extracellular membrane serine protease [1], is ubiquitously expressed in epithelial tissues such as the bladder, colon, kidney, lung, prostate, breast, skin, and placenta [2; 3]. Prostasin has been shown to play many functional roles in epithelial physiology, including activation of the epithelial sodium channel (ENaC) [4–6], suppression of in vitro invasion [7; 8], maintenance of epidermal integrity [9], and regulation of inflammation-induced gene expression [10]. The promoter of the prostasin gene is positively regulated by sterol regulatory element binding proteins (SREBP’s) and negatively regulated by SNAI family transcription factors [11; 12]. Matriptase, a type-II transmembrane extracellular serine protease coordinately expressed with prostasin in normal tissues [13], activates the prostasin zymogen by a site-specific cleavage [14]. The active prostasin is regulated by serine protease inhibitors such as the hepatocyte growth factor activator inhibitor-1B (HAI-1B) [2] and protease nexin-1 (PN-1) [15].
Down-regulation of prostasin expression is associated with high-grade and hormone refractory prostate cancers [7; 16]. In an androgen-independent human prostate carcinoma cell line PC-3, prostasin expression is greatly compromised partly due to promoter DNA hypermethylation, while re-expression of prostasin reduced cell invasion through the Matrigel without affecting cell proliferation [7; 15]. In a recent report, we have shown that the expression of several invasion-promoting molecules is regulated by prostasin re-expression in the PC-3 cells. The gene expression regulation is potentially mediated by a protein-level down-regulation of the epidermal growth factor receptor (EGFR) [17]. As a result, the cellular response to EGF was reduced as shown by the down-regulation of EGF-stimulated phosphorylation of the extracellular signal-regulated kinases (Erk1/2). The expression of SLUG, urokinase-type plasminogen activator (uPA), uPA receptor (uPAR), inducible nitric oxide synthase (iNOS), cyclooxygenase-2 (COX-2), and granulocyte-macrophage colony stimulating factor (GM-CSF) was also down-regulated by prostasin in the PC-3 cells.
EGFR, a member of the ErbB (erythroblastic leukemia viral (v-erb-b) oncogene) family of receptor tyrosine kinases (RTK’s), plays pivotal roles in many diverse cellular responses ranging from proliferation to apoptosis, migration to adhesion, and differentiation to depolarization [18; 19]. Dys-regulation of EGFR-initiated cell signaling as a result of over-expression or constitutively activating mutations is causative to at least 10 different types of solid tumor [20–24]. The PC-3 prostate cancer cells express an abundance of EGFR and its ligands [25; 26], creating an autocrine signaling loop to confer the cells with highly invasive properties. Inhibition of EGFR signaling could be the mechanism of reduced invasiveness of PC-3 cells re-expressing prostasin [17], either through down-regulation of uPA-uPAR signaling [27; 28] or up-regulation of cell adhesion molecules such as the E-cadherin [29]. The PC-3 cell line is not dependent on EGFR signaling for proliferation [30], and the prostasin re-expression had no effect on cell growth [7]. Our observations in the PC-3 cells re-expressing prostasin were consistent with a mechanism of proteolytic modification of the EGFR in its extracellular domain (ECD) because prostasin is an active extracellular serine protease. We did not observe, however, a truncated EGFR fragment in the PC-3 cells with prostasin re-expression, potentially due to rapid internalization of the cleaved receptor. In this report, we present biochemical evidence that prostasin induces site-specific cleavages of the EGFR ECD following activation by matriptase in FT-293 cells. The amino-terminally truncated EGFR fragments were characterized for their response to EGF, tyrosine phosphorylation, and impact on cell signaling.
Materials and methods
Materials
A full-length human EGFR cDNA (Clone No. PR1116_D04, corresponding to GenBankTM sequence NM_005228) was purchased from OriGene Technologies, Inc. (Rockville, MD). A Flp-In T-REx derivative of the human embryonic kidney cell line HEK-293, designated FT-293, and a recombinant human EGF were purchased from Invitrogen Corporation (Carlsbad, CA). Polyclonal antibodies against human prostasin [1] and mouse PN-1 [15] were described previously. A polyclonal antibody to EGFR (sc-03), a monoclonal antibody to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (sc-32233), and a polyclonal antibody to Erk1/2 (sc-94) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). A polyclonal antibody to phospho-EGFR (Tyr1068, #2234) and a monoclonal antibody to the HA-Tag (6E2, #2367) were purchased from Cell Signaling Technology, Inc. (Danvers, MA). A monoclonal antibody to phospho-tyrosine (PY-20, #03-7799) was purchased from Zymed Laboratories, Inc. (South San Francisco, CA). Monoclonal antibodies to phospho-Akt (#550747) and Akt (#610860) were purchased from BD Biosciences (Franklin Lakes, NJ). A polyclonal antibody to phospho-Erk1/2 (V803A) was purchased from Promega Corporation (Madison, WI). Tyrphostin AG1478 (#658548) was purchased from EMD Chemicals, Inc. (San Diego, CA). The BEAS-2B normal human lung epithelial cells were purchased from the American Type Culture Collection (Manassas, VA). The BPH-1 human benign prostate hyperplasia cells were kindly provided by Dr. Simon W. Hayward of the Vanderbilt University Medical Center (Nashville, TN). The MDA-MB-231 human breast carcinoma cells were kindly provided by Dr. Janet E. Price of the MD Anderson Cancer Center (Houston, TX).
Construction of expression plasmids
The full-length human EGFR cDNA clone from OriGene was subcloned into the pcDNA3 plasmid (Invitrogen), generating the EGFR expression plasmid pcDNA3-EGFR. The EGFR ECD deletion mutant plasmid pcDNA3-EGFRΔ2-7 was generated by polymerase chain reaction (PCR) using the following primers: 5'-TTT CTT TTC CTC CAG AGC CCG ACT CGC C -3' and 5- AAT TAT GTG GTG ACA GAT CAC GGC TCG TGC GTC -3’. The pcDNA3-EGFRΔ2-7 encodes a mutant EGFR with a deletion of exons 2–7 (amino acid residues 6-272). The prostasin expression plasmids pcDNA3-Pro and pcDNA3-ProM have been described previously [10]. The plasmid pcDNA3-ProMG, coding for a GPI-anchor-free but protease-competent human prostasin, was generated by PCR, using the following primers: 5’-GCT CGA TAC AAT AAA CGC CA -3’ and 5’-GGA AGC TTC ACC TCA GCA AGC CCT GGG -3’. A pcDNA3-HA plasmid was generated by inserting into pcDNA3 a double-stranded oligonucleotide encoding a 9-amino acid residue segment of the Influenza A Virus Hemagglutinin (HA) tag (YPYDVPDYA). A full-length human matriptase cDNA [31] was subcloned into the pcDNA3-HA plasmid to generate a carboxyl-terminally HA-tagged matriptase expression plasmid pcDNA3-Mat-HA. A plasmid encoding an active-site mutant matriptase, pcDNA3-MatM-HA was generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The serine active site was changed to an alanine in pcDNA3-MatM-HA. A plasmid, pcDNA3-EGFR-HA, encoding a carboxyl-terminally HA-tagged EGFR was also created. All plasmids were verified by DNA sequencing.
Cell culture, transfection and western blot
The FT-293 cells were cultured in the D-MEM (High Glucose) medium supplemented with 10% (v/v) fetal bovine serum (FBS, Atlanta Biologicals, Inc., Lawrenceville, GA). For transfection experiments cells were plated on poly-L-lysine (PLL)-coated 12-well plates at a density of 4 × 105 cells per well. On the second day, appropriate expression plasmids (0.8μg of total DNA per transfection) were transfected into the cells using the Lipofectamine 2000 reagent (Invitrogen, 2μl per transfection) according to the manufacturer’s protocol. The BEAS-2B cells were cultured in BEBM (Lonza, Walkersville, MD), and set in 12-well plates at a density of 3 × 105 cells per well for transfections. The BPH-1 cells were cultured in RPMI-1640 medium supplemented with 5% FBS, and set in 6-well plates at a density of 5 × 105 cells per well for transfections. The MDA-MB-231 cells were cultured in MDA medium (MEM with 5% FBS, 1x sodium pyruvate, 1x non-essential amino acids, 1x glutamine, and 2x vitamins), and set in 6-well plates at a density of 1 × 106 cells per well for transfections. At 24 hours after transfection, cells were lysed in the RIPA lysis buffer [1] supplemented with a protease inhibitor cocktail. Twenty micrograms (for FT-293 and BEAS-2B transfectants) or forty micrograms (for BPH-1 and MDA-MB-231 transfectants) of total protein per sample were subjected to western blot analysis with appropriate antibodies. Each membrane was also blotted with a GAPDH antibody as a control of protein loading.
Deglycosylation of EGFR
Deglycosylation of the recombinant EGFR expressed in FT-293 cells and those cleaved by the proteases was carried out using an Enzymatic Protein Deglycosylation Kit (E-DEGLY, Sigma-Aldrich, St. Louis, MO), following the manufacturer’s protocol. Deglycosylated EGFR was then subjected to SDS-PAGE (7.5% gel) and western blot analysis with 30μg of total protein per sample.
PN-1 cell binding assay
FT-293 cells transfected with matriptase and prostasin expression plasmids were washed with 1x PBS once, scraped off in 1x PBS and collected by centrifugation. The cell pellet was then re-suspended in 100 μl of 25 mM Tris-HCl, pH 7.6, and incubated with 5 μl of mouse seminal vesicle fluid at 37° C for 2 hours as described previously [1]. Following the incubation, the cells were pelleted and lysed with 1% SDS in 25 mM Tris-HCl, pH 7.6 for western blot analysis with a prostasin antibody [1] and a PN-1 antibody [15].
EGF treatment of cells and analysis of EGF-EGFR signaling pathway activation
FT-293 cells transfected with EGFR, matriptase, and prostasin expression plasmids were serum-starved for 24 hours, treated with 10 ng/ml of EGF for 10 minutes, or left untreated, before western blot analysis for phospho-EGFR and total EGFR, phospho-Erk1/2 and total Erk1/2, or phospho-Akt and total Akt. Each membrane was also blotted with a GAPDH antibody as a control of protein loading. For EGFR phosphorylation inhibition assays, FT-293 cells were treated with AG1478 (2 μM), or DMSO (solvent control) during transfection, serum-starvation, and EGF stimulation.
Results
Prostasin activated by matriptase induces cleavages of EGFR in FT-293 cells
In the previous study [17], a proteolytic mechanism of EGFR regulation by prostasin was suggested from the observation that re-expression of prostasin down-regulated EGFR protein expression in the PC-3 cells. To determine whether EGFR is a biological substrate of prostasin, we co-expressed EGFR, prostasin and an HA-tagged matriptase in the FT-293 cells. The FT-293 cells were derived from the human embryonic kidney cell line HEK-293 which does not express significant levels of EGFR [32; 33], prostasin [1] or matriptase (data not shown). First, we evaluated activation of prostasin co-expressed with matriptase (Mat-HA, HA-tagged, same hereon) by performing a binding assay with PN-1. The PN-1 binding assay is a reliable method for evaluating prostasin’s serine protease activation state in vitro and in vivo [1; 14; 15]. When expressed without matriptase, prostasin remained in the zymogen form and no prostasin-PN-1 complex was detected in the binding assay, indicating a lack of serine protease activity (Figure 1, Lane 2). When co-expressed with matriptase, prostasin was shown to be cleaved, apparently by matriptase, to produce a lower molecular weight prostasin, and at the same time a prostasin-PN-1 complex (Figure 1, Lane 5). The cleaved prostasin is active as shown by its ability to form the covalent complex with its cognate serpin PN-1. In the prostasin and the PN-1 western blots, the upper band indicated by an asterisk is the prostasin-PN-1 complex; other high-molecular weight bands are unidentified. The serine active-site mutant prostasin was also cleaved by matriptase but no prostasin-PN-1 complex was observed (Figure 1, Lane 6). The serine active-site mutant matriptase, expectedly, was unable to cleave and activate the prostasin zymogen (Figure 1, Lanes 8–9). We also performed western blots to evaluate the expression of Mat-HA and MatM-HA. An active form and the zymogen form were observed for Mat-HA but only the zymogen form was seen for MatM-HA (data not shown), as expected from previous results [31].
Figure 1. Prostasin is activated by matriptase in FT-293 cells.
FT-293 cells were transfected with cDNA’s encoding the wild-type prostasin (Pro, 0.5 μg), a serine active-site mutant prostasin (ProM, 0.5 μg), an HA-tagged matriptase (Mat-HA, 0.3 μg) or an HA-tagged serine active-site mutant matriptase (MatM-HA, 0.3 μg), or in combinations as indicated in the figure. Each transfection was carried out with an equal amount of total plasmid DNA (0.8 μg) using the vector plasmid pcDNA3 (Vec) as a substitute when appropriate. At 24 hours post transfection, cells were subjected to PN-1 binding assays with mouse seminal vesicle fluid followed by western blot analysis for prostasin and PN-1. Gel electrophoresis was performed with SDS under boiling and reducing conditions. The positions of the covalent prostasin-PN-1 complex (82 kDa), prostasin zymogen (40 kDa), and prostasin (38 kDa) are indicated by arrows. The specific prostasin-PN-1 complex band is indicated by the asterisk. The results shown are representative of three independent experiments. WB: western blot.
Next we examined the EGFR protein co-expressed with matriptase and prostasin by western blot analysis using a polyclonal antibody that specifically recognizes a carboxyl-terminal intracellular epitope of the human EGFR. Because matriptase and prostasin are both extracellular proteases, any proteolytic action on the EGFR protein would result in amino-terminal ECD truncation on the outside of the cell. The truncated fragments would be detectable by an antibody against the carboxyl-terminal region of the receptor. Two modified EGFR fragments were produced when matriptase was co-expressed (Figure 2A, Middle Panel, Long Exposure, Lane 4), detected at approximately 135 kDa and 110 kDa (named EGFR135 and EGFR110). Quantities of these truncated EGFR, EGFR135 and EGFR110, were greatly increased when the wild-type prostasin was co-expressed with EGFR and matriptase (Figure 2A, Lane 5). There was no increase in quantities of EGFR135 and EGFR110 when the serine active-site mutant prostasin was co-expressed with EGFR and matriptase (Figure 2A, Lane 6). The amount of uncut full-length EGFR at 170 kDa was reduced with matriptase co-expression, and to a much greater extent with the addition of the wild-type prostasin (Figure 2A, Upper Panel, Short Exposure, Lanes 4 and 5). The serine active-site mutant matriptase (MatM-HA) was unable to induce any cleavage of EGFR, with or without prostasin (Figure 2A, Lanes 7–9).
Figure 2. Prostasin and matriptase induce EGFR ECD cleavages in FT-293 cells.
(A and B) FT-293 cells were transfected with cDNA’s encoding EGFR, wild-type prostasin (Pro, 0.2 μg), serine active-site mutant prostasin (ProM, 0.2 μg), anchor-free mutant prostasin (ProMG, 0.2 μg), HA-tagged matriptase (Mat-HA, 0.1 μg) or HA-tagged serine active-site mutant matriptase (MatM-HA, 0.1 μg), or in combinations as indicated in the figure. Each sample was transfected with an equal amount of the EGFR plasmid (0.5 μg) and an equal amount of total plasmid DNA (0.8 μg) using the vector plasmid pcDNA3 (Vec) as a substitute when appropriate. At 24 hours post transfection, cells were assayed for EGFR protein expression by SDS-PAGE and western blot analysis with an anti-EGFR antibody that recognizes a carboxyl-terminal intracellular epitope of EGFR. The membrane was re-blotted with a GAPDH antibody as a sample loading control. The results shown are representative of three independent experiments. (C) Deglycosylation (DeGly) of EGFR and protease-cleaved EGFR was performed before western blot analysis with an EGFR antibody to show primary structure modifications of EGFR.
To determine whether the membrane anchorage of prostasin is required for inducing EGFR cleavage, we generated a GPI-anchor-free but protease-competent prostasin for co-expression with EGFR and matriptase. The GPI-anchor-free prostasin was secreted into the culture medium and undetectable in the cell lysate (data not shown), confirming previous findings on this type of prostasin mutant [34]. The GPI-anchor-free prostasin produced a similar pattern of EGFR cleavage as that seen with the wild-type prostasin when co-expressed with matriptase (Figure 2B, Lane 6), suggesting that the GPI membrane anchorage is not required for the active prostasin-induced EGFR cleavage.
To determine if the EGFR135 and EGFR110 fragments were the products of primary structure modification, or other potential processes, such as differential glycosylation, we performed an enzymatic deglycosylation experiment for EGFR co-expressed with matriptase-prostasin. As shown in Figure 2C, the deglycosylation step resulted in expected shifts of the apparent molecular mass for the full-length EGFR (from 170 kDa to 140 kDa), EGFR135 (to 100 kDa), and EGFR110 (to 80 kDa). Three distinct EGFR protein molecule or fragments can still be detected in the western blot after deglycosylation, ruling out differential glycosylation as a mechanism for producing EGFR135 and EGFR110 and supporting an amino-terminal ECD truncation mechanism.
EGFR110 is constitutively active
To evaluate the impact of the protease-induced EGFR ECD cleavages on cell signaling, FT-293 cells presenting uncleaved or cleaved EGFR were assayed for changes in phosphorylation states of EGFR and downstream signaling molecules Erk1/2 and Akt. The full-length EGFR (170 kDa) remained in the unphosphorylated state under the serum-free culture condition and became highly tyrosine-phosphorylated once stimulated with EGF (Figure 3A). The EGFR110 fragment was shown to be tyrosine-phosphorylated under the serum-free culture condition, i.e., in the absence of EGF (Figure 3A, Lane 2), but was not responsive to EGF stimulation (Figure 3A, Lane 4). There was an apparent reduction of tyrosine-phosphorylated EGFR110 in the EGF-treated sample. The EGFR135 fragment did not present detectable tyrosine phosphorylation under either the serum-free or the EGF-stimulated condition. Expression of EGFRΔ2–7, corresponding to the constitutively active mutant EGFRvIII [35], led to increased Erk1/2 phosphorylation under the serum-free culture condition when compared with the cells expressing the wild-type EGFR (Figure 3B, Upper Panels, Lanes 1 and 2). In cells where EGFR was co-transfected with prostasin and matriptase (i.e., presenting the tyrosine-phosphorylated EGFR110), Erk1/2 phosphorylation was increased to a similar extent as the cells expressing EGFRΔ2–7 (Figure 3B, Upper Panels, Lane 3). On the other hand, Akt phosphorylation was not affected in these cells (Figure 3B, Lower Panels).
Figure 3. EGFR110 is constitutively active and activates Erk1/2.
(A) FT-293 cells were transfected with cDNA’s encoding EGFR (0.05 μg for Lanes 1 and 3; 0.2 μg for Lanes 2 and 4), wild-type prostasin (P, 0.2 μg), or HA-tagged matriptase (M, 0.1 μg), or in combinations as indicated in the figure. Each transfection was carried out with an equal amount of total plasmid DNA (0.8 μg) using the vector plasmid pcDNA3 as a substitute when appropriate. At 24 hours after transfection, cells were placed under serum-free medium for overnight and treated with 10 ng/ml EGF for 10 minutes (Lanes 3 and 4) or left untreated (Lanes 1 and 2) before western blot analysis for total EGFR protein expression and EGFR tyrosine phosphorylation (pTyr and pEGFR/pY1068). The membrane was re-blotted with a GAPDH antibody as a sample loading control. The results shown are representative of three independent experiments. (B) FT-293 cells were transfected with cDNA’s encoding EGFRΔ2-7 or EGFR (0.3 μg), with or without wild-type prostasin (P, 0.2 μg) and HA-tagged matriptase (M, 0.1 μg) as indicated in the figure. Each transfection was carried out with an equal amount of total plasmid DNA (0.8 μg) using the vector plasmid pcDNA3 as a substitute when appropriate. At 24 hours after transfection, cells were placed under serum-free medium for 24 hours before western blot analysis for phosphorylated Erk1/2 (pErk) and Akt (pAkt), Total Erk, and Total Akt. The results shown are representative of three independent experiments.
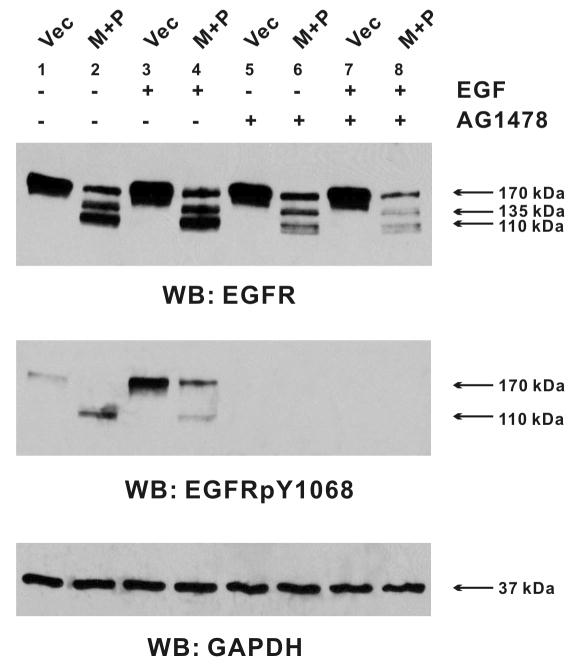
Protease-induced EGFR cleavages are not dependent on tyrosine phosphorylation
When EGFR was over-expressed in the FT-293 cells, a small portion of the receptor appeared tyrosine-phosphorylated (Figure 4, Lane 1). To determine whether this auto-phosphorylation of EGFR is required for the protease-induced cleavage, we performed co-transfection experiments in the presence of AG1478, a potent inhibitor of EGFR kinase activity and auto-phosphorylation. Incubation with AG1478 completely abolished not only the auto-phosphorylation and EGF-stimulated phosphorylation of the wild-type EGFR, but also the cleavage-induced phosphorylation of EGFR110 (Figure 4, Middle Panel, Lanes 5-8). On the other hand, the proteases induced the specific EGFR cleavages in the presence of AG1478 (Figure 4, Upper Panel, Lane 5–8), suggesting that the protease-induced EGFR cleavages are independent of receptor tyrosine phosphorylation.
Figure 4. Protease-induced EGFR cleavages are independent of EGFR phosphorylation.
FT-293 cells were treated with AG1478 (2 μM, Lanes 5-8), or with DMSO (solvent control of AG1478, Lanes 1-4) during transfection, serum-starvation, and EGF stimulation. Cells were transfected with cDNA’s encoding EGFR, wild-type prostasin (P, 0.2 μg), or HA-tagged matriptase (M, 0.1 μg), or in combinations as indicated in the figure. Each sample was transfected with an equal amount of the EGFR plasmid (0.3 μg) and an equal amount of total plasmid DNA (0.8 μg) using the vector plasmid pcDNA3 (Vec) as a substitute when appropriate. At 24 hours after transfection, cells were placed under serum-free medium for overnight and treated with 10 ng/ml of EGF for 10 minutes (Lanes 3, 4, 7 and 8) or left untreated (Lanes 1, 2, 5 and 6) before western blot analysis for EGFR and phosphorylated EGFR (EGFRpY1068). The results shown are representative of three independent experiments.
Matriptase and prostasin induce EGFR ECD cleavages in BEAS-2B, BPH-1, and MDA-MB-231 cells
To determine if the protease-induced EGFR cleavages occur in other cell lines, we performed EGFR/protease co-transfection experiments in normal human lung epithelial cells BEAS-2B, the human benign prostate hyperplasia cell line BPH-1, and the human breast carcinoma cell line MDA-MB-231. As shown in Figure 5, the actions of the matriptase and prostasin serine protease cascade produced the same EGFR ECD cleavages observed in the FT-293 cell line, generating the EGFR135 and EGFR110 fragments in BPH-1 and MDA-MB-231. The EGFR135 fragment was not detected in the BEAS-2B cells under the experiment conditions but the EGFR110 fragment was clearly present in these cells.
Figure 5. Matriptase and prostasin induce EGFR ECD cleavages in BEAS-2B, BPH-1, and MDA-MB-231 cells.
BEAS-2B, BPH-1, and MDA-MB-231 cells were transfected with cDNA’s encoding EGFR-HA (0.6 μg), wild-type prostasin (Pro, 0.4 μg), and wild-type matriptase (Mat, 0.2 μg) in combinations as indicated in the figure. Each sample was transfected with an equal amount of total plasmid DNA (1.2 μg) using the vector plasmid pcDNA3 (Vec) as a substitute when appropriate. At 24 hours post transfection, cells were assayed for EGFR-HA protein expression by SDS-PAGE and western blot analysis with an anti-HA antibody (upper panels). The membrane was re-blotted with a GAPDH antibody as a sample loading control (lower panels). The results shown are representative of three independent experiments.
Discussion
Prostasin as a GPI-anchored protein is routed to the apical membrane of the normally polarized epithelial cells [1]. EGFR and matriptase are normally routed to the basolateral sides of the cells [36; 37]. Topologically distinct but coordinate expression of matriptase and prostasin is observed in many terminally differentiated epithelial tissues, implicating coordinate roles for these two serine proteases in normal epithelial physiology [13]. But during epithelial carcinogenesis expression of matriptase and prostasin begins to show divergent patterns of change, implicating diverging and independent roles for the two serine proteases in cancer. In the prostate and the breast, prostasin is abundantly expressed in the normal tissue, but down-regulated in cancers [7; 8]. An exception may be the ovarian cancer, which is marked by an up-regulation of prostasin [38]. For matriptase, its expression is up-regulated in prostate and breast cancers [39; 40], and its mechanistic role in cancer is believed to be activating the ligands of the Ron signaling pathway [41]. In terminally differentiated epithelial tissues, EGFR is not expressed in abundance, but over-expression of EGFR is commonly associated with cancer [42–44]. In the normal tissue, prostasin and matriptase are controlled by topological separation on the plasma membrane, if not also by differential expression, i.e., a high abundance of prostasin and a low abundance of matriptase. We may view prostasin as a sensor or surveillance agent for epithelial polarity and integrity. Once an insult, e.g., inflammation, results in injury of the normal epithelium, depolarization ensues and injury repair programs are mobilized. EGFR is a key player in epithelial injury repair [45] but at some point the cells need to initiate a “stop program” of EGFR signaling to allow epithelial differentiation and re-polarization. The executor of the “stop program” appears to be prostasin or related proteases, activated or re-expressed to initiate terminal differentiation. Prostasin’s role in this regard is in complete agreement with the observation that in prostasin-knocked-out mouse skin, terminal differentiation of the skin epithelium is defective, marked by lack of tight junction formation and absence of occludin expression [9]. Also in agreement with this model is the observation that prostasin expression is down-regulated during inflammation while forced prostasin expression attenuates inflammation-induced gene expression [10]. Silencing of prostasin by epigenetic events or growth factors and cytokines [8; 10; 11; 15], would be a pre-requisite for tumorigenesis and gain of aggressive properties such as invasion and metastasis.
It is apparently paradoxical for prostasin to down-regulate EGF-EGFR signaling and suppress invasion in some cells, but produce an autoactivated EGFR110 in others with up-regulation of downstream signals. We reason that the overall cellular impact of the protease-induced EGFR cleavage is dependent on the rate of internalization for the cleaved receptor. In cells with normal or even accelerated turnover rate, such as the PC-3 [46], the net effect is inhibition of EGF-EGFR signaling due to reduced cell surface receptor presentation. We predict for cancer cells that are EGFR-dependent this will be the predominant phenotype as in these cells the EGF-EGFR signaling is tightly coupled with receptor internalization [47]. In these cells, the rapid receptor internalization could also have prevented our detection of the cleaved forms of EGFR in our previous study [17]. In cells with impeded receptor internalization, such as the HEK-293 and its derivatives (the FT-293) [48], the net effect is a constitutively activated truncated EGFR remaining on the membrane, activating the downstream signals. Cells that are EGFR-over-expressing but are not EGFR-dependent would present this phenotype.
The HA-tagged matriptase was confirmed for its ability to activate prostasin in the FT-293 cells when the two proteases were co-expressed from their full-length cDNA’s (Figure 1). Matriptase is also capable of cleaving the EGFR at apparently the same sites cleaved by prostasin, but appeared to be a less effective enzyme in this role (Figure 2A, compare Lanes 4 and 5). Matriptase action on EGFR could be amplified, however, if an abundance of matriptase is expressed, as was seen in PC-3 cells transfected with the serine active-site mutant prostasin [17]. In these cells, we now know that the mutant prostasin is incapable of cleaving EGFR, but the robustly induced matriptase certainly could. We attribute the EGFR ECD cleavages to matriptase and prostasin but we do not have evidence for a direct interaction between matriptase and EGFR, or between prostasin and EGFR. The molecular landscape of the membrane-anchored or membrane-associated serine proteases is still expanding. We are limited by our knowledge of this new family of proteases to determine whether the EGFR ECD is potentially cleaved by a downstream protease activated by prostasin. We did not observe a significant induction of matriptase expression in the FT-293 cells expressing prostasin (data not shown), ruling out a solo matriptase action on EGFR when active prostasin is also present.
The EGFR ECD contains the ligand-binding domains that form the target of the monoclonal antibody (Mab) drug ImClone C225/cetuximab/Erbitux [49], approved for advanced head-and-neck and colorectal cancers as a third-line treatment option [50; 51]. The matriptase → prostasin → EGFR cascade can play a critical role in anti-EGFR therapies for cancer using Mab drugs targeting the ECD. The cleaved EGFR, EGFR135 and EGFR110, are no longer responsive to EGF stimulation (Figure 3A), presumably due to the loss of the ligand-binding domains, all or partial. By this reasoning, Erbitux or similar Mab drugs targeting the ligand-binding domains of EGFR would no longer be effective. If the cleaved EGFR is retained on the membrane and activates downstream signaling, it will likely result in lower cell sensitivity to the Mab drugs, i.e., requiring higher doses for growth inhibition. In this scenario, protease inhibitors specific for the matriptase-prostasin cascade may be considered as an appropriate adjuvant. If the proteolytic cleavage of EGFR results in its rapid internalization and turnover [17], drug sensitivity to the Mab’s should increase, i.e., showing growth inhibition at a lower dose. In this scenario, prostasin, pre-activated or activated upon reaching the cancer cells by their endogenously over-expressed matriptase or related proteases, may be used as an adjuvant. We have shown that the role of prostasin in EGFR ECD cleavage is not dependent on its membrane anchorage via the GPI (Figure 2B), making it a suitable candidate for development of a systemically deliverable agent to treat certain cancers that are dependent on over-expressed EGFR for growth and survival signals.
Conclusions
We have identified EGFR as a biological substrate for the epithelial extracellular serine protease activation cascade involving matriptase and prostasin. The protease-induced EGFR ECD cleavage activates the receptor tyrosine kinase but the impact on cell signaling varies between different cell types, probably due to differences in receptor internalization rate. The novel protease-activated EGFR signal modulation mechanism may have clinical implications in therapies for treating cancers by targeting the EGFR ECD.
Acknowledgments
This work was supported by DOD Grants DAMD17-02-1-0032, DAMD17-02-1-0338, and Susan G. Komen for the Cure Grant BCTR0707538 (to K. X. C.), by Florida Biomedical Research Program Grant 06NIR-03 (to L-M. C.), and by NIH Grants R01-CA-104944 and R01-CA-096851 (to C.-Y. L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen LM, Skinner ML, Kauffman SW, Chao J, Chao L, Thaler CD, Chai KX. Prostasin is a glycosylphosphatidylinositol-anchored active serine protease. J Biol Chem. 2001;276:21434–21442. doi: 10.1074/jbc.M011423200. [DOI] [PubMed] [Google Scholar]
- 2.Fan B, Wu TD, Li W, Kirchhofer D. Identification of hepatocyte growth factor activator inhibitor-1B as a potential physiological inhibitor of prostasin. J Biol Chem. 2005;280:34513–34520. doi: 10.1074/jbc.M502119200. [DOI] [PubMed] [Google Scholar]
- 3.Yu JX, Chao L, Chao J. Molecular cloning, tissue-specific expression, and cellular localization of human prostasin mRNA. J Biol Chem. 1995;270:13483–13489. doi: 10.1074/jbc.270.22.13483. [DOI] [PubMed] [Google Scholar]
- 4.Adachi M, Kitamura K, Miyoshi T, Narikiyo T, Iwashita K, Shiraishi N, Nonoguchi H, Tomita K. Activation of epithelial sodium channels by prostasin in Xenopus oocytes. J Am Soc Nephrol. 2001;12:1114–1121. doi: 10.1681/ASN.V1261114. [DOI] [PubMed] [Google Scholar]
- 5.Vallet V, Chraibi A, Gaeggeler HP, Horisberger JD, Rossier BC. An epithelial serine protease activates the amiloride-sensitive sodium channel. Nature. 1997;389:607–610. doi: 10.1038/39329. [DOI] [PubMed] [Google Scholar]
- 6.Vuagniaux G, Vallet V, Jaeger NF, Pfister C, Bens M, Farman N, Courtois-Coutry N, Vandewalle A, Rossier BC, Hummler E. Activation of the amiloride-sensitive epithelial sodium channel by the serine protease mCAP1 expressed in a mouse cortical collecting duct cell line. J Am Soc Nephrol. 2000;11:828–834. doi: 10.1681/ASN.V115828. [DOI] [PubMed] [Google Scholar]
- 7.Chen LM, Hodge GB, Guarda LA, Welch JL, Greenberg NM, Chai KX. Down-regulation of prostasin serine protease: a potential invasion suppressor in prostate cancer. Prostate. 2001;48:93–103. doi: 10.1002/pros.1085. [DOI] [PubMed] [Google Scholar]
- 8.Chen LM, Chai KX. Prostasin serine protease inhibits breast cancer invasiveness and is transcriptionally regulated by promoter DNA methylation. Int J Cancer. 2002;97:323–329. doi: 10.1002/ijc.1601. [DOI] [PubMed] [Google Scholar]
- 9.Leyvraz C, Charles RP, Rubera I, Guitard M, Rotman S, Breiden B, Sandhoff K, Hummler E. The epidermal barrier function is dependent on the serine protease CAP1/Prss8. J Cell Biol. 2005;170:487–496. doi: 10.1083/jcb.200501038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LM, Wang C, Chen M, Marcello MR, Chao J, Chao L, Chai KX. Prostasin attenuates inducible nitric oxide synthase expression in lipopolysaccharide-induced urinary bladder inflammation. Am J Physiol Renal Physiol. 2006;291:F567–F577. doi: 10.1152/ajprenal.00047.2006. [DOI] [PubMed] [Google Scholar]
- 11.Chen M, Chen LM, Chai KX. Androgen regulation of prostasin gene expression is mediated by sterol-regulatory element-binding proteins and SLUG. Prostate. 2006 doi: 10.1002/pros.20325. [DOI] [PubMed] [Google Scholar]
- 12.Chen M, Chen LM, Chai KX. Mechanisms of sterol regulatory element-binding protein-2 (SREBP-2) regulation of human prostasin gene expression. Biochem Biophys Res Commun. 2006;346:1245–1253. doi: 10.1016/j.bbrc.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 13.List K, Hobson JP, Molinolo A, Bugge TH. Co-localization of the channel activating protease prostasin/(CAP1/PRSS8) with its candidate activator, matriptase. J Cell Physiol. 2007 doi: 10.1002/jcp.21115. [DOI] [PubMed] [Google Scholar]
- 14.Netzel-Arnett S, Currie BM, Szabo R, Lin CY, Chen LM, Chai KX, Antalis TM, Bugge TH, List K. Evidence for a matriptase-prostasin proteolytic cascade regulating terminal epidermal differentiation. J Biol Chem. 2006;281:32941–32945. doi: 10.1074/jbc.C600208200. [DOI] [PubMed] [Google Scholar]
- 15.Chen LM, Zhang X, Chai KX. Regulation of prostasin expression and function in the prostate. Prostate. 2004;59:1–12. doi: 10.1002/pros.10346. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi S, Suzuki S, Inaguma S, Ikeda Y, Cho YM, Hayashi N, Inoue T, Sugimura Y, Nishiyama N, Fujita T, Chao J, Ushijima T, Shirai T. Down-regulated expression of prostasin in high-grade or hormone-refractory human prostate cancers. Prostate. 2003;54:187–193. doi: 10.1002/pros.10178. [DOI] [PubMed] [Google Scholar]
- 17.Chen M, Fu YY, Lin CY, Chen LM, Chai KX. Prostasin induces protease-dependent and independent molecular changes in the human prostate carcinoma cell line PC-3. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamcr.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 19.Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31:637–643. doi: 10.1016/s1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]
- 20.Bellezza I, Bracarda S, Caserta C, Minelli A. Targeting of EGFR tyrosine kinase by ZD1839 (“Iressa”) in androgen-responsive prostate cancer in vitro. Mol Genet Metab. 2006;88:114–122. doi: 10.1016/j.ymgme.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Lo HW, Hsu SC, Hung MC. EGFR signaling pathway in breast cancers: from traditional signal transduction to direct nuclear translocalization. Breast Cancer Res Treat. 2006;95:211–218. doi: 10.1007/s10549-005-9011-0. [DOI] [PubMed] [Google Scholar]
- 22.Normanno N, De LA, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De FG, Caponigro F, Salomon DS. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed SM, Salgia R. Epidermal growth factor receptor mutations and susceptibility to targeted therapy in lung cancer. Respirology. 2006;11:687–692. doi: 10.1111/j.1440-1843.2006.00887.x. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 25.Morris GL, Dodd JG. Epidermal growth factor receptor mRNA levels in human prostatic tumors and cell lines. J Urol. 1990;143:1272–1274. doi: 10.1016/s0022-5347(17)40253-9. [DOI] [PubMed] [Google Scholar]
- 26.Ching KZ, Ramsey E, Pettigrew N, D'Cunha R, Jason M, Dodd JG. Expression of mRNA for epidermal growth factor, transforming growth factor-alpha and their receptor in human prostate tissue and cell lines. Mol Cell Biochem. 1993;126:151–158. doi: 10.1007/BF00925693. [DOI] [PubMed] [Google Scholar]
- 27.Skogseth H, Larsson E, Halgunset J. The invasive behaviour of prostatic cancer cells is suppressed by inhibitors of tyrosine kinase. APMIS. 2006;114:61–66. doi: 10.1111/j.1600-0463.2006.apm_230.x. [DOI] [PubMed] [Google Scholar]
- 28.Skogseth H, Larsson E, Halgunset J. Inhibitors of tyrosine kinase inhibit the production of urokinase plasminogen activator in human prostatic cancer cells. APMIS. 2005;113:332–339. doi: 10.1111/j.1600-0463.2005.apm_113504.x. [DOI] [PubMed] [Google Scholar]
- 29.Yates CC, Shepard CR, Stolz DB, Wells A. Co-culturing human prostate carcinoma cells with hepatocytes leads to increased expression of E-cadherin. Br J Cancer. 2007;96:1246–1252. doi: 10.1038/sj.bjc.6603700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Sheikh SS, Domin J, Abel P, Stamp G, Lalani e. Phosphorylation of both EGFR and ErbB2 is a reliable predictor of prostate cancer cell proliferation in response to EGF. Neoplasia. 2004;6:846–853. doi: 10.1593/neo.04379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberst MD, Chen LY, Kiyomiya K, Williams CA, Lee MS, Johnson MD, Dickson RB, Lin CY. HAI-1 regulates activation and expression of matriptase, a membrane-bound serine protease. Am J Physiol Cell Physiol. 2005;289:C462–C470. doi: 10.1152/ajpcell.00076.2005. [DOI] [PubMed] [Google Scholar]
- 32.Jones SM, Foreman SK, Shank BB, Kurten RC. EGF receptor downregulation depends on a trafficking motif in the distal tyrosine kinase domain. Am J Physiol Cell Physiol. 2002;282:C420–C433. doi: 10.1152/ajpcell.00253.2001. [DOI] [PubMed] [Google Scholar]
- 33.Stern KA, Visser Smit GD, Place TL, Winistorfer S, Piper RC, Lill NL. Epidermal growth factor receptor fate is controlled by Hrs tyrosine phosphorylation sites that regulate Hrs degradation. Mol Cell Biol. 2007;27:888–898. doi: 10.1128/MCB.02356-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallet V, Pfister C, Loffing J, Rossier BC. Cell-surface expression of the channel activating protease xCAP-1 is required for activation of ENaC in the Xenopus oocyte. J Am Soc Nephrol. 2002;13:588–594. doi: 10.1681/ASN.V133588. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Yuan M, Kim IA, Chang CM, Bernhard EJ, Shu HK. Mutant epidermal growth factor receptor displays increased signaling through the phosphatidylinositol-3 kinase/AKT pathway and promotes radioresistance in cells of astrocytic origin. Oncogene. 2004;23:4594–4602. doi: 10.1038/sj.onc.1207602. [DOI] [PubMed] [Google Scholar]
- 36.Hobert M, Carlin C. Cytoplasmic juxtamembrane domain of the human EGF receptor is required for basolateral localization in MDCK cells. J Cell Physiol. 1995;162:434–446. doi: 10.1002/jcp.1041620316. [DOI] [PubMed] [Google Scholar]
- 37.List K, Hobson JP, Molinolo A, Bugge TH. Co-localization of the channel activating protease prostasin/(CAP1/PRSS8) with its candidate activator, matriptase. J Cell Physiol. 2007 doi: 10.1002/jcp.21115. [DOI] [PubMed] [Google Scholar]
- 38.Mok SC, Chao J, Skates S, Wong K, Yiu GK, Muto MG, Berkowitz RS, Cramer DW. Prostasin, a potential serum marker for ovarian cancer: identification through microarray technology. J Natl Cancer Inst. 2001;93:1458–1464. doi: 10.1093/jnci/93.19.1458. [DOI] [PubMed] [Google Scholar]
- 39.Jin JS, Cheng TF, Tsai WC, Sheu LF, Chiang H, Yu CP. Expression of the serine protease, matriptase, in breast ductal carcinoma of Chinese women: correlation with clinicopathological parameters. Histol Histopathol. 2007;22:305–309. doi: 10.14670/HH-22.305. [DOI] [PubMed] [Google Scholar]
- 40.Kang JY, Dolled-Filhart M, Ocal IT, Singh B, Lin CY, Dickson RB, Rimm DL, Camp RL. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res. 2003;63:1101–1105. [PubMed] [Google Scholar]
- 41.Welm AL, Sneddon JB, Taylor C, Nuyten DS, dvanV, Hasegawa BH, Bishop JM. The macrophage-stimulating protein pathway promotes metastasis in a mouse model for breast cancer and predicts poor prognosis in humans. Proc Natl Acad Sci U S A. 2007;104:7570–7575. doi: 10.1073/pnas.0702095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rusnak DW, Alligood KJ, Mullin RJ, Spehar GM, renas-Elliott C, Martin AM, Degenhardt Y, Rudolph SK, Haws TF, Jr, Hudson-Curtis BL, Gilmer TM. Assessment of epidermal growth factor receptor (EGFR, ErbB1) and HER2 (ErbB2) protein expression levels and response to lapatinib (Tykerb, GW572016) in an expanded panel of human normal and tumour cell lines. Cell Prolif. 2007;40:580–594. doi: 10.1111/j.1365-2184.2007.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rotterud R, Fossa SD, Nesland JM. Protein networking in bladder cancer: immunoreactivity for FGFR3, EGFR, ERBB2, KAI1, PTEN, and RAS in normal and malignant urothelium. Histol Histopathol. 2007;22:349–363. doi: 10.14670/HH-22.349. [DOI] [PubMed] [Google Scholar]
- 44.Christensen ME. The EGF receptor system in head and neck carcinomas and normal tissues. Immunohistochemical and quantitative studies. Dan Med Bull. 1998;45:121–134. [PubMed] [Google Scholar]
- 45.Repertinger SK, Campagnaro E, Fuhrman J, El-Abaseri T, Yuspa SH, Hansen LA. EGFR enhances early healing after cutaneous incisional wounding. J Invest Dermatol. 2004;123:982–989. doi: 10.1111/j.0022-202X.2004.23478.x. [DOI] [PubMed] [Google Scholar]
- 46.Bonaccorsi L, Nosi D, Muratori M, Formigli L, Forti G, Baldi E. Altered endocytosis of epidermal growth factor receptor in androgen receptor positive prostate cancer cell lines. J Mol Endocrinol. 2007;38:51–66. doi: 10.1677/jme.1.02155. [DOI] [PubMed] [Google Scholar]
- 47.Carpenter G. The EGF receptor: a nexus for trafficking and signaling. Bioessays. 2000;22:697–707. doi: 10.1002/1521-1878(200008)22:8<697::AID-BIES3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 48.Johannessen LE, Haugen KE, ostvold AC, Stang E, Madshus IH. Heterodimerization of the epidermal-growth-factor (EGF) receptor and ErbB2 and the affinity of EGF binding are regulated by different mechanisms. Biochem J. 2001;356:87–96. doi: 10.1042/0264-6021:3560087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baselga J. The EGFR as a target for anticancer therapy--focus on cetuximab. Eur J Cancer. 2001;37(Suppl 4):S16–S22. doi: 10.1016/s0959-8049(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 50.Hildebrandt B, le CP, Nicolaou A, Koble K, Riess H, Dorken B. Cetuximab: appraisal of a novel drug against colorectal cancer. Recent Results Cancer Res. 2007;176:135–143. doi: 10.1007/978-3-540-46091-6_11. [DOI] [PubMed] [Google Scholar]
- 51.Merlano M, Garrone O. Treatment of advanced head and neck cancer with cetuximab. Int J Biol Markers. 2007;22:S71–S76. [PubMed] [Google Scholar]