Abstract
Myxococcus xanthus utilizes extracellular signals during development to coordinate cell movement, differentiation, and changes in gene expression. One of these signals, the C signal, regulates the expression of many genes, including Ω4400, a gene identified by an insertion of Tn5 lac into the chromosome. Expression of Tn5 lac Ω4400 is reduced in csgA mutant cells, which fail to perform C signaling, and the promoter region has several sequences similar to sequences found in the regulatory regions of other C-signal-dependent genes. One such gene, Ω4403, depends absolutely on the C signal for expression, and its promoter region has been characterized previously by mutational analysis. To determine if the similar sequences within the Ω4400 and Ω4403 regulatory regions function in the same way, deletion analysis and site-directed mutagenesis of the Ω4400 promoter region were performed. A 7-bp sequence centered at −49 bp, termed a C box, is identical in the Ω4400 and Ω4403 promoter regions, yet mutations in the individual base pairs affected expression from the two promoters very differently. Also, a single-base-pair change within a similar 5-bp element, which is centered at −61 bp in both promoter regions, had very different effects on the activities of the two promoters. Further mutational analysis showed that two regions are important for Ω4400 expression; one region, from −63 to −31 bp, is required for Ω4400 expression, and the other, from −86 to −81 bp, exerts a two- to fourfold effect on expression and is at least partially responsible for the C signal dependence of the Ω4400 promoter. Mutations in sigD and sigE, which are genes that encode σ factors, abolished and reduced Ω4400 expression, respectively. Expression of Ω4400 in actB or actC mutants correlated well with the altered levels of C signal produced in these mutants. Our results provide the first detailed analysis of an M. xanthus regulatory region that depends partially on C signaling for expression and indicate that similar DNA sequences in the Ω4400 and Ω4403 promoter regions function differently.
The gram-negative bacterium Myxococcus xanthus exhibits social behavior during multicellular development (4). When starved at a high cell density on a solid surface, rod-shaped M. xanthus cells begin to glide to foci where three-dimensional mounds, each containing approximately 105 cells, are built. Within these mounds (called fruiting bodies), some of the cells undergo morphological changes to form heat- and desiccation-resistant, spherical myxospores.
The developmental program of M. xanthus relies on a specific temporal and spatial pattern of events, the progression of which is controlled by extracellular signals (43). A defect in production of any of the signals leads to arrest at a specific juncture during development, and the defects can be complemented by codevelopment with wild-type cells (which provide the missing signal) or mutants defective in production of a different signal (11, 31). C signaling is required after 6 h of development (28) and involves the product of csgA, a 25-kDa protein that may have enzymatic activity and is believed to be cleaved to a 17-kDa form associated with the cell surface (24, 25, 30, 32, 45, 46). C signaling is essential for three behaviors exhibited by M. xanthus during development; a low level is sufficient for rippling (formation of parallel ridges that appear as traveling waves in movies made by time-lapse microscopy), a higher level is needed for aggregation in foci, and an even higher level is necessary for sporulation within the fruiting body (23, 33). Transmission of the C signal requires motility, presumably due to the need for cell-cell contact (21, 22, 26, 41). The response to C signaling involves a putative transcription factor, FruA (5, 36), which governs a branched pathway inside the recipient cell (47). One branch leads to rippling and aggregation through modification of the gliding movement of cells, which is mediated by the products of the frz operon (16, 17). A second branch includes expression of genes such as the dev operon (49) and the locus identified by insertion Ω7536 (34). This branch leads to sporulation. Expression of other genes also depends on the response to C signaling mediated by FruA (36), but some of these genes are not required for development. These genes were identified by random insertion into the M. xanthus genome of a transposon, Tn5 lac, which contains a promoterless Escherichia coli lacZ gene (27). Insertion of Tn5 lac led to transcriptional fusions between M. xanthus promoters and lacZ. To understand how C signaling regulates developmental gene expression, fusion Ω4403 (7), which depends absolutely on C signaling for expression, and fusions Ω4400 (2) and Ω4499 (6), whose expression depends partially on C signaling, have been studied previously.
The Ω4403 promoter region has been extensively mutagenized to identify the DNA elements that are important for expression (53). Three elements, the C box, a 5-bp element, and a 10-bp element, were found to be absolutely necessary for expression from the Ω4403 promoter, and similar sequences were observed in several other C-signal-dependent genes. A C box, which has the consensus sequence CAYYCCY, where Y is a pyrimidine nucleotide, is centered at −49 bp relative to the transcriptional start site in the Ω4403 promoter region (7). Interestingly, the same sequence (CATCCCT) is found at precisely the same location in the Ω4400 regulatory region (2). The 5-bp element has a consensus GAACA sequence and is located between −63 and −59 bp in the Ω4403 promoter region (53). The Ω4400 upstream region exactly matches the 5-bp element consensus sequence at −63 to −59 bp. The 10-bp element in the Ω4403 promoter region is located at −79 to −70 bp, and the Ω4400 promoter region has a sequence that matches at 6 of 10 positions and is located at −82 to −73 bp.
To determine if the three elements important for Ω4403 expression are functionally conserved in the Ω4400 upstream region and to further characterize this promoter region, we performed a mutational analysis. Our results show that the C box centered at −49 bp is absolutely required for Ω4400 expression; however, the pattern of mutational effects of the individual base pairs within the C box is different than the pattern observed for the Ω4403 promoter. The 5-bp element is also essential for Ω4400 expression, as is the entire region immediately downstream to about −31 bp. Unlike the absolutely required 10-bp element of Ω4403, the upstream region of Ω4400 seems to have a short sequence between −86 and −81 bp that exerts a two- to fourfold positive effect on expression. We concluded that the Ω4400 promoter is regulated differently than the Ω4403 promoter, and we speculated that the promoter regions are recognized by different transcription factors. Further studies indicated that the level of Ω4400 expression correlates well with the level of C signaling during development and that expression from the Ω4400 promoter is dependent on σD and σE.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids that were used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli DH5α | φ80 lacZΔM15 ΔlacU169 recA1 endA1 hsdR17 supE44 thi-1 gyrA relA1 | 12 |
| M. xanthus strains | ||
| DK1622 | Wild type | 19 |
| DK4292 | Tn5 lac (Kmr) Ω4400 | 29 |
| JPB40030 | attB::pJB40030 (pREG1727 with 297-bp XhoI-BamHI fragment from pJB40029)a | This study |
| MDY1727 | attB::pREG1727 | This study |
| MGV4400.2 | attB::pGV4400.2 (pREG1727 with 297-bp XhoI-BamHI fragment from pGV4400.1) | This study |
| MGV4400.10 | attB::pGV4400.10 (pREG1727 with 297-bp XhoI-BamHI fragment from pGV4400.5) | This study |
| MGV4400.12 | attB::pGV4400.12 (pREG1727 with 297-bp XhoI-BamHI fragment from pGV4400.7) | This study |
| MDY6 | attB::pDY6 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY6) | This study |
| MDY8 | attB::pDY8 (pREG1727 with 265-bp XhoI-BamHI fragment from pDY7) | This study |
| MDY10 | attB::pDY10 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY9) | This study |
| MDY12 | attB::pDY12 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY11) | This study |
| MDY14 | attB::pDY14 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY13) | This study |
| MDY16 | attB::pDY16 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY15) | This study |
| MDY18 | attB::pDY18 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY17) | This study |
| MDY20 | attB::pDY20 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY19) | This study |
| MDY22 | attB::pDY22 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY21) | This study |
| MDY24 | attB::pDY24 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY23) | This study |
| MDY26 | attB::pDY26 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY25) | This study |
| MDY28 | attB::pDY28 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY27) | This study |
| MDY30 | attB::pDY30 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY29) | This study |
| MDY32 | attB::pDY32 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY31) | This study |
| MDY34 | attB::pDY34 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY33) | This study |
| MDY36 | attB::pDY36 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY35) | This study |
| MDY38 | attB::pDY38 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY37) | This study |
| MDY54 | attB::pDY54 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY53) | This study |
| MDY56 | attB::pDY56 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY55) | This study |
| MDY58 | attB::pDY58 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY57) | This study |
| MDY60 | attB::pDY60 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY59) | This study |
| MDY62 | attB::pDY62 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY61) | This study |
| MDY64 | attB::pDY64 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY63) | This study |
| MDY66 | attB::pDY66 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY65) | This study |
| MDY68 | attB::pDY68 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY67) | This study |
| MDY70 | attB::pDY70 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY69) | This study |
| MDY72 | attB::pDY72 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY71) | This study |
| MDY74 | attB::pDY74 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY73) | This study |
| MDY76 | attB::pDY76 (pREG1727 with 252-bp XhoI-BamHI fragment from pDY75) | This study |
| MDY78 | attB::pDY78 (pREG1727 with 111-bp XhoI-BamHI fragment from pDY77) | This study |
| MDY80 | attB::pDY80 (pREG1727 with 297-bp XhoI-BamHI fragment from pDY79) | This study |
| MDY102 | attB::pDY102 (pREG1727 with 297-bp XhoI-BamHI fragment from pDO1) | This study |
| DK5208 | csgA::Tn5-132 (Tcr) Ω205 | 49 |
| MDY5208-30 | csgA::Tn5-132 (Tcr) Ω205 attB::pDY30 | This study |
| MDY5208-70 | csgA::Tn5-132 (Tcr) Ω205 attB::pDY70 | This study |
| MDY4400.CA | csgA::Tn5-132 (Tcr) Ω205 attB::pJB40030 | This study |
| DK10603 | ΔactB | 10 |
| MDY4400.AB | ΔactB attB::pJB40030 | This study |
| DK10604 | ΔactC | 10 |
| MDY4400.AC | ΔactC attB::pJB40030 | This study |
| ΔsigD | ΔsigD | 51 |
| MDY4400.SD | ΔsigD Tn5 lac (Kmr) Ω4400 | This study |
| ΔsigE | ΔsigE | 52 |
| MDY4400.SE | ΔsigE Tn5 lac (Kmr) Ω4400 | This study |
| Plasmids | ||
| pGEM7Zf | Aprlacα | Promega |
| pREG1727 | Apr Kmr P1-inc attP′lacZ | 7 |
| pJB40015 | Apr (pGEM7Zf); 0.64-kb ApaLI-BamHI fragment from pJB4001 | 2 |
| pJB40029 | pGEM7Zf with 267-bp EcoRI-SmaI fragment from pJB40015 | This study |
| pGV4400.1 | pJB40029 with CATCCCT-to-ACGAAAG mutation from −52 to −46 bp | This study |
| pGV4400.5 | pJB40029 with C-to-A mutation at −48 bp | This study |
| pGV4400.7 | pJB40029 with C-to-A mutation at −47 bp | This study |
| pDY5 | pJB40029 with T-to-G mutation at −50 bp | This study |
| pDY7 | pGEM7Zf with XhoI-BamHI fragment from −86 to 155 bp of Ω4400 DNA generated by PCR by using pJB40029 as the template | This study |
| pDY9 | pJB40029 with C-to-A mutation at −52 bp | This study |
| pDY11 | pJB40029 with A-to-C mutation at −51 bp | This study |
| pDY13 | pJB40029 with C-to-A mutation at −49 bp | This study |
| pDY15 | pJB40029 with C-to-T mutation at −49 bp | This study |
| pDY17 | pJB40029 with T-to-C mutation at −46 bp | This study |
| pDY19 | pJB40029 with T-to-G mutation at −46 bp | This study |
| pDY21 | pJB40029 with G-to-T mutation at −77 bp | This study |
| pDY23 | pJB40029 with T-to-G mutation at −78 bp | This study |
| pDY25 | pJB40029 with G-to-T mutation at −79 bp | This study |
| pDY27 | pJB40029 with G-to-T mutation at −80 bp | This study |
| pDY29 | pJB40029 with G-to-T mutation at −81 bp | This study |
| pDY31 | pJB40029 with G-to-T mutation at −82 bp | This study |
| pDY33 | pJB40029 with G-to-T mutation at −83 bp | This study/PICK> |
| pDY35 | pJB40029 with GAAC-to-TCCA mutation at −63 to −60 bp | This study |
| pDY37 | pJB40029 with CGGTG-to-ATTGT mutation at −74 to −70 bp | This study |
| pDY53 | pJB40029 with TACAAC-to-GCACCA mutation at −13 to −8 bp | This study |
| pDY55 | pJB40029 with AGGCGC-to-CTTATA mutation at −36 to −30 bp | This study |
| pDY57 | pJB40029 with A-to-C mutation at −53 bp | This study |
| pDY59 | pJB40029 with GTCCC-to-TGAAA mutation at −58 to −54 bp | This study |
| pDY61 | pJB40029 with A-to-C mutation at −59 bp | This study |
| pDY63 | pJB40029 with C-to-A mutation at −60 bp | This study |
| pDY65 | pJB40029 with GGGAGC-to-TTTCTA mutation at −69 to −64 bp | This study |
| pDY67 | pJB40029 with TG-to-GT mutation at −76 to −75 bp | This study |
| pDY69 | pJB40029 with GTC-to-TGA mutation at −86 to −84 bp | This study |
| pDY71 | pJB40029 with GGCGG-to-TTATT mutation at −45 to −41 bp | This study |
| pDY73 | pJB40029 with CCGG-to-AATT mutation at −40 to −37 bp | This study |
| pDY75 | pGEM7Zf with XhoI-BamHI fragment from −73 to 155 bp of Ω4400 DNA generated by PCR by using pJB40029 as the template | This study |
| pDY77 | pGEM7Zf with XhoI-BamHI fragment from −86 to 25 bp of Ω4400 DNA generated by PCR by using pJB40029 as the template | This study |
| pDY79 | pJB40029 with GGGGGTG-to-TTTTTGT mutation at −83 to −77 bp | This study |
| pDO1 | pJB40029 with T-to-C mutation at −50 bp | This study |
Where possible, the plasmid description is given in parentheses after the strain description.
Growth and development.
E. coli DH5α strains were grown at 37°C in Luria-Bertani medium (42) containing 50 μg of ampicillin per ml. M. xanthus strains were grown at 32°C in CTT broth or agar (1.5% agar) plates (14) (1% Casitone, 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4 [final pH 7.6]). When necessary, 40 μg of kanamycin per ml was used for selection. Fruiting body development was performed on TPM agar plates (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4, 1.5% agar [final pH 7.6]) as described previously (29).
Construction of plasmids.
An EcoRI-SmaI restriction fragment containing the Ω4400 promoter region from −101 to 155 bp relative to the start site of transcription was purified from pJB40015 and ligated into pGEM7Zf to form pJB40029. Additional deletion constructs were created by PCR by using pJB40029 as a template and primers designed to produce a product with an XhoI restriction site at the upstream end and a BamHI restriction site at the downstream end. PCR products were then restricted with XhoI and BamHI, gel purified, and ligated into pGEM7Zf, and the ligation products were electroporated into E. coli DH5α. Ampicillin-resistant (Apr) transformants were selected, and plasmid DNA was sequenced at the Michigan State University Genomics Technology Support Facility to confirm the sequence and end points of the M. xanthus DNA insert.
A Quikchange site-directed mutagenesis kit (Stratagene) was used to create mutations in the Ω4400 promoter region that, in most cases, were A↔C or T↔G single-base-pair or multiple-base-pair transversion mutations. In addition, three mutations that were T↔C transition mutations were created (Table 2). Plasmid pJB40029 described above was used as a template in PCRs with various combinations of mutagenic primers. The M. xanthus DNA insert was sequenced at the Michigan State University Genomics Technology Support Facility to ensure that only the proper mutations had been created.
TABLE 2.
Activities of mutant Ω4400 promoter regions
| Promoter assayed | Avg maximum β-galactosidase sp act during developmenta | % of wild-type activity measured in the same exptb |
|---|---|---|
| Vector (no insert) | 11 ± 5 | |
| Wild-type Ω4400 (−101 to 155 bp) | 210 ± 52 | |
| Deletions | ||
| −86 to 155 bp | 186 ± 24 | 93 ± 8 |
| −73 to 155 bp | 160 ± 19 | 60 ± 7 |
| −86 to 25 bp | 146 ± 31 | 54 ± 12 |
| Mutationsc | ||
| TACAAC to GCACCA (−13 to −8 bp) | 5 ± 2 | 0 ± 0.4 |
| AGGCGC to CTTATA (−36 to −31 bp) | 19 ± 0 | 5 ± 1 |
| CCGG to AATT (−40 to −37 bp) | 12 ± 1 | 2 ± 0.4 |
| GGCGG to TTATT (−45 to −41 bp) | 25 ± 6 | 9 ± 3 |
| CATCCCT to ACGAAAG (−52 to −46 bp) | 16d | 5 |
| T to G (−46 bp) | 364 ± 90 | 210 ± 53 |
| T to C (−46 bp) | 273 ± 55 | 156 ± 33 |
| C to A (−47 bp) | 15 ± 1 | 4 ± 0.3 |
| C to A (−48 bp) | 208 ± 140 | 109 ± 76 |
| C to A (−49 bp) | 25 ± 3 | 9 ± 2 |
| C to T (−49 bp) | 17 ± 5 | 4 ± 3 |
| T to G (−50 bp) | 150 ± 41 | 50 ± 4 |
| T to C (−50 bp) | 276 ± 38 | 158 ± 23 |
| A to C (−51 bp) | 184 ± 11 | 98 ± 4 |
| C to A (−52 bp) | 16 ± 2 | 4 ± 1 |
| A to C (−53 bp) | 54 ± 18 | 15 ± 8 |
| GTCCC to TGAAA (−58 to −54 bp) | 15 ± 0.2 | 3 ± 1 |
| A to C (−59 bp) | 12 ± 1 | 2 ± 0.2 |
| GAAC to TCCA (−63 to −60 bp) | 12 ± 4 | 2 ± 2 |
| C to A (−60 bp) | 12 ± 2 | 1 ± 1 |
| GGGAGC to TTTCTA (−69 to −64 bp) | 167 ± 4 | 87 ± 2 |
| CGGTG to ATTGT (−74 to −70 bp) | 214 ± 7 | 89 ± 3 |
| TG to GT (−76 to −75 bp) | 176 ± 47 | 92 ± 26 |
| GGGGGTG to TTTTTGT (−83 to −77 bp) | 102 ± 1 | 51 ± 0.4 |
| G to T (−77 bp) | 271 ± 15 | 116 ± 7 |
| T to G (−78 bp) | 220 ± 25 | 92 ± 12 |
| G to T (−79 bp) | 199 ± 11 | 82 ± 5 |
| G to T (−80 bp) | 192 ± 6 | 79 ± 3 |
| G to T (−81 bp) | 130 ± 30 | 57 ± 15 |
| G to T (−82 bp) | 176 ± 53 | 71 ± 24 |
| G to T (−83 bp) | 285 ± 51 | 122 ± 24 |
| GTC to TGA (−86 to −84 bp) | 61 ± 16 | 26 ± 7 |
The maximum β-galactosidase specific activity (in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein; average ± standard deviation) is shown for three independently isolated M. xanthus transformants (one determination each) in the case of mutant promoter regions and for one isolate (13 determinations) in the case of the wild-type promoter and vector controls. Samples were assayed at 0, 6, 12, 18, 24, 30, 36, and 48 h during development.
The wild-type promoter and vector-only strains were included in each experiment. The maximum value for each mutant promoter region is expressed as a percentage of the maximum value observed for the wild-type promoter in the same experiment, after the maximum value observed for the vector-only control in that experiment was subtracted from both values. The values are averages ± standard deviations. A zero indicates that the expression from the mutant promoter region was equal to or slightly less than that observed for the vector-only control.
For example, mutant TACAAC to GCACCA (−13 to −8 bp) had a mutation that changed TACAAC at positions −13 to −8 to GCACCA, and mutant T to G (−50 bp) had a mutation that changed T at position −50 to G.
Only one determination was made for this mutation.
Each mutant derivative of pJB40029 was restricted with XhoI and BamHI, gel purified, and ligated into pREG1727 previously cut with the same enzymes. The ligation products were introduced into E. coli DH5α by electroporation, and Apr transformants were selected. A transformant containing the mutant Ω4400 plasmid was identified by using colony PCR with primers to ensure proper orientation. The transformants containing the mutated Ω4400 promoter regions were then used to prepare plasmid DNA for introduction into M. xanthus.
Construction of M. xanthus strains and determination of lacZ expression during development.
Strains containing pREG1727 derivatives integrated at the Mx8 phage attachment site (designated attB in Table 1) were constructed by electroporation (20) of M. xanthus, and transformants were selected on CTT agar plates containing kanamycin. Based on previous experience in our laboratory (2, 6, 7), the majority of the transformants had a single copy of the plasmid integrated at attB. To eliminate colonies with unusual developmental lacZ expression, we screened at least 10 transformants on TPM agar plates containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml. Any colonies with unusual expression of lacZ were discarded, and three of the remaining independent isolates of each mutant construct were chosen for development. In all cases, the three transformants gave similar results (Table 2) when developmental β-galactosidase activity was measured as described previously (29).
To transduce Tn5 lac Ω4400 into M. xanthus sigD and sigE mutants, Mx4 phage stocks (3, 8, 18) were prepared with M. xanthus DK4292 and used to infect the mutants at multiplicities of 2.0, 1.0, 0.5, and 0.1. Transductants were selected on CTT agar plates containing kanamycin. Developmental β-galactosidase activity was determined as described previously (29) for three transductants.
RESULTS
Effects of mutations in a C box centered at −49 bp.
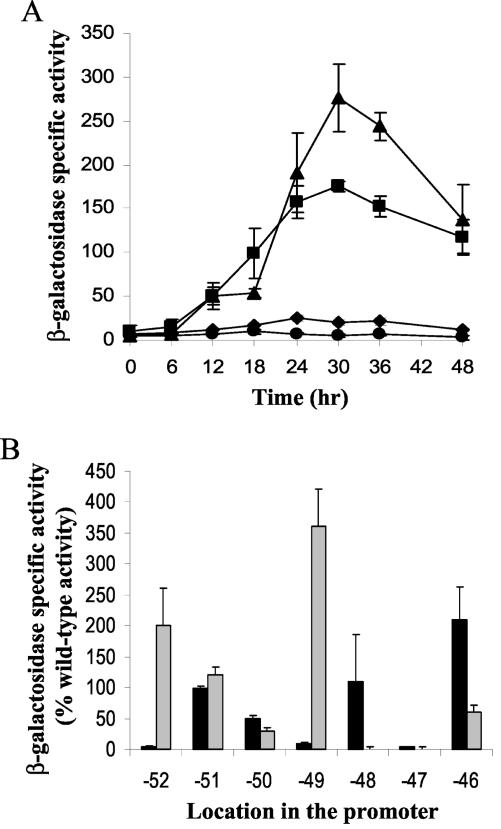
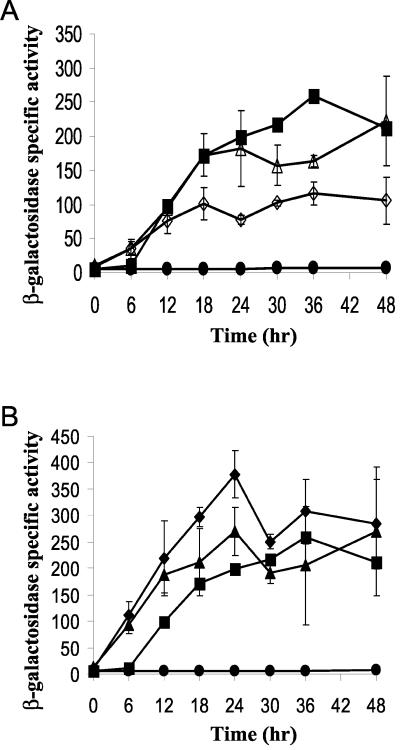
A conserved 7-bp sequence (CATCCCT), termed a C box (6), is centered at −49 bp in both the Ω4400 (2) and Ω4403 (7) promoter regions. The effects of single-base-pair changes in this C box in the Ω4403 promoter region have been established previously (53). If the C box centered at −49 bp functions in the same way in both promoter regions, the effects of mutations should be the same. To test this prediction, we created a plasmid that contained the Ω4400 wild-type promoter region (−101 to 155 bp) and used site-directed mutagenesis to create A↔C and T↔G transversion mutations at each of the base pairs within the C box centered at −49 bp (Table 2). In addition, we constructed three mutations which had T↔C transitions at −50, −49, and −46 bp and a multiple-base-pair change of the entire C box (Table 2). These mutant promoter regions were subcloned directly upstream of the E. coli lacZ gene in pREG1727, and the resulting plasmids were transformed into M. xanthus wild-type strain DK1622 for determination of lacZ expression during development (see Materials and Methods). A strain bearing a pREG1727 derivative containing the Ω4400 wild-type promoter region served as a positive control, and a strain containing only the pREG1727 vector (without a promoter) served as a negative control. Table 2 shows the average maximum activity and the percentage of wild-type activity for each strain. The complete developmental lacZ expression data for the controls and for mutants with a T-to-C transition at −50 bp or a C-to-A transversion at −49 bp are shown in Fig. 1A. Five of the eleven mutations showed a strong (more-than-10-fold) decrease in the maximum β-galactosidase specific activity, including the multiple-base-pair change of the entire C box, the transversions at −52, −49, and −47 bp, and the transition at −49 bp (Table 2 and Fig. 1). Transversion mutations at −51 or −48 bp had no significant effect on lacZ expression (Table 2), while mutations at −46 bp increased the maximum activity. The T-to-C and T-to-G changes at −50 bp caused a small increase and a small decrease, respectively, in activity. Taken together, the results show that certain base pairs in the C box centered at −49 bp are critical for developmental expression from the Ω4400 promoter.
FIG. 1.
Mutational analysis of the C box centered at −49 bp in the Ω4400 promoter region and comparison with the Ω4403 promoter region. (A) Developmental lacZ expression was determined for three independent isolates for each strain. Symbols: ▪, Ω4400 wild-type promoter (−101 to 155 bp), which served as a positive control; •, vector without insert negative control; ▴, T-to-C single-base-pair change at −50 bp; ⧫, C-to-A single-base-pair change at −49 bp. The average β-galactosidase activity is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. The error bars indicate one standard deviation. (B) Summary of the effects of transversion mutations at each base pair of the C box centered at −49 bp in both the Ω4400 (solid bars) and Ω4403 (gray bars) promoter regions. The bars indicate the average maximum β-galactosidase specific activity during a 48-h time course, expressed as a percentage of the maximum value observed for the corresponding wild-type promoter. The error bars indicate one standard deviation for data taken from Table 2 and published previously (53).
The patterns of the effects of transversion mutations in the C boxes centered at −49 bp in the Ω4400 and Ω4403 (53) promoter regions are compared in Fig. 1B. A very different effect was observed at four of the seven positions. The most profound difference was at −49 bp, where in the Ω4400 promoter region the mutation led to a 90% decrease in activity, while in the Ω4403 promoter region it led to a 360% increase in activity. Because the patterns of mutational effects for the two C boxes were markedly different, we concluded that they function differently. For example, if these sequences are recognized by transcription factors, our results suggest that different proteins bind to the two promoter regions.
Effects of mutations in the C box centered at −80 bp.
The presence of a second C box, centered at −80 bp in the Ω4400 promoter region, was noted previously (6). To determine if this C box is important for Ω4400 promoter activity, a multiple-base-pair change from GGGGGTG (note that the sequence of the opposite DNA strand, CACCCCC, matches the C box consensus sequence) to TTTTTGT was tested as described above. This mutation resulted in a 50% decrease in developmental promoter activity (Table 2). The effects of single-base-pair changes in this C box were also examined. None of the mutations showed as strong an effect as the multiple-base-pair change, although the G-to-T change at position −81 bp did decrease activity by about 40% (Table 2). We concluded that the C box centered at −80 bp in the Ω4400 regulatory region is not essential for developmental expression, although it does exert an approximately twofold positive effect. This region is different from the Ω4403 regulatory region, which has an essential 10-bp element between −79 and −70 bp.
Deletion analysis of the Ω4400 promoter region.
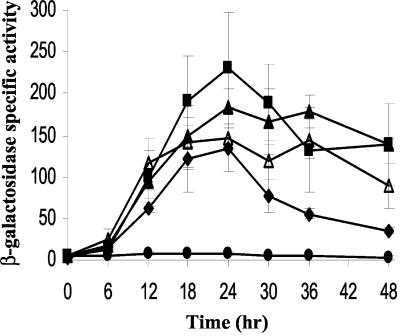
Brandner and Kroos (2) previously identified the transcriptional start site for the Ω4400 promoter and reported that a fragment spanning from −101 to 455 bp could drive developmental lacZ expression comparable to that of the original Tn5 lac Ω4400 insertion in the M. xanthus chromosome, but a 5′ deletion to −73 bp (erroneously reported as −76 to 455 bp) with the same downstream end lost all activity. All the mutations described above were tested in the context of Ω4400 DNA from −101 to 155 bp, because this fragment produced levels of β-galactosidase activity during development (Fig. 2) similar to those of the construct from −101 to 455 bp (2). In order to determine whether a smaller region is sufficient for full expression of the Ω4400 promoter, a series of deletions (both 5′ and 3′) were constructed, fused to lacZ in pREG1727, transformed into M. xanthus DK1622, and tested for developmental production of β-galactosidase. A 5′ deletion that contained −86 to 155 bp exhibited activity comparable to that of the segment from −101 to 155 bp (Table 2 and Fig. 2), indicating that the region between −101 and −86 bp is not necessary for expression. A 5′ deletion containing −73 bp to 155 bp showed a 40% decrease in the average maximum activity, which is similar to the effect of the multiple-base-pair change in the C box centered at −80 bp or the single-base-pair change at −81 bp (Table 2 and Fig. 2). This result was surprising, because, as noted above, the segment from −73 to 455 bp was reported previously to be inactive (2). The fact that considerable activity was observed for the segment from −73 to 155 bp (Table 2 and Fig. 2) suggests that a potential negative regulatory element lies between 155 and 455 bp. However, negative regulation was observed only in the absence of sufficient upstream DNA (i.e., beyond −73 bp) because no significant difference in activity was observed for two constructs with the same 5′ end at −101 bp and different downstream ends at 155 bp (Table 2 and Fig. 2) and 455 bp (2).
FIG. 2.
Deletion analysis of the Ω4400 promoter region. The 5′ deletion constructs contained Ω4400 DNA from −86 to 155 bp (▴) or from −73 to 155 bp (▵), whereas the 3′ deletion construct contained Ω4400 DNA from −86 to 25 bp (⧫). The Ω4400 promoter region from −101 to 155 bp served as the positive control in these experiments (▪). The vector was the no-insert negative control (•). The average β-galactosidase activity of at least three independent isolates is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. The error bars indicate one standard deviation.
To determine the role, if any, of the sequence between 25 and 155 bp, a 3′ deletion containing the region from −86 to 25 bp was tested. This deletion reduced the developmental promoter activity nearly twofold (Table 2 and Fig. 2), suggesting that the region between 25 and 155 bp plays a weak positive role in expression from the Ω4400 promoter. Because DNA between −86 and 25 bp relative to the Ω4400 transcriptional start site displayed considerable activity, we focused our efforts on identifying and characterizing cis-regulatory elements within this region by testing the effects of additional mutations in the context of Ω4400 DNA from −101 to 155 bp.
Effects of mutations between −86 and −64 bp.
To test the importance of the region surrounding the C box centered at −80 bp, we constructed several multiple-base-pair mutations in this region (Table 2 and Fig. 3). Mutation of GTC to TGA at −86 to −84 bp resulted in only 26% of the wild-type activity. We do not understand why this particular mutation impaired developmental expression more than the 5′ deletion to −73 bp, but both results support the idea that there is a positive regulatory element in this region. We also created a dinucleotide TG-to-GT mutation at −76 to −75 bp because this region was shown to be essential in the Ω4403 promoter region (53). This mutation had little effect on Ω4400 promoter activity (Table 2 and Fig. 3). Likewise, neither a CGGTG-to-ATTGT mutation centered at −72 bp nor a GGGAGC-to-TTTCTA mutation spanning from −69 to −64 bp had much effect on expression. These results, together with the mutations in the C box centered at −80 bp and the 5′ deletion to −73 bp, suggest that there is a positive regulatory element that exerts a two- to fourfold effect on developmental lacZ expression between −86 and −81 bp in the Ω4400 regulatory region.
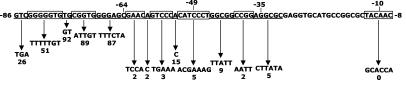
FIG. 3.
Summary of mutational effects on developmental expression from the Ω4400 promoter. The promoter region from −86 to −8 bp is shown. The downward arrows indicate decreased developmental lacZ expression caused by the mutations shown, and the numbers indicate the relative amounts of β-galactosidase specific activity observed for the mutants, expressed as percentages of the wild-type promoter activity measured in the same experiment (Table 2). Mutations are alternatively underlined and enclosed in boxes.
Effects of mutations in the 5-bp element.
Another conserved sequence that is found in the Ω4400 and Ω4403 promoter regions, as well as the Ω4499, fruA, and csgA promoter regions, has been termed the 5-bp element (53). In all of these regulatory regions, a 5-bp sequence with the consensus sequence GAACA can be found approximately 5 to 7 bp upstream of a C box sequence. In the Ω4400 promoter region, the sequence is GAACA at −63 to −59 bp. In the case of Ω4403, the sequence is GACCG at −63 to −59 bp, and this element appears to be essential for activity of the Ω4403 promoter (53). A single-base-pair change at any position except the C at −60 bp greatly impaired or abolished expression. To determine if this element is important for expression from the Ω4400 promoter, we first constructed a strain with a 4-bp mutation from GAAC to TCCA at −63 to −60 bp. This change led to complete loss of promoter activity (Table 2 and Fig. 3). A single-base-pair mutation of A to C at −59 bp also caused a complete loss of activity (Table 2 and Fig. 3). Because the fourth base pair of the 5-bp element is the most conserved yet when this base pair was mutated in the Ω4403 promoter region a nearly twofold increase in developmental lacZ expression was observed (53), we tested the effect of making the same change in the Ω4400 promoter region. Changing C to A at −60 bp abolished Ω4400 promoter activity (Table 2). We concluded that base pairs in the position −60 region are essential for activity of both the Ω4400 and Ω4403 promoters, but the effects of changing C to A at −60 bp are quite different for the two promoters, which is consistent with the notion that these promoter regions may be recognized by different transcription factors.
Effects of mutations between −58 and −53 bp.
Two mutations were created in the region between the 5-bp element and the C box centered at −49 bp. A multiple-base-pair mutation of GTCCC to TGAAA centered at −56 bp led to a complete loss of promoter activity (Table 2 and Fig. 3). In contrast, a comparable mutation in the corresponding region of the Ω4403 promoter region caused a 1.6-fold increase in activity (53). A single base change at −53 bp from A to C caused a strong decrease in Ω4400 promoter activity (Table 2 and Fig. 3), which was comparable to the effect of a T-to-G change at −53 bp in the Ω4403 promoter region (53). As summarized in Table 2 and Fig. 3, the region between −64 and −46 bp contains many base pairs that are vital for expression of Ω4400.
Effects of mutations downstream of −46 bp.
We constructed two mutations between the C box centered at −49 bp and the promoter −35 region. Changing GGCGG at −45 to −41 bp to TTATT resulted in a strong decrease in developmental expression, as did changing CCGG at −40 to −37 bp to AATT (Table 2 and Fig. 3). We noted that both of these mutations not only changed the DNA sequence but also altered the local G+C content of the DNA, although this was also the case when we changed GGGAGC at −69 to −64 bp to TTTCTA, CGGTG at −74 to −70 bp to ATTGT, and GGGGGTG at −83 to −77 bp to TTTTTGT, yet these mutations had a less-than-twofold effect on Ω4400 expression (Table 2 and Fig. 3).
The Ω4400 promoter has a −10 region with the sequence TACAAC (Fig. 3), which resembles the E. coli σ70 consensus sequence TATAAT (35). However, the sequence of the −35 region of the Ω4400 promoter (AGGCGC) does not match the σ70 consensus sequence (TTGACA) (35). To determine the effects of mutating these regions, we created two mutations, a TACAAC-to-GCACCA mutation at −13 to −8 bp and a AGGCGC-to-CTTATA mutation at −36 to −31 bp. In both cases, we observed a complete loss of promoter activity (Table 2 and Fig. 3).
To summarize the results of our mutational analyses, the Ω4400 promoter −10 region and DNA spanning at least from the −35 promoter region to −60 bp are critical for developmental expression, and DNA extending from −81 bp to approximately −86 bp stimulates expression two- to fourfold.
C signal dependence of the Ω4400 promoter.
The Ω4400 promoter exhibits partial dependence on extracellular C signaling; a threefold decrease in developmental expression was observed in a csgA mutant that was unable to make C signal, but expression was restored in the csgA background upon codevelopment with wild-type cells, which provided C signal (2). Since our mutational analysis suggested that a positive regulatory element between −86 and −81 bp stimulates Ω4400 promoter activity two- to fourfold, we hypothesized that this element might mediate the partial C signal dependence of the promoter. If this hypothesis is correct, mutations in the region from −86 to −81 bp might reduce or eliminate dependence on C signaling. We transformed pDY70 containing the GTC-to-TGA mutation at −86 to −84 bp into csgA mutant DK5208 cells and measured developmental lacZ expression (Fig. 4A). The activity of the mutant promoter in the csgA mutant background was not significantly different than the activity in the wild-type background. Addition of wild-type DK1622 cells to the csgA mutant bearing the mutant promoter region did not alter developmental lacZ expression. These results indicate that the mutant regulatory region is C signal independent and are consistent with the idea that the region from −86 to −84 bp mediates the partial C signal dependence of the Ω4400 promoter.
FIG. 4.
C signal dependence of mutant Ω4400 promoter regions: developmental lacZ expression of pDY70 (A) or pDY30 (B), integrated at attB of wild-type DK1622 (⧫) or csgA mutant DK5208 in the absence (◊) or in the presence (▴) of an equal number of DK1622 cells (lacking lacZ but capable of C signaling). The average β-galactosidase activity of at least three independent isolates is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. The error bars indicate one standard deviation. Single isolates with pJB40030 (wild-type Ω4400 promoter from −101 to 155 bp) (▪) or pREG1727 (vector without insert) (•) integrated at attB were included as controls.
We also transformed pDY30, which has a G-to-T mutation at −81 bp, into csgA mutant cells and carried out a similar experiment (Fig. 4B). This mutant promoter showed about one-half as much activity in the csgA mutant as in the wild-type background, suggesting that there was some residual dependence on C signaling. However, expression of the mutant promoter in the csgA background did not increase significantly upon codevelopment with wild-type DK1622 cells. This mutant promoter appeared to be less responsive to C signaling than the wild-type Ω4400 promoter, further supporting the notion that the partial C signal dependence of the Ω4400 promoter is mediated, at least in part, through the region from −86 to −81 bp.
Expression of Ω4400 in act mutants.
Gronewold and Kaiser previously identified the act operon, which controls the timing and level of CsgA production in M. xanthus (10). In-frame deletions in actA or actB reduced the amount of CsgA accumulated during development. An in-frame deletion in actC caused earlier accumulation of CsgA during development, whereas an insertion mutation in actD delayed the normal rise in the CsgA level. Expression of several C-signal-dependent genes correlated with the timing and level of CsgA production in the act mutants (9). To test whether Ω4400 expression behaves similarly, we transformed both an actB mutant, DK10603, and an actC mutant, DK10604, with plasmid pJB40030, which contained the Ω4400 wild-type promoter region fused to lacZ, and measured developmental lacZ expression.
Figure 5 shows that expression of Ω4400 correlated with CsgA production. In the actB mutant, expression was reduced 50% (Fig. 5A), as observed for two other developmental promoters (Ω4414 and Ω4499) that depend partially on C signaling for expression (9). In the actC mutant, Ω4400 expression increased 6 h earlier than it increased in the wild-type background (Fig. 5B). This correlates with the earlier rise in the CsgA level in the actC mutant and matches the behavior of several other developmental reporters (Ω4414, Ω4499, Ω7536) in the actC background (9). If expression of Ω4400 correlates with CsgA production in the act mutants only because of the role that CsgA plays in extracellular C signaling, it might be possible to restore Ω4400 expression in the act mutant to the wild-type pattern by codevelopment with wild-type cells. Figure 5A shows that wild-type cells restored the normal level of developmental lacZ expression to Ω4400 in the actB mutant. For the actC mutant, codevelopment with wild-type cells produced little change in the pattern of lacZ expression during the first 12 h of development (Fig. 5B), but at 18 and 24 h Ω4400 expression was more similar to the expression in the wild-type background than to the expression in the actC mutant without codevelopment with wild-type cells. Apparently, at a ratio of 1:1 in the mixture, wild-type cells cannot compensate for the excess CsgA produced by the actC mutant early in development, but as aggregation and mound formation progress later in development, the wild-type cells appear to dilute C signaling interactions and partially restore Ω4400 expression to the normal, lower levels. Taken together, these results demonstrate that expression from the Ω4400 promoter responds to the timing and level of CsgA production. Moreover, the defects in CsgA production in actB and actC mutants can be complemented extracellularly by codevelopment with wild-type cells, which restores Ω4400 expression to nearly normal levels by 18 h during development.
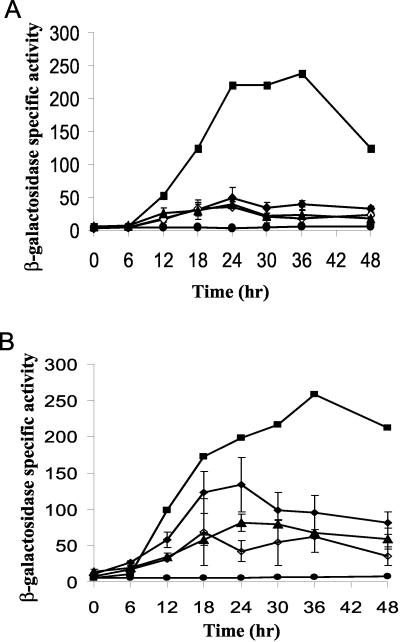
FIG. 5.
Effects of actB and actC mutations on expression of the Ω4400 promoter and extracellular complementation of the defects. The Ω4400 wild-type promoter fused to lacZ in pJB40030 was integrated into the Mx8 phage attachment site of DK10603 (ΔactB) and DK10604 (ΔactC). (A) Developmental lacZ expression in the actB mutant alone (◊) or upon codevelopment with wild-type DK1622 (▵). (B) Expression in the actC mutant alone (⧫) or upon codevelopment with DK1622 (▴). In both panels, the average β-galactosidase activity is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. The error bars indicate one standard deviation. Single isolates with pJB40030 (wild-type Ω4400 promoter from −101 to 155 bp) (▪) or pREG1727 (vector without insert) (•) integrated at attB were included as controls.
Expression of Ω4400 in sigD and sigE mutants.
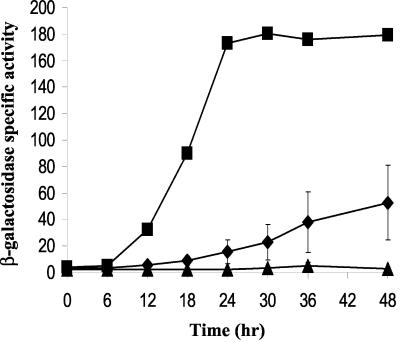
The form of RNA polymerase responsible for transcription from the Ω4400 promoter is unknown. σA RNA polymerase, the major form in growing cells (1), was unable to produce transcripts from the Ω4400 promoter in vitro (D. Biran and L. Kroos, unpublished data). Brandner and Kroos showed previously that null mutations in the sigB gene (encoding σB) or the sigC gene (encoding σC) did not affect the expression of Ω4400 (2). To investigate if the remaining σ70 sigma family members that have been described (51, 52) directly or indirectly control the expression of Ω4400, we used Mx4 phage to transduce two M. xanthus strains that contain a null allele of the sigD gene (encoding σD) or the sigE gene (encoding σE) with the original Tn5 lac insertion Ω4400 from DK4292. Transductants containing a mutation in the sigD gene failed to express β-galactosidase from the Ω4400 promoter (Fig. 6), suggesting that σD RNA polymerase activity is directly or indirectly required for Ω4400 expression. In a sigE mutant, the expression of Ω4400 was reduced and did not reach the maximum wild-type activity level by 48 h of development (Fig. 6). This suggests that σE RNA polymerase is not solely responsible of transcription of Ω4400, although it may be partially responsible or it may indirectly affect Ω4400 expression.
FIG. 6.
Expression of Ω4400 in sigD and sigE mutants. Developmental β-galactosidase activity was determined for Tn5 lac Ω4400 transduced into sigD (▵) and sigE (⧫) mutant backgrounds. The average β-galactosidase activity of three independent isolates is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. The error bars indicate one standard deviation. A single isolate of DK4292 bearing Tn5 lac Ω4400 in an otherwise wild-type background was included as a positive control (▪).
DISCUSSION
Our characterization of the Ω4400 regulatory region provides the first comprehensive examination of a partially C-signal-dependent promoter region in M. xanthus. The mutational analysis of the Ω4400 promoter region indicated that the C box centered at −49 bp functions differently than the same sequence in the absolutely C-signal-dependent Ω4403 promoter region. Also, the C box centered at −80 bp in the Ω4400 regulatory region is not essential for expression, unlike the C boxes centered at −49 bp in the Ω4400 and Ω4403 promoter regions. The picture that emerges from our mutational analysis is that, in addition to the −10 region, there are two regions important for expression of the Ω4400 promoter. One region spans from at least −60 bp through the promoter −35 region and is essential for expression. The other region lies between −86 and −81 bp and only exerts a two- to fourfold positive effect on expression. This picture is quite different from the one that emerged from a mutational analysis of the Ω4403 promoter region (53), as discussed further below. Because these two promoters exhibit different degrees of dependence on C signaling (2, 7), our findings provide the first insight into how differential regulation of C-signal-dependent genes is achieved. Moreover, our results show that the region responsible for conveying partial C signal dependence in the Ω4400 promoter includes, but may not be limited to, the region from −86 to −81 bp. We also showed that expression of the Ω4400 promoter tracks with the levels of CsgA expression in actB and actC mutants and that the effects of the mutations on expression can be rescued by mixing with wild-type cells, demonstrating that the Ω4400 promoter is very responsive to the level of C signaling. Finally, we showed that Ω4400 expression is completely dependent on sigD and partially dependent on sigE.
The C box element has been found in the regulatory regions of several C-signal-dependent genes, including csgA, Ω4499, and fruA (6). However, when the patterns of mutational effects on C boxes of identical sequences located at −49 bp in the Ω4400 and Ω4403 promoter regions were compared, striking differences were observed at four of the seven positions (Fig. 1B). If these C boxes are bound by transcription factors, as seems likely, the results suggest that different proteins bind in different ways to the identical sequence in the two promoter regions. Alternatively, a single protein might bind differently to the C boxes in the two promoter regions by adopting different conformations, possibly due to interactions with other proteins or with DNA surrounding the C boxes. In either case, the protein(s) involved seems most likely to be a transcriptional activator(s) rather than a σ factor(s), as the C boxes centered at −49 bp are located farther upstream than the regions typically recognized by σ. Another possibility is that C boxes function in a manner analogous to UP elements, which are AT-rich sequences typically located between −60 and −40 bp that interact with the C-terminal domain of the α subunit of RNA polymerase (39, 40). According to this model, the M. xanthus α subunit would have to interact differently with the C boxes in the two promoters in order to explain our results.
A 7-bp mutation of the C box centered at −80 bp in the Ω4400 regulatory region indicated that this element is not essential for expression (Fig. 3). Furthermore, none of the single-base-pair mutations within this C box showed even a twofold effect on expression (Table 2). The 3-bp sequence directly upstream of this C box appeared to have more of an effect when it was mutated, although it still exhibited only a fourfold decrease in expression (Fig. 3). It is possible that a transcriptional activator binds to this region. This putative activator may mediate the response of the Ω4400 promoter to C signaling, because a 3-bp change at −86 to −84 bp made the promoter oblivious (Fig. 4A) to the normal threefold reduction in expression caused by a csgA mutation (2). In contrast to the partial dependence on C signaling of the wild-type Ω4400 promoter, expression of the Ω4403 promoter depends absolutely on C signaling (7). Interestingly, in the Ω4403 regulatory region, a 10-bp element from −79 to −70 bp is absolutely required for expression (53). Perhaps this element mediates the absolute dependence of Ω4403 expression on C signaling. For example, the same or a different transcriptional activator may bind differentially in response to C signaling to the 10-bp element in the Ω4403 promoter region and to the region from −86 to −81 bp upstream of Ω4400. As noted previously, the region from −84 to −73 bp upstream of Ω4400 matches the Ω4403 10-bp element at only six positions, but it matches a sequence between −72 and −61 bp in the partially C-signal-dependent Ω4499 promoter at 9 of 12 positions (53).
Deletion analysis of the Ω4400 promoter region suggested that there is surprising complexity in the transcriptional regulation of this gene. In addition to the 86 bp of upstream DNA that is required for full expression, an element between 25 and 155 bp exerts a weak positive effect (Fig. 2). Also, a strong negative element between 155 and 455 bp acts only in the absence of the region between −86 and −73 bp (Fig. 2) (2). Above, we speculated that the region between −86 and −81 bp interacts with a transcriptional activator that mediates the response to C signaling. Perhaps the transcription complex assembled in the presence of this putative activator is more resistant to premature termination as RNA polymerase traverses the region from 155 to 455 bp. Participation of an upstream sequence in transcriptional antitermination would be unusual since such mechanisms in prokaryotes typically involve sequences downstream of the transcriptional start site (13).
The developmentally regulated fruA promoter of M. xanthus also has a downstream regulatory sequence that acts negatively; however, this negative element functions with a heterologous promoter (15), suggesting that it does not require a particular upstream sequence, as appears to be the case for the Ω4400 promoter.
The Ω4400 regulatory region has a 5-bp element similar to that found centered at −61 bp in the Ω4403 promoter region (53). As shown in Fig. 3, a 4-bp mutation from −63 to −60 bp eliminated activity, as did a mutation at −59 bp. The loss of activity is in accordance with the effect obtained by mutating the 5-bp element in the Ω4403 promoter region (53). However, in the Ω4400 promoter region, a single-base change at −60 bp (the most conserved of all five bases in the 5-bp element) from C to A caused a complete loss of activity (Table 2). In the Ω4403 promoter region, the same mutation increased activity 1.8-fold (53). The differential effects of mutations within the 5-bp element further support the conclusion that the Ω4400 promoter is regulated differently than the Ω4403 promoter.
Additional evidence for differences between the Ω4400 and Ω4403 promoter regions comes from a comparison of the effects of mutations between the 5-bp elements and C boxes centered at −49 bp and a comparison of the effects of mutations in the −35 regions. A mutation of the 5-bp sequence spanning from −58 to −54 bp led to a strong decrease in activity of the Ω4400 promoter (Fig. 3). In contrast, a mutation spanning from −58 to −54 bp led to a 1.6-fold increase in Ω4403 promoter activity (53). Whereas the region from at least −41 to −36 bp (and perhaps as large as the region from −45 to −31 bp) was shown to be essential for expression of the Ω4400 promoter (Fig. 3), a mutant with a multiple-base-pair change in the −35 region of the Ω4403 promoter was shown to retain 60% of the activity (53). Sequence analysis of the Ω4400 regulatory region revealed an imperfect inverted repeat from −48 to −27 bp that could potentially be a recognition site for a dimeric DNA-binding protein that activates transcription. Classic examples of this type of regulator include the cI protein of phage λ, which binds in a dimeric fashion to the region from −51 to −35 bp upstream of the PRM promoter (37), and the cyclic AMP receptor protein of E. coli, which binds in a dimeric fashion to a similar region upstream of the melR and galP1 class II promoters (38).
One similarity between the Ω4400 and Ω4403 promoter regions is that in both cases a single-base-pair change at −53 bp strongly decreased promoter activity (Fig. 3) (53). This position was not included in the C box consensus sequence because the nucleotide found at this position was variable in the nine sequences used to generate the consensus (6). Four of the nine C box sequences have been subjected to single-base-pair changes, and in each case the pattern of effects on promoter activity is different (Fig. 1B and Table 2) (unpublished data). Clearly, these C boxes are not bound by a protein(s) in the same way. While the C box consensus sequence has been useful in identifying regions important for developmental promoter activity, the concept of a C box adhering to the initially proposed CAYYCCY consensus sequence (6) or even the more degenerate (C/A)(A/C)Y(C/A)CC(T/G) consensus sequence proposed subsequently (53) no longer appears to be useful.
Regulation of the Ω4400 promoter was also studied by measuring expression in mutants. We found that expression of Ω4400 correlated with the timing and level of CsgA production in actB and actC mutants (Fig. 5). This is consistent with the behavior of two other partially C-signal-dependent promoters, Ω4414 and Ω4499, and also with the behavior of the absolutely C-signal-dependent Ω7536 promoter (9). We also showed that the defects in Ω4400 expression in the act mutants could be corrected by codevelopment with wild-type cells. This is the first demonstration of extracellular complementation of act mutants. The results indicate that the actB and actC genes do not affect Ω4400 promoter expression in a cell-autonomous fashion. Rather, actB and actC affect Ω4400 expression by altering extracellular C signaling. Likewise, other defects observed for act mutants, such as blocked or reduced sporulation (10), may be due to altered C signaling, and it may be possible to rescue these defects by codevelopment with wild-type cells.
The Ω4400 promoter was not expressed in a sigD mutant (Fig. 6). The product of sigD, σD, is known to function during the transition between growth and development (51), although its exact role has yet to be elucidated. The sigD mutant did not aggregate under the conditions which we used, suggesting that there was an early block in development. Hence, it seems likely that the effects of the sigD mutation on Ω4400 promoter expression are indirect. However, we cannot rule out the possibility of a direct effect, especially since our mutational analysis showed that the promoter −35 and −10 regions are essential for expression, suggesting that a σ70 family member (such as σD) recognizes this promoter. The Ω4400 promoter in a sigE mutant exhibited severely reduced expression (Fig. 6), although this mutant seemed to aggregate normally. The effect of the sigE mutation could indicate that σE RNA polymerase is partially responsible for Ω4400 expression. The σB and σC sequences are similar to the σE sequence, and there may be functional redundancy among these σ factors (52). It is also possible that the sigE mutation affects Ω4400 expression by an indirect mechanism.
The results of this study should facilitate identification of proteins that regulate expression of the Ω4400 promoter during development. Very few developmental transcription factors have been identified in M. xanthus. They include ActB (10), MrpC (48), protein X (15), and FruA (36). ActB probably does not bind to the Ω4400 promoter region since expression of Ω4400 in an actB mutant was restored to the normal level upon codevelopment with wild-type cells (Fig. 5A). MrpC binds to two sets of inverted repeats in the region from −154 to −107 bp upstream of fruA (50). The sequences to which MrpC binds in the fruA promoter region are not found in the Ω4400 promoter region, providing no indication that this protein binds to the Ω4400 regulatory region. Likewise, the Ω4400 regulatory region does not exhibit the short sequence found at 78 to 94 bp downstream of the fruA transcription start site, which is bound by protein X (15). The best candidate for a protein that binds to the region upstream of the Ω4400 promoter is FruA, a putative response regulator with no known sensor kinase (5, 36). Expression of Ω4400 is absolutely dependent on fruA (L. Sogaard-Andersen, personal communication), but FruA has not yet been reported to bind DNA. Of course, it is also possible that the transcription factors that directly regulate the Ω4400 promoter have yet to be identified.
Acknowledgments
We thank G. Velicer, J. Brandner, and D. Oluwole for constructing the pGV, pJB, and pDO plasmids, respectively, listed in Table 1. We are grateful to D. Kaiser, Y. Cheng, T. Ueki, and S. Inouye for providing bacterial strains. We thank P. Viswanathan and D. Srinivasan for critical reading of the manuscript.
This research was supported by NSF grant MCB-0090478 and by the Michigan Agricultural Experiment Station.
REFERENCES
- 1.Biran, D., and L. Kroos. 1997. In vitro transcription of Myxococcus xanthus genes with RNA polymerase containing σA, the major sigma factor in growing cells. Mol. Microbiol. 25:463-472. [DOI] [PubMed] [Google Scholar]
- 2.Brandner, J. P., and L. Kroos. 1998. Identification of the Ω4400 regulatory region, a developmental promoter of Myxococcus xanthus. J. Bacteriol. 180:1995-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos, J. M., and D. R. Zusman. 1975. Regulation of the development in Myxococcus xanthus: effect of 3′:5′-cyclic AMP, ADP, and nutrition. Proc. Natl. Acad. Sci. USA 72:518-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dworkin, M. 1996. Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 60:70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellehauge, E., M. Norregaard-Madsen, and L. Sogaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol. Microbiol. 30:807-817. [DOI] [PubMed] [Google Scholar]
- 6.Fisseha, M., D. Biran, and L. Kroos. 1999. Identification of the Ω4499 regulatory region controlling developmental expression of a Myxococcus xanthus cytochrome P-450 system. J. Bacteriol. 181:5467-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisseha, M., M. Gloudemans, R. Gill, and L. Kroos. 1996. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J. Bacteriol. 178:2539-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geisselsoder, J., J. M. Campos, and D. R. Zusman. 1978. Physical characterization of bacteriophage Mx4, a generalized transducing phage for Myxococcus xanthus. J. Mol. Biol. 119:179-189. [DOI] [PubMed] [Google Scholar]
- 9.Gronewold, T. M., and D. Kaiser. 2002. act operon control of developmental gene expression in Myxococcus xanthus. J. Bacteriol. 184:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gronewold, T. M., and D. Kaiser. 2001. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol. Microbiol. 40:744-756. [DOI] [PubMed] [Google Scholar]
- 11.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557. [DOI] [PubMed] [Google Scholar]
- 13.Henkin, T. M., and C. Yanofsky. 2002. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24:700-707. [DOI] [PubMed] [Google Scholar]
- 14.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of motility in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. USA 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horiuchi, T., T. Akiyama, S. Inouye, and T. Komano. 2003. Regulation of FruA expression during vegetative growth and development of Myxococcus xanthus. J. Mol. Microbiol. Biotechnol. 5:87-96. [DOI] [PubMed] [Google Scholar]
- 16.Jelsbak, L., and L. Sogaard-Andersen. 1999. The cell surface-associated intercellular C-signal induces behavioral changes in individual Myxococcus xanthus cells during fruiting body morphogenesis. Proc. Natl. Acad. Sci. USA 96:5031-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jelsbak, L., and L. Sogaard-Andersen. 2002. Pattern formation by a cell surface-associated morphogen in Myxococcus xanthus. Proc. Natl. Acad. Sci. 99:2032-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser, D. 1984. Genetics of myxobacteria, p. 163-184. In E. Rosenberg (ed.), Myxobacteria: development and cell interactions. Springer-Verlag, New York, N.Y.
- 19.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashefi, K., and P. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthux frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S. K., and D. Kaiser. 1990. Cell alignment required in differentiation of Myxococcus xanthus. Science 249:926-928. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. K., and D. Kaiser. 1990. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 4:896-905. [DOI] [PubMed] [Google Scholar]
- 23.Kim, S. K., and D. Kaiser. 1991. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J. Bacteriol. 173:1722-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19-26. [DOI] [PubMed] [Google Scholar]
- 25.Kim, S. K., and D. Kaiser. 1990. Purification and properties of Myxococcus xanthus C-factor, an intercellular signaling protein. Proc. Natl. Acad. Sci. USA 87:3635-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroos, L., P. Hartzell, K. Stephens, and D. Kaiser. 1988. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev. 2:1677-1685. [DOI] [PubMed] [Google Scholar]
- 27.Kroos, L., and D. Kaiser. 1984. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 81:5816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroos, L., and D. Kaiser. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1:840-854. [DOI] [PubMed] [Google Scholar]
- 29.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 30.Kruse, T., S. Lobedanz, N. M. Berthelsen, and L. Sogaard-Andersen. 2001. C-signal: a cell surface-associated morphogen that induces and co-ordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol. Microbiol. 40:156-168. [DOI] [PubMed] [Google Scholar]
- 31.LaRossa, R., J. Kuner, D. Hagen, C. Manoil, and D. Kaiser. 1983. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J. Bacteriol. 153:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, B.-U., K. Lee, J. Mendez, and L. Shimkets. 1995. A tactile sensory system of Myxococcus xanthus involves an extracellular NAD(P)+-containing protein. Genes Dev. 9:2964-2973. [DOI] [PubMed] [Google Scholar]
- 33.Li, S.-F., B. Lee, and L. J. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 34.Licking, E., L. Gorski, and D. Kaiser. 2000. A common step for changing cell shape in fruiting body and starvation-independent sporulation of Myxococcus xanthus. J. Bacteriol. 182:3553-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lisser, S., and H. Margalit. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22:757-767. [DOI] [PubMed] [Google Scholar]
- 37.Ptashne, M. 1992. A genetic switch: phage λ and higher organisms, 2nd ed. Cell Press and Blackwell Scientific Publications, Cambridge, Mass.
- 38.Rhodius, V., D. West, C. Webster, S. Busby, and N. Savery. 1997. Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res. 25:326-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross, W., A. Ernst, and R. L. Gourse. 2001. Fine structure of E. coli RNA polymerase-promoter interactions: alpha subunit binding to the UP element minor groove. Genes Dev. 15:491-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 41.Sager, B., and D. Kaiser. 1994. Intercellular C-signaling and the traveling waves of Myxococcus. Genes Dev. 8:2793-2804. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Shimkets, L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 44.Shimkets, L. J., and S. J. Asher. 1988. Use of recombination techniques to examine the structure of the csg locus of Myxococcus xanthus. Mol. Gen. Genet. 211:63-71. [DOI] [PubMed] [Google Scholar]
- 45.Shimkets, L. J., and H. Rafiee. 1990. CsgA, an extracellular protein essential for Myxococcus xanthus development. J. Bacteriol. 172:5299-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sogaard-Andersen, L., M. Overgaard, S. Lobedanz, E. Ellehauge, L. Jelsbak, and A. A. Rasmussen. 2003. Coupling gene expression and multicellular morphogenesis during fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 48:1-8. [DOI] [PubMed] [Google Scholar]
- 47.Sogaard-Andersen, L., F. Slack, H. Kimsey, and D. Kaiser. 1996. Intercellular C-signaling in Myxococcus xanthus involves a branched signal transduction pathway. Genes Dev. 10:740-754. [DOI] [PubMed] [Google Scholar]
- 48.Sun, H., and W. Shi. 2001. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J. Bacteriol. 183:4786-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thony-Meyer, L., and D. Kaiser. 1993. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J. Bacteriol. 175:7450-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ueki, T., and S. Inouye. 2003. Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100:8782-8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueki, T., and S. Inouye. 1998. A new sigma factor, SigD, essential for stationary phase is also required for multicellular differentiation in Myxococcus xanthus. Genes Cells 3:371-385. [DOI] [PubMed] [Google Scholar]
- 52.Ueki, T., and S. Inouye. 2001. SigB, SigC, and SigE from Myxococcus xanthus homologous to σ32 are not required for heat shock response but for multicellular differentiation. J. Mol. Microbiol. Biotechnol. 3:287-293. [PubMed] [Google Scholar]
- 53.Viswanathan, P., and L. Kroos. 2003. cis elements necessary for developmental expression of a Myxococcus xanthus gene that depends on C signaling. J. Bacteriol. 185:1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]