Abstract
Membrane-associated serine protease matriptase has been implicated in human diseases, and might be a drug target. In the present study, a novel class of matriptase inhibitors targeting zymogen activation is developed by a combination of the screening of compound library using a cell-based matriptase activation assay and a computer-aided search of commercially available analogs of a selected compound. Four structurally related compounds are identified that can inhibit matriptase activation with IC50 at low μM in both intact-cell and cell-free systems, suggesting that these inhibitors target the matriptase autoactivation machinery rather than the intracellular signaling pathways. These activation inhibitors can also inhibit prostasin activation, a downstream event that occurs in lockstep with matriptase activation. In contrast, the matriptase catalytic inhibitor CVS-3983 at a concentration 300-fold higher than its Ki fails to inhibit activation of either protease. Our results suggest that inhibiting matriptase activation is an efficient way to control matriptase function.
Introduction
Inappropriate proteolysis has been found to be critical component of various pathological processes, such as carcinogenesis, metastasis, inflammation, hypertension, skin disease, and osteoarthritis. Proteolytic activities are, therefore, considered to be promising targets for drug development1. Matriptase, a type II transmembrane serine protease2, has been implicated in many disease processes, and this enzyme may be an attractive drug target for treating various human diseases. Increased matriptase expression and an imbalance between matriptase and its endogenous inhibitor hepatocyte growth factor activator inhibitor 1 (HAI-1), a Kunitz-type serine protease inhibitor, are commonly observed in a wide variety of primary human carcinomas3, 4. In some cases, this dysregulation is associated with poor patient outcome5–7. Evidence for the oncogenic potential and a pro-metastatic role of matriptase has been provided by studies using animal models, including matriptase transgenic mice and tumor xenograft studies in nude mice8, 9. Increased matriptase zymogen activation has also been seen in several distinct human skin diseases, and keratinocytes exhibiting increased matriptase activation have been observed in close proximity to areas of inflammation10. In addition to expression in epithelial and carcinoma cells, matriptase is also expressed by chondrocytes11, monocytes12–14, and mast cells15. Matriptase activity in chondrocytes may play an important role in the degradation of cartilage matrix and contribute to osteoarthritis11. In atherosclerotic lesions, monocytic matriptase may stimulate endothelial cells to release pro-inflammatory cytokines and thereby contribute to the disease14. Expression of matriptase in mast cells that play important roles in allergy-related diseases, such as asthma, suggests that the membrane protease may have the potential to contribute to these diseases as well15.
Traditionally, strategies to control protease activity have been targeted directly against the proteolytic mechanism of the enzymes using synthetic inhibitors. Several catalytic inhibitors of matriptase have been developed, including small molecule and peptide-based inhibitors, which exhibit great potency against matriptase in in vitro assays that, in most cases, have utilized a recombinant serine protease domain of matriptase16–21. The specificity of these inhibitors has generally been investigated by testing them against a relatively small number of commonly available serine proteases. Targeting matriptase activity through the use of catalytic inhibitors presents several challenges, some of which relate to this approach in general, and some of which derive from the unique dynamics of the matriptase activation system. There are a great number of serine proteases, and there is significant structural homology between the catalytic domains of these enzymes, many of which exhibit overlapping specificity. This makes the task of designing truly specific catalytic inhibitors very difficult, and the goal of demonstrating that they are indeed specific almost impossible. The unusually tight control of the cellular matriptase proteolytic activity presents additional challenges. Matriptase, like most serine proteases, is synthesized as a zymogen and acquires its full proteolytic activity only after undergoing zymogen activation through a cleavage of the enzyme at the canonical activation motif22–25. Instead of relying on other active proteases for the activating cleavage, as occurs during the activation of most serine proteases, matriptase undergoes autoactivation26. This autoactivation process requires interactions between the matriptase zymogen molecules and HAI-1, and probably involves other proteins yet to be identified. The zymogen form of matriptase possesses unusually high intrinsic activity, which is characterized by a maximal activity at pH 6.0 and the inhibition by increased concentrations of sodium chloride27. Both of these biochemical features mirror the key features of the induction of matriptase activation in cells, suggesting that this intrinsic activity is responsible for matriptase zymogen activation28. The endogenous matriptase inhibitor HAI-1 is involved in matriptase autoactivation and has direct access to the nascent active matriptase29. The tight coupling of matriptase zymogen activation with HAI-1-mediated inhibition means that the uncomplexed, free active matriptase is an extremely short-lived species30. The events associated with matriptase processing, activation, and inhibition are summarized in Figure 1. Interestingly, even with such rapid inhibition, matriptase is still able to activate its physiological substrate, prostasin, a glycosylphosphatidylinositol (GPI)-anchored serine protease10. It seems that the activation of matriptase zymogen, the activation of prostasin by active matriptase, and the inhibition of active matriptase by HAI-1 take place at essentially the same time10. As a result, the scarcity of free active matriptase as a target for catalytic inhibitors represents a major challenge that will limit the potential utility of matriptase catalytic inhibitors as a means to control matriptase function. The catalytic inhibitors may not find the desired target, the scarce active matriptase and significant dose-escalation may be required to produce an effect, which might produce off-target effects on other serine proteases. For example, the matriptase inhibitor CJ-730 was tested for its ability to inhibit cellular activation of pro-hepatocyte growth factor (HGF), a well known matriptase substrate31, 32. In spite of the high potency of this inhibitor which has a Ki of 40 nM against matriptase19, inhibition of pro-HGF activation required an inhibitor concentration of 50 μM, more than 1,000-fold higher than the Ki33. Likewise, in the case of a matriptase inhibitory antibody which has a Ki of 15 pM against matriptase, more than 10,000-fold of the Ki (200 nM) was used to inhibit the P1-Arg proteolytic activity on the cell surface associated with several matriptase-expressing cancer cells34. Furthermore, the inhibition of active matriptase by catalytic inhibitors may be too late to effectively suppress matriptase's biological action, since much of the function of active matriptase, such as the activation of prostasin, occurs at essentially the same time as the generation of active matriptase and its inhibition through binding to HAI-1.
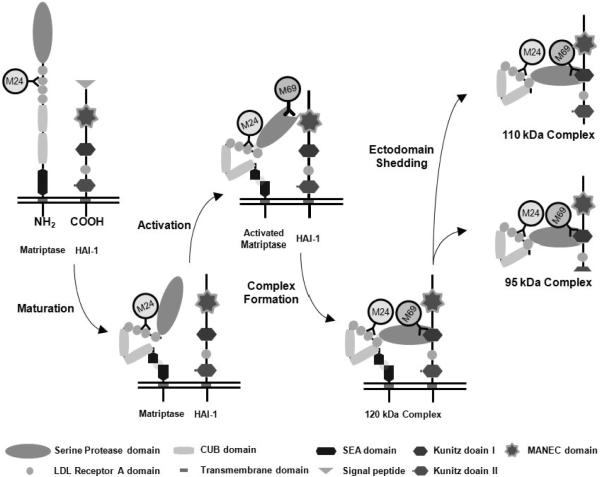
Figure 1. The matriptase-HAI-1 life cycle and the specificity of the various matriptase mAbs.
Both matriptase and HAI-1 are membrane-bound proteins with multiple distinct domains and modules, as indicated. Matriptase is synthesized as a full length zymogen which is rapidly converted into an activation-competent, mature form through a cleavage within the SEA domain (N-terminal process)26, 49. The signal peptide of HAI-1 is removed during the maturation process. Through autoactivation, latent matriptase is converted into active matriptase with full proteolytic activity. Active matriptase is rapidly inactivated by binding to its endogenous inhibitor HAI-1. Matriptase-HAI-1 complexes then are shed as complexes of two different sizes, depending on where the HAI-1 is cleaved. The mAb M24 recognizes a non-catalytic domain of matriptase, though the exact location of the epitope has not been determined. This matriptase mAb can detect both latent and activated forms of matriptase, as indicated. The mAb M69 recognizes an epitope on the serine protease domain that is only present in activated matriptase. This mAb is, therefore, able to distinguish activated matriptase from latent matriptase.
In the light of the apparent disadvantages of matriptase catalytic inhibitors, we intend to develop a novel strategy to control matriptase activity with the goal of translating it into effective interventions in human diseases in the future. In the present study, a novel scheme, consisting of a cell-based, ELISA-like screening assay and a computational search approach, was established to identify novel small molecule inhibitors of matriptase, targeting its zymogen activation rather than the catalytic activity. Four lead compounds were identified and validated to target the activation mechanism directly, rather than the intracellular signaling that triggers activation. These matriptase activation inhibitors effectively inhibit not only matriptase activation but also matriptase-mediated prostasin activation. In contrast, the matriptase catalytic inhibitor CVS-398317 inhibits neither event. These data support our hypothesis that inhibition of matriptase activation would be a more efficient approach to suppress the actions of matriptase than directly inhibiting matriptase catalytic activity. Our compounds may have potential for development into drugs for the treatment of human diseases associated with dysregulated matriptase.
Results
Cell-based, ELISA-like matriptase activation assay
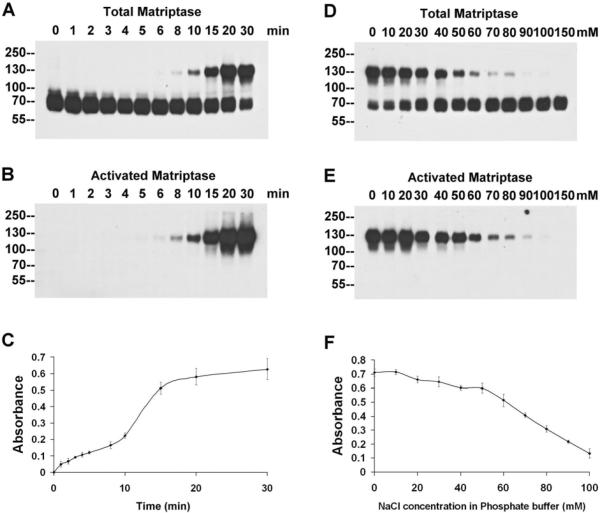
In order to identify small molecule inhibitors that can suppress matriptase zymogen activation, a cell-based, ELISA-like matriptase activation assay was developed. This activation assay combines a system in which robust matriptase zymogen activation is induced in whole cells by exposing them to a pH 6.0 buffer followed by formalin fixation, with a highly specific and sensitive method for detecting the level of activated matriptase, based on the ability of a unique matriptase monoclonal antibody M69 that specifically recognizes activated matriptase, but not latent matriptase. The optimal conditions for the whole-cell activation assay were first assessed using immunoblot assays. As shown in Figure 2, prior to induction of matriptase zymogen activation in the 184 A1N4 cells, matriptase is detected as a 70-kDa latent form in immunoblots of cell lysates probed with the total matriptase mAb (Fig. 2A, 0 min) and the cells are devoid of the activated form of matriptase, as no protein band is detected by the activated matriptase-specific mAb M69 (Fig. 2B, 0 min). When the cells are exposed to a pH 6.0 buffer and incubated at room temperature, matriptase activation is rapidly initiated within few minutes and 70-kDa latent matriptase is rapidly converted into 120-kDa activated matriptase-HAI-1complexes, in a time-dependent manner (Fig. 2 A and B). The appearance of the 120-kDa complexes and disappearance of the 70-kDa matriptase can be detected by the total matriptase mAb (Fig. 2A). The activation and the appearance of 120-kDa matriptase-HAI-1 complex can also be detected by the activated matriptase mAb M69 (Fig. 2B). After 20 min, a large portion of the latent matriptase had been converted into activated matriptase, detected in the 120-kDa complexes. The magnitude and the rapidness of matriptase activation observed in these immunoassays assure the likelihood of being able to specifically determining the matriptase activation event in a 96-well plate assay with a high signal-to-noise ratio.
Figure 2. Development of the cell-based, ELISA-like assay for matriptase activation.
The kinetics of matriptase activation: Mammary epithelial 184 A1N4 cells were incubated with a pH 6.0 buffer at room temperature for the indicated times, and the kinetics of the acid-induced matriptase activation were analyzed by immunoblotting with the mAb M24 (against total matriptase, A) and the mAb M69 (against activated matriptase, B), and using the cell-based, ELISA-like assay with the mAb M69 (C). The dose-dependent inhibition of matriptase activation by NaCl: 184 A1N4 cells were incubated with pH 6.0 buffers containing the indicated concentrations of NaCl at room temperature for 20 min, and matriptase activation was analyzed by immunoblotting with the mAb M24 (against total matriptase, D) and the mAb M69 (against activated matriptase, E), and using the cell-based, ELISA-like assay with the mAb M69 (F).
We then set out to convert the immunoblot-based activation assay into an ELISA-like assay, which can be further configured into a high-throughput screening format. The assay capitalizes on the ability of the mAb M69 to specifically detect activated matriptase in formalin-fixed cells. This allows us to build an assay that does not require cell lysis, with all of the technical challenges that this process would present for the development of a high-throughput assay. The cells were grown in 96-well plates and following the induction of matriptase activation by exposure to the pH 6.0 buffer, the cells were washed and fixed using formaldehyde. The level of activated matriptase was determined using the M69 mAb as the primary antibody, HRP-labeled anti-mouse IgG as the secondary and the chromogenic peroxidase substrate 3,3',5,5' tetramethyl benzidine (TMB). Cellular levels of matriptase activation are reported as the absorbance at 450 nm. Quantitation of acid-induced matriptase activation using this ELISA-like assay exhibits the same kinetic patterns as demonstrated in the Western blot-based assays (Fig. 2B and 2C), indicating the successful adaptation of the assay system.
One of the unique features of the acid-induced matriptase activation is its sensitivity to sodium chloride28. The NaCl-mediated inhibition of acid-induced matriptase activation was initially observed in a cell-free, in vitro activation system and may be due to the suppression of the intrinsic matriptase zymogen activity27. Results from experiments performed with immunoblots showed that the NaCl-mediated inhibition of matriptase activation also occurred in the whole living cells activation system and that the inhibition occurred in a dose-dependent manner (Fig. 2D and 2E). Using the ELISA-like assay, the NaCl-mediated inhibition showed the same dose-dependent profile as seen with the immunoblot analysis (Fig. 2F). Sodium chloride (150mM) was, therefore, used as the positive control for inhibition in our screening.
Characterization of the ELISA-like assay
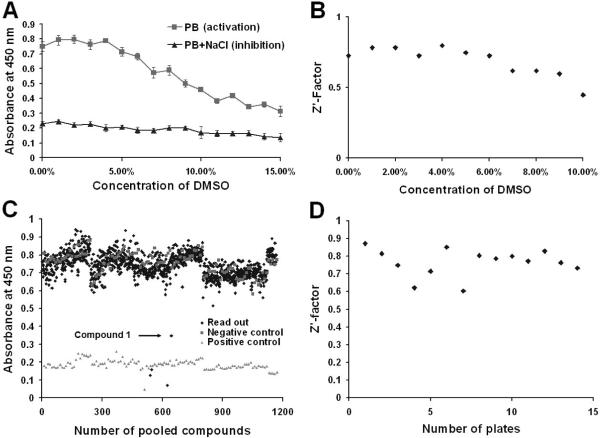
The successful development of the cell-based, ELISA-like assay enables us to conduct screenings for small molecule inhibitors of matriptase activation using a random compound library (ChemDiv, San Diego, CA). Since the compounds in the library were dissolved in dimethyl sulfoxide (DMSO), the effects of DMSO on the assay and matriptase activation were determined by incorporating 0 to 15% DMSO into the activation induction buffer. The impact of DMSO on matriptase activation was evaluated using Z'-factor as an indicator. As shown in Figures 3A and 3B, DMSO had no significant influence on the detection of either matriptase activation or the sodium chloride-mediated inhibition when present at concentrations below 4 %, and the Z'-factors were all above 0.5, indicating an excellent high-throughput screening capability in this range37.
Figure 3. Screening for small molecule inhibitors of matriptase activation.
The effect of DMSO on the ELISA-like assay: 184 A1N4 cells were seeded in 96-well plates, and matriptase activation was induced by exposure to phosphate buffer (PB) pH 6.0, containing increasing concentrations of DMSO, in the presence or absence of 150 mM NaCl. The levels of activated matriptase were determined by the ELISA-like assay (A). The Z'-factors were also calculated as described in the Materials and Methods and plotted with their corresponding concentrations of DMSO (B). Screening of the compound libraries. Chemical libraries were screened using the ELISA-like assay for inhibitors of matriptase activation, and the readouts of each test sample from a random selection of 14 plates of the 96-well are displayed (C). The pH 6.0 buffer alone was used as negative inhibition control while the buffer containing 150 mM NaCl was used as a positive control for inhibition. One lead compound 1 was identified as indicated. Z'-factors of these selected plates were calculated to evaluate for the quality of the screening (D).
Screening and computational search for inhibitors of matriptase activation
A screen for small molecule inhibitor of matriptase activation was then performed using the cell-based, ELISA-like assay described above. In order to increase the efficiency of screening, 4 different compounds were pooled as a testing sample, with an approximate final concentration for each compound in the assay of about 5 μM in 1% DMSO. A total of 20,000 compounds from the ChemDiv 40,000 library were tested and a selection of the readouts of these assays, and the negative and positive controls are presented in Figure 3C. The 450 nm absorbance readouts for the negative controls fluctuated between 0.6 and 0.9 absorbance units, while that of the positive controls with 0.15 M NaCl were approximately 0.2 (Fig. 3A). The vast majority of the test compounds showed no inhibition of matriptase activation and yielded readouts similar to that of the negative controls. Several compounds that caused the detachment of the cells from the plates gave false positive readouts giving absorbance values even lower than the positive controls. The Z'-factors of randomly selected plates were calculated based on the positive and negative controls and all were above 0.6, further confirming the quality of this high-throughput screening assay (Fig. 3D). One pooled compound mixture with inhibitory activity of matriptase activation was identified (Fig. 3C) and after testing the individual compounds making up the pool, the active compound 3-hydroxy-1-(thiazol-2-yl)-5-(thiophen-2-yl)-4-(thiophene-2-carbonyl)-1H-pyrrol-2(5H)-one (A2844/119997) was identified.
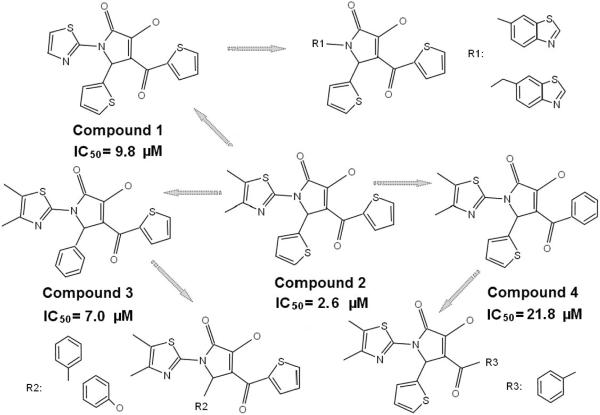
To identify additional matriptase activation inhibitors, a chemical fingerprint-based similarity search was performed against the structure of the active compound 1 (A2844/119997). One hundred and forty compounds with the structures similar to 1 were identified based on a Tanimoto index of 0.86 or greater in combination with the BIT-MACCS fingerprints38, 39. Of these compounds 61 were purchased based on the availability and tested in a follow-up screen. Three more active compounds were identified, two of which exhibit improved inhibitory potency when compared with1. The IC50 of these four compounds were determined using the ELISA-like assay to be 2.6 μM for 1-(4,5-dimethylthiazol-2-yl)-3-hydroxy-5-(thiophen-2-yl)-4-(thiophene-2-carbonyl)-1H-pyrrol-2(5H)-one (F3226-1198), 7.0 μM for 1-(4,5-dimethylthiazol-2-yl)-3-hydroxy-5-phenyl-4-(thiophene-2-carbonyl)-1H-pyrrol-2(5H)-one (F3226-1197), 9.8 μM for 1 and 21.8 μM for 4-benzoyl-1-(4,5-dimethylthiazol-2-yl)-3-hydroxy-5-(thiophen-2-yl)-1H-pyrrol-2(5H)-one (STOCK3S-92907 ) (Fig. 4).
Figure 4. Determination of the IC50 for the lead compounds.
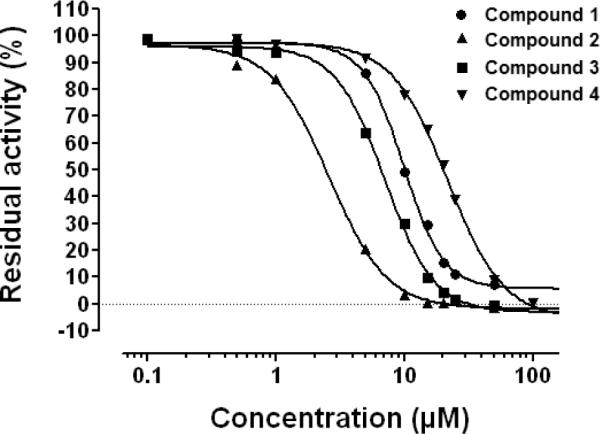
184 A1N4 cells were seeded in 96-well plates and treated with varying concentrations of the four matriptase activation inhibitors. The levels of matriptase activation were determined by the ELISA-like assay. The inhibition of matriptase activation by these inhibitors is presented as the residual activity versus their concentrations and IC50 values were calculated using Graphpad Prism 5 (Graphpad Software Inc.).
As shown in Figure 5, these four compounds share the same 3-hydroxy-1H-pyrrol-2(5H)-one scaffold. Notably, modifications on the substituents on this scaffold affect their ability to inhibit matriptase activation. The initial hit compound 1 with 1-thiazol, 4-thiophen, and 5-thiophen substituents exhibited a micromolar inhibition. Introduction of two methyl groups onto the 1-thiazol moiety (compound 2 (F3226-1198)) improves the inhibitory capacity; alternatively increasing the size of the 1-position substituent diminished the potency. Based on the structure of compound 2, replacing either of the 4- or 5-thiophen substituents with a benzene group, yielding compounds 3 (F3226-1197) and 4 (STOCK3S-92907), respectively, affected the inhibitory activity. Compounds with a benzene group on both of the 4- and 5-position, or with even larger substituents on either position totally lost their inhibitory potency at the tested concentration. These results suggest that both the size and polarity of the 1-, 4- and 5-substituents are crucial toward optimizing the inhibitory effect of the compounds.
Figure 5. The structural and functional relationships among the matriptase activation inhibitors.
The chemical structures of the four matriptase activation inhibitors are presented and compared in relation to their IC50 values. Some of the analogs which showed no inhibition of matriptase activation (without measurable IC50 values) are also presented and compared in order to show the negative impact of the modifications in the surrounding rings (R1, R2 and R3) on the inhibitory activity. The arrows indicate a decreasing in inhibition potency with the structure changes.
Validation of the active compounds
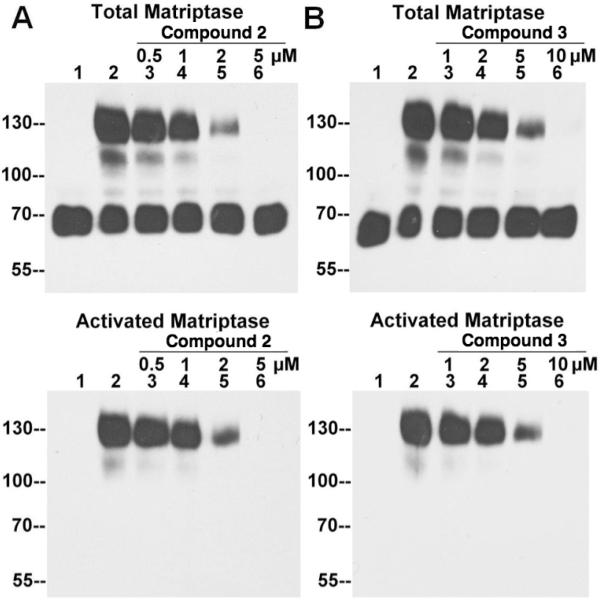
Although the amount of activated matriptase detected by the M69 mAb in the ELISA-like assay is a good indication for the levels of activation, a decrease in the readout, independent of the effects on matriptase activation, might result from the loss of total matriptase due to enhanced shedding. One way to control for this potential confounding effect is to assess the total matriptase which can easily be achieved using immunoblot assays. All the compounds that scored positively for the suppression of matriptase activation in the ELISA-like screens were, therefore, validated using immunoblot assays. As shown in Figure 6A, prior to the induction of activation, all of the matriptase is in the 70kDa latent form (Fig 6A Total Matriptase lane 1). After the induction of matriptase activation, but in the absence of the compound 2, the majority of matriptase was detected in the 120-kDa complex (Fig. 6A, Total Matriptase, lane 2). With increasing concentrations of the compound, the levels of 120-kDa matriptase-HAI-1 complex gradually decreased (Fig. 6, Total Matriptase, lane 3–6) and the levels of 70-kDa latent matriptase increased. In the presence of 5 μM compound 2, there was no detectable 120-kDa complex (Fig. 6A, Total Matriptase, lane 6), and the level of 70-kDa latent matriptase became comparable to the control (Fig. 6A, Total Matriptase, lane 1). The same pattern of inhibition was observed with increasing concentrations of compounds 3 (Fig. 6B), 1 and 4 (data not shown). The inhibition of matriptase activation by these compounds was also confirmed by the reduced signal from the mAb M69 specific for activated matriptase (Fig. 6, lower panels). These data confirmed that all four active compounds are inhibitors of matriptase activation.
Figure 6. Validation of lead compound 2 and 3 by immunoblot assay.
184 A1N4 cells were incubated with PBS (lanes 1, as a non-activation control) or phosphate buffer pH 6.0 alone (lanes 2, as an activation control), or phosphate buffer pH 6.0 with the matriptase inhibitors 2 (A) and 3 (B) at the indicated concentrations (lanes 3–6) for 30 min at 4 °C, followed by the induction of matriptase activation at room temperature for 20 min. Cell lysates were subjected to immunoblot analyses for total matriptase with the mAb M24 and activated matriptase with the mAb M69.
Mechanistic study
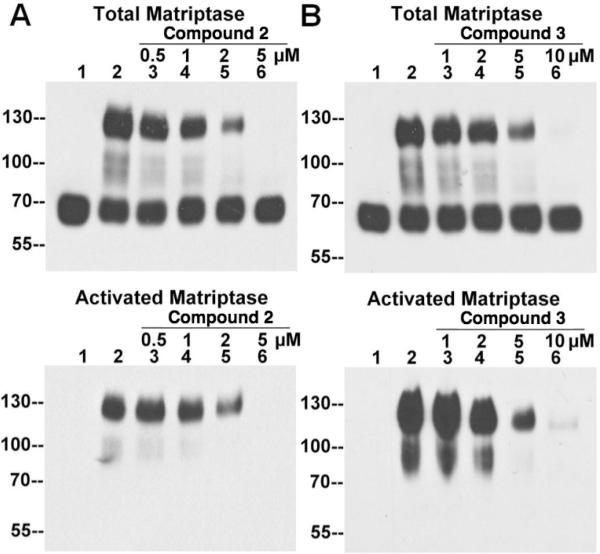
As described above, the inhibitory effect of the lead compounds on the acid-induced matriptase activation was identified and validated using intact cells. A question remains as to whether the inhibition results from a direct action against the matriptase activation process or is mediated indirectly through the blocking of some intracellular signal pathways. While it is very likely that the activation of matriptase caused by mild acid exposure bypasses the intracellular signal pathways and directly acts on the activation machinery through the significant enhancement of the intrinsic catalytic activity of the matriptase zymogen under acidic conditions, the possibility remains that the compounds may suppress signal pathways involved in the control of matriptase activation. We previously demonstrated that the acid-induced matriptase activation also occurs in a cell-free setting, in which the insoluble fractions of the cell homogenates are used in the absence of soluble cytosolic factors28. In such a cell-free setting, any intracellular signaling pathways are likely either absent or incomplete. Thus, to determine whether the inhibitory effect of the active compounds was the result of directly targeting the activation machinery or involved in inhibiting the intracellular signaling pathways, the ability of the compounds to inhibit matriptase activation was tested using the cell-free system. The insoluble fractions of 184 A1N4 cell homogenates were preincubated with the compounds, and then activation of matriptase was induced. Immunoblot analyses using the M24 and M69 mAbs show that these compounds can suppress matriptase activation in a dose-dependent manner (2 and 3 Fig. 7, 1 and 4 not shown). In spite of the fact that the activation is less robust in the cell-free setting when compared with that in the intact-cell setting, the inhibitory efficiency achieved was almost the same in both systems (Figs. 6 and 7), suggesting that all of the inhibitions are likely to be directly against the activation machinery, rather than signal pathways.
Figure 7. Inhibition of matriptase activation by the inhibitors using an in vitro activation assay.
184 A1N4 cells were homogenized in PBS and the insoluble fractions were collected by centrifugation. The insoluble fractions were incubated with PBS (lanes 1, as non-activation control) or phosphate buffer (PB) pH 6.0 alone (lanes 2, as activation control), or PB pH 6.0 with the compound 2 (A) and 3 (B) at the indicated concentrations (lanes 3–6) for 30 min at 4 °C, followed by induction of matriptase activation at room temperature for 20 min. The insoluble fractions were lysed and subjected to immunoblot analyses for total matriptase using the mAb M24 and activated matriptase using the mAb M69.
Inhibition of the activation of a matriptase substrate
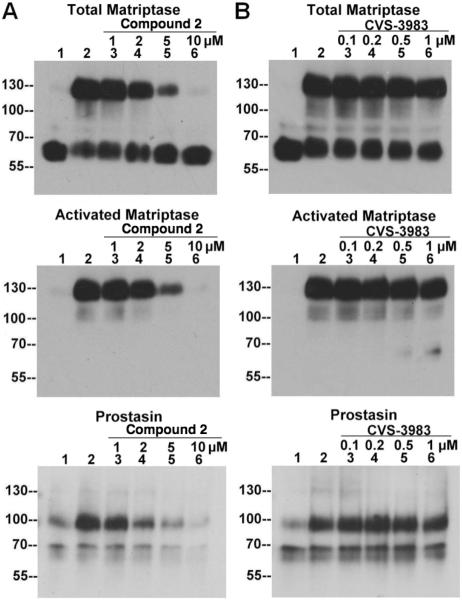
One of the most remarkable features of matriptase regulation is that the activation of the enzyme is rapidly followed by HAI-1-mediated inhibition. It is this rapid HAI-1-mediated inhibition of active matriptase that may reduce the effectiveness of the catalytic inhibitors for the control of matriptase function and that led to our attempts to develop the activation inhibitors. With the activation inhibitors available, we set out to test our hypothesis that inhibition of matriptase activation would prove to be a more effective means of controlling matriptase function than the catalytic inhibitors. In our previous study10, we demonstrated that even in the face of the rapid HAI-1-mediated inhibition of matriptase immediately following activation, activated matriptase is still able to activate a physiological substrate, prostasin, a GPI-anchored serine protease. When HaCaT human keratinocytes were exposed to a pH 6.0 buffer, activation of matriptase was induced, followed by the rapid HAI-1-mediated inhibition of active matriptase and the formation of 120-kDa matriptase-HAI-1 complexes, similar to what is seen with 184 A1N4 cells. The activation of matriptase and the appearance of 120-kDa matriptase-HAI-1 complex in the keratinocytes can be demonstrated using the total matriptase mAb, the activated matriptase mAb, and the HAI-1 mAb (Fig. 8, comparing lanes 2 with lanes 1). Activation of prostasin also occurred during the course of matriptase activation and inhibition. Since HAI-1 also rapidly inhibits newly activated prostasin, as we have previously shown10, prostasin activation can be assessed by the appearance of the prostasin-HAI-1 complexes near the 100-kDa molecular weight marker using the HAI-1 mAb M19 (Fig. 8, HAI-1, comparing lanes 2 with lanes 1) and a prostasin antibody (Fig. 8, Prostasin, comparing lanes 2 with lanes 1). The acid-induced activation of both matriptase and prostasin in the keratinocytes was clearly inhibited by the activation inhibitor 2 in a dose-dependent manner as shown in Figure 8A. In contrast, acid-induced activation of matriptase and prostasin was not inhibited by the matriptase catalytic inhibitor CVS-3983 even at concentrations as high as 1 μ M, 60-fold higher than the IC50 (17 nM) or 300-fold higher than its Ki (3.3 nM)17 against matriptase proteolytic activity (Fig. 8B). The same pattern of inhibition was obtained using the compounds 3, 1 and 4 (data not shown). The inhibition of prostasin activation by the inhibitors of matriptase activation and the inactivity of the catalytic inhibitor suggest that matriptase activation would be a better target than its active site in controlling matriptase function.
Figure 8. Inhibition of the matriptase-prostasin cascade by the matriptase activation inhibitors but not by the catalytic inhibitor CVS-3983.
HaCaT cells were incubated with PBS (lanes 1, as the non-activation control), or phosphate buffer (PB) pH 6.0 alone (lanes 2, as activation control), or PB pH 6.0 with matriptase activation inhibitor compound 2 or catalytic inhibitor CVS-3983 at the indicated concentrations (lanes 3–6, μM) for 30 min at 4 °C followed by the induction of matriptase activation at room temperature for 20 min. Cell lysates were subjected to immunoblot analyses for total matriptase using the mAb M24, activated matriptase using the mAb M69, HAI-1 using the mAb M19, and prostasin with a prostasin polyclonal antibody.
Discussion and Conclusions
Proteolytic activity can be controlled by a variety of different strategies. The use of small molecule and peptide-based inhibitors directly targeting the catalytic site of a given protease has, for many years, been the mainstream strategy. Several matriptase inhibitors of such kind have been developed with excellent potency and good specificity. With an understanding of the complex regulatory mechanisms governing matriptase activation and inhibition, it has become clear that matriptase catalytic inhibitors may, however, not be efficient in the control of matriptase function. In the current study, a new strategy of targeting matriptase zymogen activation rather than its proteolytic activity to control matriptase function is established. This proof-of-concept study encompasses the development and optimization of an approach involving compound screening, computational search for the analogs of the screening hits, and the validation of the inhibitors identified using an alternative assay. This successful scheme led to the identification and validation of 4 inhibitors of matriptase activation with the same backbone structure. This novel class of matriptase inhibitor can successfully suppress not only the acid-induced matriptase activation but also the activation of the physiologically relevant down-stream matriptase substrate prostasin, in an assays system using living cells under conditions wherein free active matriptase is a very short- lived species due to its rapid inhibition by HAI-1. These matriptase activation inhibitors appear to be much more efficient than matriptase catalytic inhibitors in terms of their ability to suppress matriptase activation and prostasin activation by active matriptase.
The ability to efficiently control matriptase function has potentially important clinical implications, and although matriptase is broadly expressed by epithelial tissues throughout the body40, there is reason to believe that inhibiting the enzyme is feasible from a toxicity stand point. Individuals with inherited matriptase frame-shift mutations at the splice site appear to be healthy with clinical consequences mainly for the skin, but which are not serious or life-threatening41. Blockage of matriptase would not, therefore, be expected to cause catastrophic mechanism-based toxicity. With respect to efficacy, due to the complexity of protease signaling pathways, blockade of a protease cascade at an early stage should be more efficient than blocking one of the later stages. Our previous discoveries have demonstrated that matriptase is at the pinnacle of protease cascades based on our observations that matriptase undergoes autoactivation in response to non-protease stimuli26, 42 and that several downstream protease substrates have been identified, including uPA and prostasin10, 31, 43. Therefore, targeting matriptase would lead to more effective blockade of the entire protease cascade than could be achieved by targeting the individual downstream proteases, and would likely require a lower dose of an inhibitor to achieve comparable effectiveness, which should reduce safety and toxicity concerns.
In general, proteases and their cognate inhibitors are typically expressed by different cell types, with the result that it is likely that active proteases will have enough time to act on their substrates prior to being inactivated by their inhibitors. For example, in colorectal and breast carcinomas uPA is expressed by fibroblasts44–46 and the uPA inhibitor PAI-1 is expressed by endothelial cells47. In contrast, the relationship between matriptase and its endogenous inhibitor HAI-1 is relatively unusual, since both matriptase and HAI-1 are found to be almost ubiquitously co-expressed and co-distributed along the secretory pathway in many cells48. Furthermore, previous studies have indicated that HAI-1 plays important roles in the regulation of matriptase at multiple levels. The interaction between matriptase and HAI-1 is crucial for matriptase expression and intracellular trafficking29. HAI-1 is also involved in matriptase zymogen activation26 and is responsible for the rapid inhibition of both matriptase and its substrate prostasin10. This tight relationship among matriptase zymogen activation, prostasin activation and their inhibition by HAI-1 represents a major challenge for the development of catalytic inhibitors that are able to effectively control matriptase function. This unique feature of matriptase regulation may also provide an explanation for the inefficiency of the catalytic inhibitor CVS-3983 in controlling matriptase function (Fig. 8), as there is very limited free active matriptase or limited time for CVS-3983 to exhibit its inhibitory effect.
In summary, by directly targeting the matriptase zymogen activation machinery, a novel class of small molecule inhibitors was developed and validated in both cell-based and cell-free systems. The correlation between the structures and activities of these compounds suggested that their inhibition likely results from specific interactions with the matriptase zymogen activation machinery. Compared with the catalytic inhibitor CVS-3983, these compounds showed profound inhibitory effects not only on matriptase activation, but also on the matriptase-mediated activation of prostasin. These results demonstrate that these small molecule inhibitors may be more potent and specific than classic protease inhibitors in blocking both matriptase activity and the downstream consequences of matriptase activation. Considering the important role that matriptase plays in human carcinomas, skin disorders, and other diseases, these inhibitors have the potential to serve as lead compounds of an important new class of therapeutic agents.
Experimental procedure
Chemicals and reagents
Chemical compounds were obtained from Ambinter (Paris, France). The selected compounds are ≥95% purity by HPLC analysis conducted by the Biopolymer/Genomics Core Facility, University of Maryland School of Medicine with the help of Dr. Pat Campbell. Media, supplements, buffers, and general chemicals used in these experiments were purchased from Sigma, unless otherwise specified.
Cell lines and culture conditions
The immortalized human mammary epithelial cells 184 A1N4 were cultured in a 50:50 mixture of Dulbecco's Modified Eagle's medium/Ham's F12 medium (DMEM-F12 50/50, Mediatech Inc.) supplemented with 0.5% fetal bovine serum (FBS) (Gemini Bio-Products), 5 μg/ml recombinant human insulin (rh-insulin) (Invitrogen), 0.5 μg/ml hydrocortisone, 10 ng/ml recombinant human epidermal growth factor (rhEGF) (Promega), 100 U/ml penicillin, and 100 μg/ml streptomycin (1% pen-strep) (Mediatech Inc.). The immortalized human keratinocytes HaCaT were cultured in DMEM (Mediatech Inc.) supplemented with 10% FBS (Gemini Bio-Products) and 1% penstrep (Mediatech Inc.).
Monoclonal antibodies
Human matriptase was detected using the monoclonal antibody (mAb) M24, which recognizes both latent and activated matriptase. The epitope recognized by the M24 mAb likely lies in the non-catalytic domains of matriptase. The mouse mAb M69 recognizes an epitope present only in the activated form of matriptase and can specifically detect activated matriptase without cross reaction with latent matriptase25. In Figure 1, we summarize the interactions of the matriptase antibodies with various matriptase species. Prostasin was detected using a polyclonal antibody recognizing both activated prostasin in complexes with HAI-1 and uncomplexed prostasin. Interestingly, this antibody appears to recognize the prostasin-HAI-1 complex much better than latent prostasin10.
Acid-driven matriptase activation
Acid-induced matriptase activation using intact cells and cell-free extracts was carried out as described previously28, 36. Briefly, cells or the insoluble fractions of cell homogenates were exposed to 150 mM sodium phosphate pH 6.0 for 20 min at room temperature to induce matriptase activation. Phosphate buffered saline (PBS) containing 1mM 5, 5'-dithiobis-(2-nitrobenzoic acid) (DTNB) and 1% triton X-100 was then used to lyse the cells. For the inhibition tests, cells or the insoluble fractions of cell homogenates were pre-treated with indicated compounds at 4 °C, and then subjected to acid-driven matriptase activation.
Immunoblotting
The protein concentration was determined using the Bradford protein assay reagents according to the manufacturer's protocol. Protein samples for Western blotting were diluted in 5x sample buffer. The sample buffer did not contain a reducing agent, and the samples were not boiled. Proteins were resolved by electrophoresis on 7.5% sodium dodecyl sulfate polyacrylamide gels (SDS-PAGE), transferred to nitrocellulose membranes (Pall Corp., Pensacola, FL), and probed with the antibodies described above. Binding of the primary antibody was detected with the use of horseradish peroxidaseconjugated secondary antibodies (Kirkegaard & Perry Laboratories, Gaithersburg, MD) and visualized using the Western Lightening Chemiluminescence Reagent Plus (Perkin-Elmer, Boston, MA). All experiments were performed in triplicate.
Cell-based, ELISA-like assay
The 184 A1N4 cells were seeded at 8×104 cells/well in 96-well plate 24 h prior to the treatment. After induction of matriptase activation, the cells were washed with PBS, and fixed with 4 % formaldehyde in 0.1% Triton for 20 min. The cellular levels of activated matriptase were determined using mAb M69 as the primary antibody, HRP-labeled anti-mouse IgG as the secondary antibody, and the chromogenic peroxidase substrate 3,3',5,5' tetramethyl benzidine (TMB). Signals were measured as the absorbance at 450 nm. All experiments were performed in triplicate.
Screening of compound libraries
The compounds screened were obtained from the ChemDiv 40,000 library which contains 40,000 diverse chemical compounds purchased from Chemical Diversity Labs (ChemDiv). The compounds in the library were selected from ChemDiv's ~600,000 compound collection to represent maximal chemical and structural diversity. Compound libraries were stored as 10 mM stock solutions in DMSO obtained from the High-Throughput Screening center, University of Maryland at Baltimore. Mixtures of four individual compounds were used as single samples in the screening assay, and the final concentration of each compound was 5 μM in a buffer containing 1% DMSO and 150 mM sodium phosphate, pH 6.0. The pH 6.0 buffer alone was used as negative control for inhibition, while the pH 6.0 buffer with 150 mM NaCl was used as positive control. Each 96-well working plate contained 80 wells for samples, 8 wells for the positive control and 8 wells for the negative control. Briefly, 184 A1N4 cells were seeded at 8×104 cells/well in 96-well plate 24 h prior to the treatment with the chemicals. The cells were incubated with control or compound-containing buffers at 4 °C for 30 min, and then at room temperature for 20 min. The level of matriptase activation in each well was then determined as described above. The inhibition of matriptase activation caused by the compounds was calculated as a percentage of the absorbance in the test compound wells relative to that in the positive controls after subtracting the absorbance of the negative control wells. Compound mixtures showing greater than 50% inhibition were selected and the individual compounds were then assessed independently using the same cell-based ELISA-like assay.
Z'-factor
Z'-factor was calculated for assay optimization and quality assessment, which is defined by four parameters: the means and standard deviations of both the positive (p) and negative (n) controls (μp, σp, and μn, σn). Given these values, the Z'-factor was calculated as: 1− 3(σp + σn)/| μp- μn|37.
Acknowledge
This study was supported by NIH R01-CA-123223 (M.D. Johnson and C.-Y. Lin) the Maryland Cigarette Restitution Fund Program (C.-Y. Lin), NIH P30 pilot grant (C.-Y. Lin), the University of Maryland Computer-Aided Drug Design Center, and the Greenebaum Cancer Center High Throughput Screening Shared Service.
Abbreviation
- DMEM
Dulbecco's Modified Eagle's medium
- DMSO
Dimethyl sulfoxide
- DTNB
5, 5'-dithiobis-(2-nitrobenzoic acid)
- ELISA
Enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- GPI
glycosylphosphatidylinositol
- HAI-1
hepatocyte growth factor activator inhibitor 1
- HGF
hepatocyte growth factor
- mAb
monoclonal antibody
- PBS
Phosphate buffered saline
- rhEGF
recombinant human epidermal growth factor
- TMB
3,3',5,5' tetramethyl benzidine
References
- 1.Drag M, Salvesen GS. Emerging principles in protease-based drug discovery. Nat. Rev. Drug Discov. 2010;9:690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bugge TH, Antalis TM, Wu Q. Type II transmembrane serine proteases. J. Biol. Chem. 2009;284:23177–23181. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhland K. Matriptase and its putative role in cancer. Cell Mol. Life Sci. 2006;63:2968–2978. doi: 10.1007/s00018-006-6298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.List K. Matriptase: a culprit in cancer? Future. Oncol. 2009;5:97–104. doi: 10.2217/14796694.5.1.97. [DOI] [PubMed] [Google Scholar]
- 5.Kang JY, Dolled-Filhart M, Ocal IT, Singh B, Lin CY, Dickson RB, Rimm DL, Camp RL. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res. 2003;63:1101–1105. [PubMed] [Google Scholar]
- 6.Saleem M, Adhami VM, Zhong W, Longley BJ, Lin CY, Dickson RB, Reagan-Shaw S, Jarrard DF, Mukhtar H. A novel biomarker for staging human prostate adenocarcinoma: overexpression of matriptase with concomitant loss of its inhibitor, hepatocyte growth factor activator inhibitor-1. Cancer Epidemiol Biomarkers Prev. 2006;15:217–227. doi: 10.1158/1055-9965.EPI-05-0737. [DOI] [PubMed] [Google Scholar]
- 7.Oberst MD, Johnson MD, Dickson RB, Lin CY, Singh B, Stewart M, Williams A, al Nafussi A, Smyth JF, Gabra H, Sellar GC. Expression of the serine protease matriptase and its inhibitor HAI-1 in epithelial ovarian cancer: correlation with clinical outcome and tumor clinicopathological parameters. Clin Cancer Res. 2002;8:1101–1107. [PubMed] [Google Scholar]
- 8.List K, Szabo R, Molinolo A, Sriuranpong V, Redeye V, Murdock T, Burke B, Nielsen BS, Gutkind JS, Bugge TH. Deregulated matriptase causes ras-independent multistage carcinogenesis and promotes ras-mediated malignant transformation. Genes Dev. 2005;19:1934–1950. doi: 10.1101/gad.1300705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ihara S, Miyoshi E, Ko JH, Murata K, Nakahara S, Honke K, Dickson RB, Lin CY, Taniguchi N. Prometastatic effect of N-acetylglucosaminyltransferase V is due to modification and stabilization of active matriptase by adding beta 1–6 GlcNAc branching. J Biol. Chem. 2002;277:16960–16967. doi: 10.1074/jbc.M200673200. [DOI] [PubMed] [Google Scholar]
- 10.Chen YW, Wang JK, Chou FP, Chen CY, Rorke EA, Chen LM, Chai KX, Eckert RL, Johnson MD, Lin CY. Regulation of the matriptase-prostasin cell surface proteolytic cascade by hepatocyte growth factor activator inhibitor-1 (HAI-1) during epidermal differentiation. J Biol. Chem. 2010;285:31755–31762. doi: 10.1074/jbc.M110.150367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milner JM, Patel A, Davidson RK, Swingler TE, Desilets A, Young DA, Kelso EB, Donell ST, Cawston TE, Clark IM, Ferrell WR, Plevin R, Lockhart JC, Leduc R, Rowan AD. Matriptase is a novel initiator of cartilage matrix degradation in osteoarthritis. Arthritis Rheum. 2010;62:1955–1966. doi: 10.1002/art.27476. [DOI] [PubMed] [Google Scholar]
- 12.Kilpatrick LM, Harris RL, Owen KA, Bass R, Ghorayeb C, Bar-Or A, Ellis V. Initiation of plasminogen activation on the surface of monocytes expressing the type II transmembrane serine protease matriptase. Blood. 2006;108:2616–2623. doi: 10.1182/blood-2006-02-001073. [DOI] [PubMed] [Google Scholar]
- 13.Tseng IC, Chou FP, Su SF, Oberst M, Madayiputhiya N, Lee MS, Wang JK, Sloane DE, Johnson M, Lin CY. Purification from human milk of matriptase complexes with secreted serpins: mechanism for inhibition of matriptase other than HAI-1. Am. J. Physiol Cell Physiol. 2008;295:C423–C431. doi: 10.1152/ajpcell.00164.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seitz I, Hess S, Schulz H, Eckl R, Busch G, Montens HP, Brandl R, Seidl S, Schomig A, Ott I. Membrane-type serine protease-1/matriptase induces interleukin-6 and -8 in endothelial cells by activation of protease-activated receptor-2: potential implications in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2007;27:769–775. doi: 10.1161/01.ATV.0000258862.61067.14. [DOI] [PubMed] [Google Scholar]
- 15.Cheng MF, Jin JS, Wu HW, Chiang PC, Sheu LF, Lee HS. Matriptase expression in the normal and neoplastic mast cells. Eur. J Dermatol. 2007;17:375–380. doi: 10.1684/ejd.2007.0233. [DOI] [PubMed] [Google Scholar]
- 16.Long YQ, Lee SL, Lin CY, Enyedy IJ, Wang S, Li P, Dickson RB, Roller PP. Synthesis and evaluation of the sunflower derived trypsin inhibitor as a potent inhibitor of the type II transmembrane serine protease, matriptase. Bioorg. Med Chem. Lett. 2001;11:2515–2519. doi: 10.1016/s0960-894x(01)00493-0. [DOI] [PubMed] [Google Scholar]
- 17.Galkin AV, Mullen L, Fox WD, Brown J, Duncan D, Moreno O, Madison EL, Agus DB. CVS-3983, a selective matriptase inhibitor, suppresses the growth of androgen independent prostate tumor xenografts. Prostate. 2004;61:228–235. doi: 10.1002/pros.20094. [DOI] [PubMed] [Google Scholar]
- 18.Forbs D, Thiel S, Stella MC, Sturzebecher A, Schweinitz A, Steinmetzer T, Sturzebecher J, Uhland K. In vitro inhibition of matriptase prevents invasive growth of cell lines of prostate and colon carcinoma. Int J Oncol. 2005;27:1061–1070. [PubMed] [Google Scholar]
- 19.Steinmetzer T, Schweinitz A, Sturzebecher A, Donnecke D, Uhland K, Schuster O, Steinmetzer P, Muller F, Friedrich R, Than ME, Bode W, Sturzebecher J. Secondary amides of sulfonylated 3-amidinophenylalanine. New potent and selective inhibitors of matriptase. J Med. Chem. 2006;49:4116–4126. doi: 10.1021/jm051272l. [DOI] [PubMed] [Google Scholar]
- 20.Desilets A, Longpre JM, Beaulieu ME, Leduc R. Inhibition of human matriptase by eglin c variants. FEBS Lett. 2006;580:2227–2232. doi: 10.1016/j.febslet.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 21.Enyedy IJ, Lee SL, Kuo AH, Dickson RB, Lin CY, Wang S. Structure-based approach for the discovery of bis-benzamidines as novel inhibitors of matriptase. J Med Chem. 2001;44:1349–1355. doi: 10.1021/jm000395x. [DOI] [PubMed] [Google Scholar]
- 22.Lin CY, Anders J, Johnson M, Sang QA, Dickson RB. Molecular cloning of cDNA for matriptase, a matrix-degrading serine protease with trypsin-like activity. J Biol. Chem. 1999;274:18231–18236. doi: 10.1074/jbc.274.26.18231. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi T, Shuman MA, Craik CS. Reverse biochemistry: use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11054–11061. doi: 10.1073/pnas.96.20.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim MG, Chen C, Lyu MS, Cho EG, Park D, Kozak C, Schwartz RH. Cloning and chromosomal mapping of a gene isolated from thymic stromal cells encoding a new mouse type II membrane serine protease, epithin, containing four LDL receptor modules and two CUB domains. Immunogenetics. 1999;49:420–428. doi: 10.1007/s002510050515. [DOI] [PubMed] [Google Scholar]
- 25.Benaud C, Dickson RB, Lin CY. Regulation of the activity of matriptase on epithelial cell surfaces by a blood-derived factor. Eur. J Biochem. 2001;268:1439–1447. doi: 10.1046/j.1432-1327.2001.02016.x. [DOI] [PubMed] [Google Scholar]
- 26.Oberst MD, Williams CA, Dickson RB, Johnson MD, Lin CY. The activation of matriptase requires its noncatalytic domains, serine protease domain, and its cognate inhibitor. J Biol. Chem. 2003;278:26773–26779. doi: 10.1074/jbc.M304282200. [DOI] [PubMed] [Google Scholar]
- 27.Inouye K, Yasumoto M, Tsuzuki S, Mochida S, Fushiki T. The Optimal Activity of a Pseudozymogen Form of Recombinant Matriptase under the Mildly Acidic pH and Low Ionic Strength Conditions. J. Biochem. 2009;147:485–492. doi: 10.1093/jb/mvp190. [DOI] [PubMed] [Google Scholar]
- 28.Lee MS, Tseng IC, Wang Y, Kiyomiya K, Johnson MD, Dickson RB, Lin CY. Autoactivation of matriptase in vitro: requirement for biomembrane and LDL receptor domain. Am. J. Physiol Cell Physiol. 2007;293:C95–C105. doi: 10.1152/ajpcell.00611.2006. [DOI] [PubMed] [Google Scholar]
- 29.Oberst MD, Chen LY, Kiyomiya KI, Williams CA, Lee MS, Johnson MD, Dickson RB, Lin CY. Hepatocyte growth factor activator inhibitor 1 (HAI-1) regulates activation and expression of matriptase, a membrane-bound serine protease. Am J Physiol Cell Physiol. 2005;289:C462–C470. doi: 10.1152/ajpcell.00076.2005. [DOI] [PubMed] [Google Scholar]
- 30.Lee M-S, Kiyomiya K, Benaud C, Dickson RB, Lin CY. Simultaneous activation and HAI-1-mediated inhibition of matriptase induced at activation foci in immortal human mammary epithelial cells. Am J Physiol Cell Physiol. 2005;288:C932–C941. doi: 10.1152/ajpcell.00497.2004. [DOI] [PubMed] [Google Scholar]
- 31.Lee SL, Dickson RB, Lin CY. Activation of hepatocyte growth factor and urokinase/plasminogen activator by matriptase, an epithelial membrane serine protease. J Biol. Chem. 2000;275:36720–36725. doi: 10.1074/jbc.M007802200. [DOI] [PubMed] [Google Scholar]
- 32.Fang JD, Chou HC, Tung HH, Huang PY, Lee SL. Endogenous expression of matriptase in neural progenitor cells promotes cell migration and neuron differentiation. J Biol. Chem. 2010;286:5667–5679. doi: 10.1074/jbc.M110.153866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owen KA, Qiu D, Alves J, Schumacher AM, Kilpatrick LM, Li J, Harris JL, Ellis V. Pericellular activation of hepatocyte growth factor by the transmembrane serine proteases matriptase and hepsin, but not by the membrane-associated protease uPA. Biochem. J. 2010;426:219–228. doi: 10.1042/BJ20091448. [DOI] [PubMed] [Google Scholar]
- 34.Darragh MR, Schneider EL, Lou J, Phojanakong PJ, Farady CJ, Marks JD, Hann BC, Craik CS. Tumor detection by imaging proteolytic activity. Cancer Res. 2010;70:1505–1512. doi: 10.1158/0008-5472.CAN-09-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirchhofer D, Peek M, Li W, Stamos J, Eigenbrot C, Kadkhodayan S, Elliott JM, Corpuz RT, Lazarus RA, Moran P. Tissue expression, protease specificity, and Kunitz domain functions of hepatocyte growth factor activator inhibitor-1B (HAI-1B), a new splice variant of HAI-1. J Biol Chem. 2003;278:36341–36349. doi: 10.1074/jbc.M304643200. [DOI] [PubMed] [Google Scholar]
- 36.Tseng IC, Xu H, Chou FP, Li G, Vazzano AP, Kao JP, Johnson MD, Lin CY. Matriptase activation, an early cellular response to acidosis. J. Biol. Chem. 2010;285:3261–3270. doi: 10.1074/jbc.M109.055640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 38.Butina D. Unsupervised Data Base Clustering Based on Daylight's Fingerprint and Tanimoto Similarity: A Fast and Automated Way To Cluster Small and Large Data Sets. J. Chem. Inf. Comput. Sci. 1999;39:747–750. [Google Scholar]
- 39.Godden JW, Stahura FL, Bajorath J. Anatomy of fingerprint search calculations on structurally diverse sets of active compounds. J Chem. Inf. Model. 2005;45:1812–1819. doi: 10.1021/ci050276w. [DOI] [PubMed] [Google Scholar]
- 40.Oberst MD, Singh B, Ossandon M, Dickson RB, Johnson MD, Lin CY. Characterization of matriptase expression in normal human tissues. J Histochem. Cytochem. 2003;51:1017–1025. doi: 10.1177/002215540305100805. [DOI] [PubMed] [Google Scholar]
- 41.Alef T, Torres S, Hausser I, Metze D, Tursen U, Lestringant GG, Hennies HC. Ichthyosis, follicular atrophoderma, and hypotrichosis caused by mutations in ST14 is associated with impaired profilaggrin processing. J. Invest Dermatol. 2009;129:862–869. doi: 10.1038/jid.2008.311. [DOI] [PubMed] [Google Scholar]
- 42.Benaud C, Oberst M, Hobson JP, Spiegel S, Dickson RB, Lin CY. Sphingosine 1-phosphate, present in serum-derived lipoproteins, activates matriptase. J Biol. Chem. 2002;277:10539–10546. doi: 10.1074/jbc.M109064200. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi T, Harris JL, Huang W, Yan KW, Coughlin SR, Craik CS. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J Biol. Chem. 2000;275:26333–26342. doi: 10.1074/jbc.M002941200. [DOI] [PubMed] [Google Scholar]
- 44.Grondahl-Hansen J, Ralfkiaer E, Kirkeby LT, Kristensen P, Lund LR, Dano K. Localization of urokinase-type plasminogen activator in stromal cells in adenocarcinomas of the colon in humans. Am. J. Pathol. 1991;138:111–117. [PMC free article] [PubMed] [Google Scholar]
- 45.Pyke C, Kristensen P, Ralfkiaer E, Grondahl-Hansen J, Eriksen J, Blasi F, Dano K. Urokinase-type plasminogen activator is expressed in stromal cells and its receptor in cancer cells at invasive foci in human colon adenocarcinomas. Am. J. Pathol. 1991;138:1059–1067. [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen BS, Sehested M, Duun S, Rank F, Timshel S, Rygaard J, Johnsen M, Dano K. Urokinase plasminogen activator is localized in stromal cells in ductal breast cancer. Lab Invest. 2001;81:1485–1501. doi: 10.1038/labinvest.3780363. [DOI] [PubMed] [Google Scholar]
- 47.Pyke C, Kristensen P, Ralfkiaer E, Eriksen J, Dano K. The plasminogen activation system in human colon cancer: messenger RNA for the inhibitor PAI-1 is located in endothelial cells in the tumor stroma. Cancer Res. 1991;51:4067–4071. [PubMed] [Google Scholar]
- 48.Wang JK, Lee MS, Tseng IC, Chou FP, Chen YW, Fulton A, Lee HS, Chen CJ, Johnson MD, Lin CY. Polarized epithelial cells secrete matriptase as a consequence of zymogen activation and HAI-1-mediated inhibition. Am. J. Physiol Cell Physiol. 2009;297:C459–C470. doi: 10.1152/ajpcell.00201.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho EG, Kim MG, Kim C, Kim SR, Seong IS, Chung C, Schwartz RH, Park D. N-terminal processing is essential for release of epithin, a mouse type II membrane serine protease. J Biol. Chem. 2001;276:44581–44589. doi: 10.1074/jbc.M107059200. [DOI] [PubMed] [Google Scholar]