Abstract
The structures of glycans N-linked to Arabidopsis proteins have been fully identified. From immuno- and affinodetections on blots, chromatography, nuclear magnetic resonance, and glycosidase sequencing data, we show that Arabidopsis proteins are N-glycosylated by high-mannose-type N-glycans from Man5GlcNAc2 to Man9GlcNAc2, and by xylose- and fucose (Fuc)-containing oligosaccharides. However, complex biantenary structures containing the terminal Lewis a epitope recently reported in the literature (A.-C. Fitchette-Lainé, V. Gomord, M. Cabanes, J.-C. Michalski, M. Saint Macary, B. Foucher, B. Cavalier, C. Hawes, P. Lerouge, and L. Faye [1997] Plant J 12: 1411–1417) were not detected. A similar study was done on the Arabidopsis mur1 mutant, which is affected in the biosynthesis of l-Fuc. In this mutant, one-third of the Fuc residues of the xyloglucan has been reported to be replaced by l-galactose (Gal) (E. Zablackis, W.S. York, M. Pauly, S. Hantus, W.D. Reiter, C.C.S. Chapple, P. Albersheim, and A. Darvill [1996] Science 272: 1808–1810). N-linked glycans from the mutant were identified and their structures were compared with those isolated from the wild-type plants. In about 95% of all N-linked glycans from the mur1 plant, l-Fuc residues were absent and were not replaced by another monosaccharide. However, in the remaining 5%, l-Fuc was found to be replaced by a hexose residue. From nuclear magnetic resonance and mass spectrometry data of the mur1 N-glycans, and by analogy with data reported on mur1 xyloglucan, this subpopulation of N-linked glycans was proposed to be l-Gal-containing N-glycans resulting from the replacement of l-Fuc by l-Gal.
Mutants have been widely used for biochemical studies of N-glycosylation in yeast and mammals. Although a rapidly increasing number of mutants are available in several plant species and particularly in Arabidopsis, only one of them presents a clearly identified mutation that affects the biosynthesis of N-linked glycans. This Arabidopsis cgl mutant lacks the N-acetyl glucosaminyltransferase I and is unable to synthesize complex-type N-glycans (von Schaewen et al., 1993). Another Arabidopsis mutant, mur1, does not synthesize l-Fuc in the leaves (Reiter et al., 1993). This mutant was found to be affected in the gene encoding for a GDP-d-Man-4,6-dehydratase, a key enzyme in the biosynthesis of l-Fuc (Bonin et al., 1997). The structure of cell wall polysaccharides from plants carrying the mur1 mutation has been investigated recently (Zablackis et al., 1996) and compared with cell wall polysaccharides from wild-type plants (Zablackis et al., 1995). This study has shown that in the xyloglucan of this Fuc-deficient mutant, l-Fuc is replaced by l-Gal, a monosaccharide structurally similar to l-Fuc, without affecting the biological activity of Fuc-containing oligosaccharides derived from these polymers.
l-Fuc is a monosaccharide that is found not only in primary cell wall polysaccharides but also in the plant N-linked oligosaccharides. Consequently, the study of N-glycosylation in the mur1 mutant could provide information on the biosynthesis and roles of glycans N-linked to proteins when l-Fuc is not available in Arabidopsis. In plants the N-glycosylation of proteins starts by the transfer in the ER of the oligosaccharide precursor Glc3Man9GlcNAc2, which is subsequently modified during the transport of the glycoprotein by glycosidases and glycosyltransferases located in the ER, the Golgi apparatus, and the vacuole. These enzymes convert the precursor to high-mannose-type N-glycans ranging from Man9GlcNAc2 to Man5GlcNAc2 and eventually to complex-type N-glycans having an α(1,3)-Fuc and/or a β(1,2)-Xyl residue (for review, see Lerouge et al., 1998). Recently, some complex N-linked glycans have been described as having two antennae containing a Lewis a epitope that is usually found on cell-surface mammalian glycoconjugates (Fitchette-Lainé et al., 1997; Melo et al., 1997). These structures result from the elongation of the intermediate complex-type N-glycan by action of α(1,4)-fucosyltransferase and β(1,3)-galactosyltransferase. Complex-type N-glycans have been described as being N-linked to extracellular glycoproteins. In contrast, vacuolar glycoproteins are N-glycosylated by modified oligosaccharides having only an α(1,3)-Fuc and/or a β(1,2)-Xyl residue linked to the core Man3GlcNAc2 and are named paucimannosidic-type N-glycans (Lerouge et al., 1998). These oligosaccharides arise from the elimination from complex-type N-glycans of terminal residues in the vacuole (Vitale and Chrispeels, 1984).
We report in this paper the first complete structural analysis, to our knowledge, of N-linked glycans isolated from leaves of Arabidopsis. The strategy developed in the first part of our study was applied to the N-glycosylation analysis of proteins isolated from the mur1 mutant to analyze the effects of the absence of l-Fuc on the structure and the biosynthesis of N-linked oligosaccharides. We show that in wild-type Arabidopsis, the proteins are N-glycosylated by high-mannose-type N-glycans and by Xyl- and Fuc-containing N-glycans previously described in other plants. Complex biantennary structures containing terminal Lewis a epitopes were not detected. Furthermore, as found previously in the cell wall xyloglucan (Zablackis et al., 1996), the analytical data suggest a partial but less prominent replacement of l-Fuc by l-Gal in the core of the N-glycans in the mur1 mutant.
MATERIALS AND METHODS
Enzymes, Chemicals, and Plant Material
PNGase A from almond was purchased from Boehringer-Mannheim. Pepsin and β-N-acetylglucosaminidase from Jack bean were from Sigma. BioGel P4 and AG50W-X2 were from Bio-Rad. C18 Bond Elut cartridges were from Varian (Sugarland, TX). 2-aminopyridine (99% purity) was from Aldrich.
Wild-type Arabidopsis and mutant plants were grown under long-day conditions in the greenhouse, and leaf material was harvested from plants in early bolting stages.
SDS-PAGE and Western-Blot Analyses
Proteins isolated from leaves (as described in the next paragraph) were resolved by SDS-PAGE in a homogeneous 15% polyacrylamide gel under reducing conditions according to the method of Laemmli (1970), and electrophoretically transferred to nitrocellulose membrane. Affinodetection using the concanavalin A/peroxidase method (Faye and Chrispeels, 1985) and immunodetection with antibodies directed against Fuc- or Xyl-containing plant N-glycans were carried out, respectively, as described in Faye et al. (1993).
Preparation of N-Linked Glycans from Protein Extracts Isolated from Wild-Type and mur1 Plants
Protein extracts from Arabidopsis were obtained by homogenizing 100 g of freeze-dried leaf material in 2 L of 50 mm Hepes (pH 7.5) and 2 mm sodium bisulfite containing 0.1% SDS. Insoluble material was eliminated by centrifugation at 4400g for 15 min at 4°C. A second protein extraction was performed using the pellet. The supernatants were pooled and filtered through Miracloth (Calbiochem). Proteins were precipitated at 0°C overnight with 12.5% TCA (final concentration), and the pellet was then washed three times with 90% (v/v) acetone. The proteins were digested with pepsin in 30 mL of 0.01 n HCl, pH 2.2, at 37°C for 48 h. Pepsin (20 mg) was dissolved in 1 mL of buffer and one-half of the enzyme solution was added at 0 and 24 h, respectively. The pepsin digest was neutralized with NaOH, heated at 100°C for 5 min, and lyophilized. The sample was desalted on a BioGel P4 column (70 × 2.6 cm) equilibrated with 0.1 n acetic acid. A portion of each fraction was assayed for monosaccharide composition. Glycopeptide fractions were pooled and then deglycosylated with 1 milliunit of PNGase A in a 100 mm sodium acetate buffer (pH 5.0) overnight at 37°C (Tomiya et al., 1987). N-glycans were purified by successive elutions through a AG50W-X2 column (5 × 1 cm) and a C18 Bond Elut cartridge. Complex-type N-glycans were separated from high-mannose-type oligosaccharides by affinity chromatography on a concanavalin A-Sepharose 4B column as described in Montreuil et al. (1986).
Fluorescent Derivatization of N-Glycans with 2-Aminopyridine
Freeze-dried N-linked oligosaccharides were coupled to 2-aminopyridine as described previously by Hase et al. (1984). PA oligosaccharides were separated from the excess of reagent by gel permeation on a BioGel P4 column (40 × 1.6 cm) equilibrated in 50 mm ammonium acetate buffer, pH 6.0.
HPAEC Analysis
Chromatography of N-linked glycans was achieved by HPAEC-PAD. Analytical HPAEC-PAD was performed on a DX500 system equipped with a GP 40 gradient pump, an ED 40 detector, and a CarboPac PA1 column (250 × 4.6 mm, Dionex, Sunnyvale, CA). Analysis of the monosaccharide composition was carried out by hydrolysis of the glycan fraction with 2 n trifluoroacetic acid at 100°C for 2 h, followed by analysis of the resulting monosaccharides using 16 mm NaOH at 1 mL min−1 as described in Hardy (1989). Elution of the high-mannose-type N-glycans was carried out using a linear gradient from 0 to 100 mm NaOAc in 100 mm NaOH at 1 mL min−1 over 30 min.
HPAEC of PA oligosaccharides was achieved by dissolving the sample in 0.1 n NaOH and elution on a CarboPac PA1 column using a gradient from 0 to 100 mm NaOAc in 100 mm NaOH at 1 mL min−1 over 60 min. PA oligosaccharides were detected using a FL 2000 system (Spectra-Physics, San Jose, CA) fluorescence detector with excitation and emission wavelengths at 320 and 400 nm, respectively. PA oligosaccharides were collected and desalted by gel filtration on a Sephadex G-10 column (30 × 1 cm) equilibrated and eluted in 50 mm sodium acetate, pH 6.0.
Reverse-Phase HPLC
A liquid chromatograph equipped with a SFM-25 fluorescence spectrophotometer (Kontron, Milan, Italy) was used. PA oligosaccharides were separated on a Spherisorb ODS2 C18 column (250 × 4.6 mm, Prolabo, Creteil, France). Elution of PA oligosaccharides was performed at a flow rate of 1 mL min−1 at 55°C using solvent A (10 mm sodium phosphate buffer, pH 3.8), and solvent B (10 mm sodium phosphate buffer, pH 3.8, 0.5% 1-butanol) (Tomiya et al., 1988). After injection of the sample, the ratio of solvent B to A increased with a linear gradient from 20:80 to 50:50 over 60 min. PA oligosaccharides were detected by fluorescence using excitation and emission wavelengths of 320 and 400 nm, respectively.
β-N-Acetylglucosaminidase Digestion of PA Oligosaccharides
PA oligosaccharides were isolated by HPAEC and then digested with 1 unit of β-N-acetylglucosaminidase in a 50 mm sodium acetate buffer, pH 5.0, at 25°C. The digest was analyzed by HPAEC with fluorescence detection after 1 and 18 h of incubation time.
NMR
1H-NMR was performed at 600 MHz on a spectrometer (model DMX600, Bruker, Billerica, MA). The spectrum was recorded at 297 K in 2H oxide (2H2O 99.97). Chemical shifts were expressed in parts per million downfield from internal tetrain etnyl silare and with an 2H water presaturation sequence.
Electrospray MS
The electrospray ionization mass experiments were carried out on a triple-quadrupole mass spectrometer (Quattro II, Micromass, Manchester, UK). Samples were dissolved in water:methanol:acetic acid (49:49:2) at a concentration of approximatively 50 pmol μL−1 and the solution was infused into the electrospray ion source by a syringe pump (Harvard Apparatus, South Natick, MA). Optimization of the parameters was performed both in the MS and MS-MS mode using (Glc)n-PA. Typically, best conditions were defined for a cone voltage of about 60 V, a collision energy range between 30 and 40 eV, and an Ar gas pressure at 4.0 mTorr. Spectra were recorded at a scan speed of 10 s and were smoothed 1 time.
RESULTS
Western-Blot Analysis of Protein Extracts from Wild-Type and mur1 Arabidopsis Plants
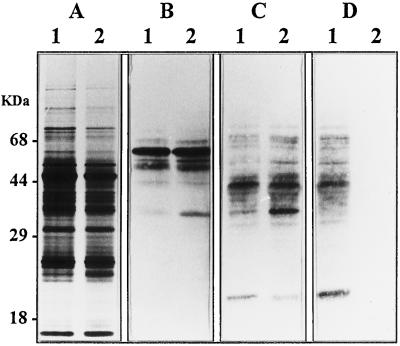
A first analysis of the N-linked glycans associated to proteins isolated from leaves of the wild-type and the mur1 mutant of Arabidopsis was obtained by affino- and immunodetections on blots using specific probes. We used antibodies of well-established specificities for the different oligosaccharide epitopes that are found in plant N-glycans, i.e. the β(1,2)-Xyl residue and the α(1, 3)-Fuc residue linked to the chitobiose unit (Faye et al., 1993). Glycoproteins from wild-type plants were detected on western blots by affinodetection using concanavalin A (Faye and Chrispeels, 1985) (Fig. 1B, lane 1), as well as by immunoblotting with antibodies directed against the β(1,2)-Xyl residue (Fig. 1C, lane 1) or directed against the α-1,3 Fuc residue (Fig. 1D, lane 1) linked to the core Man3GlcNAc2 (Faye et al., 1993). Similar results were obtained on the protein extract from mur1 plants, except that no protein was detected by the antibodies specific for the α(1,3)-Fuc residue linked to the proximal glucosamine unit (Fig. 1D, lane 2). Some differences in the detection level of some glycoproteins (see the glycoprotein band at 35 kD) were observed, but the identity of these glycoproteins was not investigated further. Protein extracts from both the wild-type and mur1 plants were also analyzed by immunoblotting with antibodies directed against Lewis a epitopes (data not shown). In contrast to results obtained in other plants, only very weak signals were detected in both extracts, confirming that such epitopes are almost completely absent in Arabidopsis leaves, as previously reported in Fitchette-Lainé et al. (1997). As a consequence, these results indicate that Arabidopsis leaf proteins are N-glycosylated by both high-mannose and paucimannosidic or complex-type N-glycans of limited size (no Lewis a-containing N-glycans). Similar results were obtained with the mur1 mutant, except for the absence of detection of the α(1,3)-Fuc epitope as expected from the previous results on this mutant (Reiter et al., 1993).
Figure 1.
SDS-PAGE and western-blot analysis of protein extracts from leaves of wild-type Arabidopsis (lanes 1) or mur1 mutant (lanes 2). A, Silver staining; B, affinodetection with concanavalin A; C, immunodetection with antibodies specific for β(1,2)-Xyl-containing plant N-glycans; D, immunodetection with antibodies specific for α(1,3)-Fuc-containing plant N-glycans.
Structural Identification of N-glycans Synthesized by Arabidopsis Wild-type and mur1 Plants
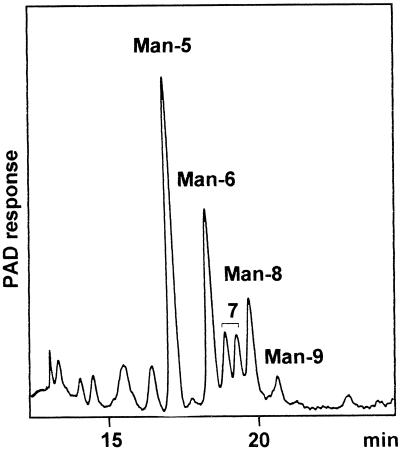
The complete structural analysis of N-linked glycans associated with proteins from leaves of wild-type Arabidopsis and mur1 plants was performed to investigate whether the partial replacement of l-Fuc by l-Gal observed in the cell wall xyloglucan (Zablackis et al., 1996) also occurs in the glycoproteins of the mur1 mutant. Since the N-glycosylation of proteins in the wild-type Arabidopsis has not yet, to our knowledge, been investigated in detail, N-linked glycans from both the wild-type and the Fuc-deficient mutant were analyzed. After TCA precipitation proteins from both wild-type and mur1 leaves were digested with pepsin. The resulting glycopeptides were isolated by size-exclusion chromatography on a BioGel P4 column and the fraction corresponding to glycopeptides from the wild-type and the mur1 plants were further treated with PNGase A. PNGase A from sweet almond is able to release both high-mannose-type and modified plant N-glycans presenting or not presenting an α(1,3)-Fuc residue linked to the proximal glucosamine unit (Taga et al., 1984). The reducing N-linked oligosaccharides released from the wild-type Arabidopsis and mur1 leaves were fractionated by affinity chromatography on a concanavalin A-Sepharose 4B column to separate paucimannosidic and complex-type N-glycans (concanavalin A− fraction) from high-mannose-type N-glycans (concanavalin A+ fraction). The HPAEC-PAD profile of the concanavalin A+ fraction isolated from the wild-type plants (Fig. 2) shows a mixture of high-mannose-type N-glycans, which were identified as structures from Man5GlcNAc2 to Man9GlcNAc2 (Man-5 to Man-9, see Fig. 3) by comparison of their retention times in HPAE-PAD chromatography with standard oligosaccharides from porcin thyroglobulin as previously described (Rayon et al., 1996). The high-mannose-type N-glycans isolated from the mur1 plants were found to be identical and in the same ratio as those of the wild-type plants (data not shown).
Figure 2.
HPAEC-PAD profile of high-mannose-type N-glycans Man5GlcNAc2 to Man9GlcNAc2 (Man-5 to Man-9) isolated from wild-type Arabidopsis leaves. Corresponding structures are represented in Figure 3.
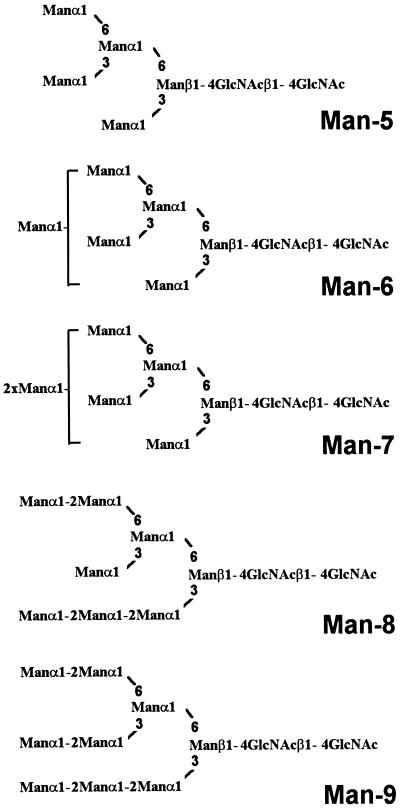
Figure 3.
Abbreviations and structures of high-mannose-type N-glycans Man5GlcNAc2 to Man9GlcNAc2 (Man-5 to Man-9) isolated from wild-type Arabidopsis leaves.
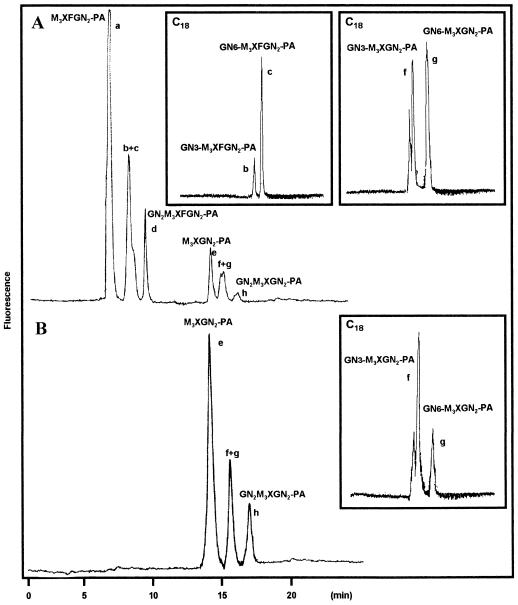
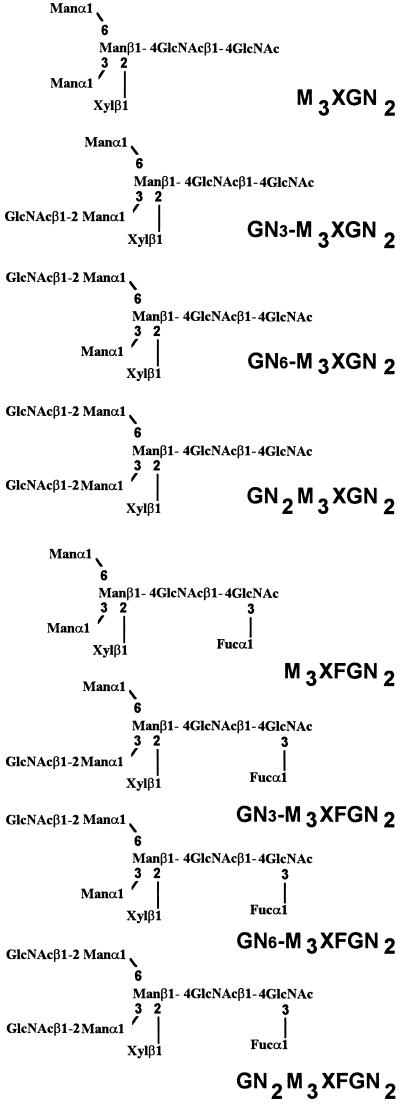
The N-glycans contained in the concanavalin A− fraction from both the wild-type and the mur1 plants were analyzed by HPAEC-PAD. The profiles were composed of a complex mixture of unresolved peaks. As a consequence, these oligosaccharides were reductively aminated with 2-aminopyridine, and the resulting PA oligosaccharides were then analyzed with HPAEC using a fluorescence detector. Under these conditions all labeled glycans from both preparations were found to be well resolved (Fig. 4). The elution profile of the PA oligosaccharides obtained from wild-type plants showed the presence of six major compounds (Fig. 4A). The structures of these oligosaccharides were identified by comparison with standard glycans and by glycosidase sequencing. Compounds a through h were collected separately. Glycans a and e were identified as the PA derivatives of Man3XylFucGlcNAc2 (M3XFGN2, Fig. 5) and Man3XylGlcNAc2 (M3XGN2, Fig. 5) oligosaccharides, respectively, by comparison of the retention time with reference standards. The structures of the other oligosaccharides were further determined after digestion with β-N-acetylglucosaminidase, an enzyme that specifically releases terminal β-GlcNAc residues. The material collected in peak d was found to be progressively converted to compounds b+c and then to compound a during β-N-acetylglucosaminidase treatment (Fig. 6). β-N-acetylglucosaminidase action on the material in peak b+c released a single β-GlcNAc residue, leading to compound a. As a consequence, compounds b+c and d were identified as Xyl and Fuc-containing oligosaccharides bearing one or two terminal glucosamine residues, respectively. Peaks f+g and h were analyzed in the same manner and were found to correspond to Xyl-containing glycans bearing one and two terminal glucosamine residues, respectively. Peaks b+c and f+g were subjected to a second chromatography analysis using a reverse-phase C18 column. In this case, both fractions were shown to be split further into two components (Fig. 4A, including boxed insets). PA derivatives of plant complex-type N-glycans bearing a single terminal GlcNAc residue have been described to be efficiently separated on a reverse-phase column. The isomers having the glucosamine unit associated to the mannose arm α(1,3)-linked to the β-mannose residue (GN3-M3XFGN2 and GN3-M3XGN2) are eluted sooner in these conditions than N-linked glycans having a terminal GlcNAc residue on the α(1,6)-antennae (Takahashi et al., 1986; Hayashi et al., 1990). In conclusion, paucimannosidic and complex-type oligosaccharides N-linked to glycoproteins from Arabidopsis can be classified into two groups: Xyl- and Fuc-containing oligosaccharides having zero, one, or two terminal glucosamine residues (M3XFGN2 to GN2M3XFGN2, Fig. 5) and analogous nonfucosylated structures (M3XGN2 to GN2M3XGN2, Fig. 5).
Figure 4.
HPAEC profiles of the PA derivatives of paucimannosidic and complex-type N-glycans. N-glycans isolated from wild-type Arabidopsis (A) and mur1 mutant (B) leaves. The chromatograms presented in the boxed insets correspond to the C18 reverse-phase profiles of peaks b+c and f+g purified by HPAEC. Abbreviations refer to N-linked glycans represented in Figure 5.
Figure 5.
Abbreviations and structures of paucimannosidic and complex-type N-glycans isolated from Arabidopsis.
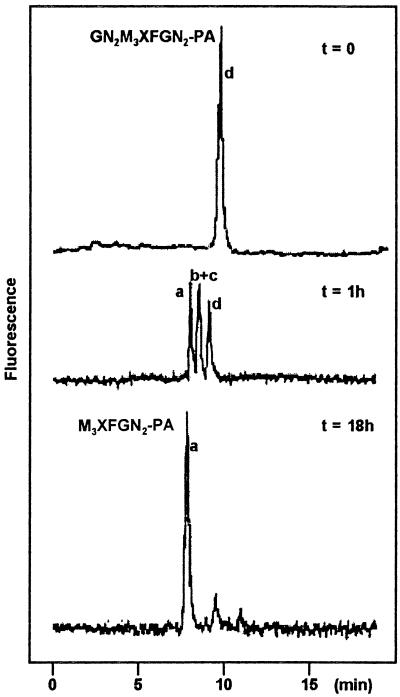
Figure 6.
HPAEC profiles of the β-N-acetylglucosaminidase digest of compound d after different incubation times. Peak a, PA derivative of M3XFGN2; peak b+c, PA derivatives of GN6-M3XFGN2 and GN3-M3XFGN2 (see Fig. 5).
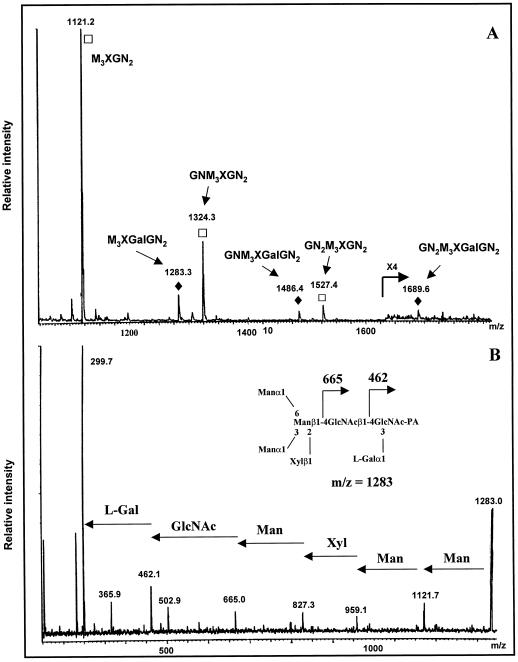
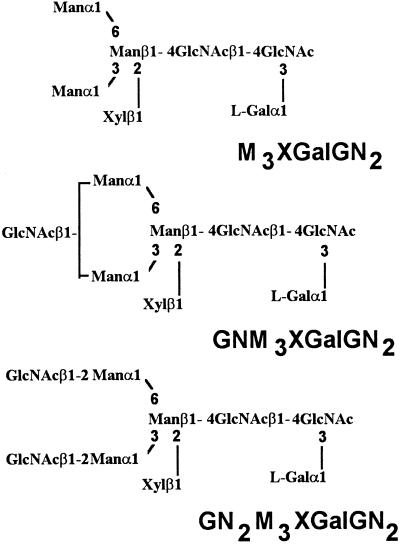
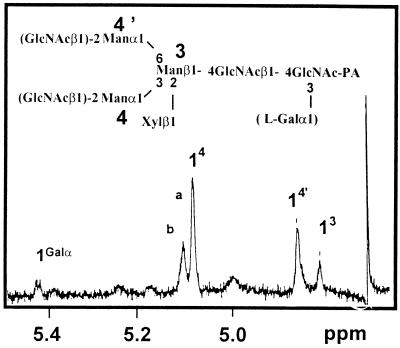
Paucimannosidic and complex-type N-glycans isolated from the mur1 plants were submitted to the same analysis. The nonfucosylated PA structures e to h (Fig. 4B), identical to those of the minor group found in the wild-type extract, were detected and identified using the same approach as M3XGN2 to GN2M3XGN2 oligosaccharides represented in Figure 5. To investigate the presence in the pool of mur1 N-glycans of minor oligosaccharides co-migrating with the xylosylated oligosaccharides and containing an additional hexose residue, the PA derivatives of complex and paucimannosidic-type N-glycans were then analyzed by electrospray MS (Fig. 7A). Three major (M+H)+ ions at m/z = 1121, 1324, and 1527 were assigned to the major xylosylated PA oligosaccharides M3XGN2 to GN2M3XGN2 identified by chromatography. However, a second family of ions was also detected at m/z = 1283, 1486, and 1689, having an additional 162 mass units. These ions were not detected in the wild type (data not shown) and were estimated to represent about 5% to 10% of the total oligosaccharides (Fig. 7A). These ions were supposed to correspond to (M+H)+ ions of the PA l-Gal-containing N-glycans M3XGalGN2 to GN2M3XGalGN2 represented in Figure 8. To demonstrate that these ions arise from the replacement of l-Fuc by l-Gal, the most abundant (M+H)+ at m/z = 1283 was submitted to a CID-MS-MS analysis (Fig. 7B). This MS analysis allows the observation of fragment ions specifically arising from the cleavage of glycosidic linkages of the selected (M+H)+ ion of PA oligosaccharides. As a consequence, some information on the oligosaccharide sequence can be deduced from the CID-MS-MS spectrum (Gu et al., 1994). In the CID-MS-MS of the ion at m/z = 1283, the successive loss of three mannose residues and one Xyl residue from the nonreducing end is observed. Furthermore, the detection of the fragment ion at m/z = 462 is strong evidence that the additional hexose unit is linked to the proximal GlcNAc residue (Fig. 7B). This fragmentation pattern is consistent with data reported in the literature on fragmentation of PA N-glycans either having or not having an additional residue linked to the proximal glucosamine residue (Gu et al., 1994). The mixture of labeled glycans was then analyzed by 1H-NMR. In addition to the H-1 signal of the β-Xyl at 4.43 ppm, the NMR spectrum shows, in the 4.5 to 5.2 ppm region, four signals that were assigned to the H-1 of mannose residues of the different N-glycans (Fig. 9). In addition to these signals, a minor doublet at 5.422 ppm (J = 3.5 Hz) was assigned to the H-1 of the α anomer of the additional hexose residue detected by MS. The chemical shift of this signal is in agreement with NMR data on terminal α-Galp residues (Zablackis et al., 1996). The amount of l-Gal containing N-glycans was estimated to represent about 5% by integration of the H-1 signal of l-Gal to H-1 signal of Man-4 (Fig. 9, signals a and b).
Figure 7.
MS analysis of the mur1 N-glycans. A, Electrospray mass spectrum of the PA derivatives of the paucimannosidic and complex-type N-glycans isolated from the mur1 mutant. □, (M+H)+ ions of the PA derivatives of M3XGN2, GNM3XGN2, and GN2M3XGN2 (see Fig. 5). ♦, (M+H)+ ions of the PA derivatives of the l-Gal-containing oligosaccharides M3XGalGN2, GNM3XGalGN2, and GN2M3XGalGN2 (see Fig. 8). B, CID-MS-MS of the (M+H)+ ion of the PA derivative of M3XGalGN2 (m/z = 1283).
Figure 8.
Abbreviations and structures of l-Gal-containing pauci-mannosidic and complex-type N-glycans isolated from the mur1 mutant.
Figure 9.
1H-NMR spectrum of the mixture of PA derivatives of the paucimanosidic and complex-type N-glycans isolated from the mur1 mutant leaves. Numbers and letters refer to the corresponding protons (for example, 14 = signal of H-1 proton of residue 4). A, H1 signal of the nonsubstituted Man 4 of M3XGN2 and GN6-M3XGN2. b, H1 signal of substituted Man 4 of GN3-M3XGN2 and GN2M3XGN2.
Taken together, these results indicate that the second family of N-linked glycans detected in the mur1 mutant contains an α-Gal linked to the proximal GlcNAc residue. The presence of Gal in the mur1 N-glycan preparation was confirmed by sugar composition analysis. However, due to the low amount of material, the absolute configuration of the Gal and its location on the GlcNAc were not determined. However, since l-Fuc was found to be replaced by l-Gal in the mur1 cell wall xyloglucan, the MS and NMR data strongly suggest that the same process occurs in the N-glycans of this mutant. As a consequence, the partial replacement of l-Fuc by l-Gal leads to the formation of plant N-glycans with a novel core structure represented in Figure 8. Thus, we conclude that the mur1 mutation results in the accumulation of major nonfucosylated structures (95%) and minor l-Gal-containing-oligosaccharides (5%) without affecting, in a detectable manner, the processing of N-linked glycans.
DISCUSSION
The structures of N-linked glycans associated to proteins isolated from Arabidopsis have been elucidated by affino- and immunodetection on blots with specific probes, as well as by structural identification of reducing oligosaccharides released from glycoproteins by treatment with a specific peptide N-glycosidase. High-mannose-type N-glycans from Man-5 to Man-9 have been identified. Furthermore, paucimannosidic and complex-type N-glycans having an α(1,3)-Fuc, a β(1,2)-Xyl residues have been fully identified. However, the total N-glycosylation in Arabidopsis presents a lower diversity than observed in other plants (Takahashi et al., 1986; Fitchette-Lainé et al., 1997). No complex-type N-glycans containing terminal Lewis a epitopes have been detected, indicating that these glycans either are not synthesized in Arabidopsis plants, or they are present only in very low amounts in this plant or only in some tissues.
N-linked glycans associated to proteins from the Fuc-deficient mur1 mutant have been isolated and identified using the same approach as was developed for the wild-type plants. This study has shown that mostly nonfucosylated oligosaccharides are N-linked to proteins of the mur1 mutant. No modification in the processing of the N-glycans was observed in the mutant. Major nonfucosylated oligosaccharide structures have been identified, bearing zero, one, or two terminal glucosamine residues in a ratio similar to that found in wild-type plants. This result confirms that the addition of the α(1,3)-Fuc residue is a late event in the processing of plant N-linked glycans. As a consequence, the absence of l-Fuc transfer does not disturb the biosynthesis of these oligosaccharides. In addition to the nonfucosylated oligosaccharides, a second family of oligosaccharides was detected by MS in the total protein extract from the mur1 plants. From analytical data these oligosaccharides, representing about 5% of the N-glycans, were found to have an additional hexose residue (α-galactose from NMR data) linked to the proximal glucosamine residue. Since the chitobiose unit of N-glycans in plants and other organisms has been reported so far as only substituted by Fuc residues, such data strongly suggest that the minor population of N-glycans, identified by MS, result from the mur1 mutation. By analogy with previously reported data on the mur1 xyloglucan (Zablackis et al., 1996), we propose that the minor 5% subpopulation of N-glycans found in this mutant are new plant N-glycans having α(1,3)-l-Gal linked to the proximal glucosamine and resulting from the replacement of l-Fuc by l-Gal.
The replacement of one-third of the terminal l-Fuc residues in the xyloglucan by l-Gal, a monosaccharide structurally related to l-Fuc, indicates that the α(1,2)-fucosyltransferase involved in the xyloglucan biosynthesis is able to transfer l-Gal from GDP-l-Gal to the xyloglycan backbone in the absence of GDP-l-Fuc. This phenomenon also seems to occur with the α(1,3)-fucosyltransferase involved in N-linked glycan biosynthesis, which is localized in the same compartment as the xyloglucan α(1,2)-fucosyltransferase, i.e. the trans cisternae of the Golgi apparatus (Driouich et al., 1993; Fitchette-Lainé et al., 1994). However, the difference in level of l-Gal incorporation suggests that the two fucosyltransferases show different efficiencies in the replacement of l-Fuc by l-Gal.
Abbreviations:
- CID-MS-MS
collision-induced dissociation MS
- HPAEC
high pH anion-exchange chromatography
- HPAEC-PAD
HPAEC coupled to pulsed amperometric detection
- PA
pyridylamino derivative
- PNGase A
peptide N-glycosidase A
Footnotes
This work was supported in France by CNRS (no. ESA 6037), the University of Rouen, and the M.E.N.E.S.R. (Actions Concertées Coordonnées des Sciences du Vivant ACCSV 14, réseau G-Trec), and by grants from the Région Haute-Normandie. This work was supported in the United States by the Department of Energy Biosciences Program (grant no. DE-FG02-95ER20203). C.R. and M.C.-M. are recipients of Region Haute-Normandie and Biopole fellowships, respectively.
LITERATURE CITED
- Bonin CP, Potter I, Vanzin GF, Reiter WD. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-l-fucose. Proc Natl Acad Sci USA. 1997;94:2085–2090. doi: 10.1073/pnas.94.5.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich A, Faye L, Staehelin A. The plant Golgi apparatus: a factory for complex polysaccharides and glycoproteins. Trends Biochem Sci. 1993;18:210–214. doi: 10.1016/0968-0004(93)90191-o. [DOI] [PubMed] [Google Scholar]
- Faye L, Chrispeels MJ. Characterization of N-linked oligosaccharides by affinoblotting with concanavalin A-peroxidase and treatment of the blots with glycosidases. Anal Biochem. 1985;118:131–137. doi: 10.1016/0003-2697(85)90498-1. [DOI] [PubMed] [Google Scholar]
- Faye L, Gomord V, Fitchette-Lainé A-C, Chrispeels MJ. Affinity purification of antibodies specific for Asn-linked glycans containing α 1-3 fucose or β 1-2 xylose. Anal Biochem. 1993;109:104–108. doi: 10.1006/abio.1993.1088. [DOI] [PubMed] [Google Scholar]
- Fitchette-Lainé A-C, Gomord V, Cabanes M, Michalski J-C, Saint Macary M, Foucher B, Cavelier B, Hawes C, Lerouge P, Faye L. N-glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J. 1997;12:1411–1417. doi: 10.1046/j.1365-313x.1997.12061411.x. [DOI] [PubMed] [Google Scholar]
- Gu J, Hiraga T, Wada Y. Electrospray ionization mass spectrometry of pyridylamino oligosaccharide derivatives: sensibility and in-source fragmentation. Biol Mass Spectrom. 1994;23:212–217. doi: 10.1002/bms.1200230405. [DOI] [PubMed] [Google Scholar]
- Hardy MR. Monosaccharide analysis of glycoconjugates by high-performance anion-exchange chromatography with pulsed amperometric detection. Methods Enzymol. 1989;179:76–82. doi: 10.1016/0076-6879(89)79115-1. [DOI] [PubMed] [Google Scholar]
- Hase S, Ibuki T, Ikenaka T. Reexamination of the pyridylamination used for fluorescence labeling of oligosaccharides and its application to glycoproteins. J Biochem. 1984;95:197–203. doi: 10.1093/oxfordjournals.jbchem.a134585. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Tsuru A, Mitsui T, Takahashi N, Hanzawa H, Arata Y, Akazawa T. Structure and biosynthesis of the xylose-containing carbohydrate moiety of rice α-amylase. Eur J Biochem. 1990;191:287–295. doi: 10.1111/j.1432-1033.1990.tb19122.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–687. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lerouge P, Cabanes-Macheteau M, Rayon C, Fitchette-Lainé A-C, Gomord V, Faye L. N-glycoprotein biosynthesis in plants: recent development and future trends. Plant Mol Biol. 1998;38:31–48. [PubMed] [Google Scholar]
- Melo NS, Nimtz M, Conradt HS, Fevereiro PS, Costa J. Identification of the human Lewisa carbohydrate motif in a secretory peroxidase from a plant cell suspension culture (Vaccinium myrtillus L.) FEBS Lett. 1997;415:186–191. doi: 10.1016/s0014-5793(97)01121-6. [DOI] [PubMed] [Google Scholar]
- Montreuil J, Bouquelet S, Debra H, Fournet B, Spick G, Strecker G. Carbohydrate analysis: a pratical approach. In: Chapelin MF, Kennedy JF, editors. Glycoproteins. Oxford University Press, New York: IRL Press; 1986. pp. 143–204. [Google Scholar]
- Rayon C, Gomord V, Faye L, Lerouge P. N-glycosylation of phytohemagglutinin expressed in bean cotyledons or in transgenic tobacco cells. Plant Physiol Biochem. 1996;34:273–281. [Google Scholar]
- Reiter WD, Chapple CCS, Somerville CR. Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science. 1993;261:1032–1035. doi: 10.1126/science.261.5124.1032. [DOI] [PubMed] [Google Scholar]
- Taga EM, Waheed A, Van Etten RL. Structural and chemical characterization of a homogeneous peptide N-glycosidase from almond. Biochemistry. 1984;23:815–822. doi: 10.1021/bi00300a006. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Hotta T, Ishihara H, Mori M, Tejima S, Bligny R, Akasawa T, Endo S, Arata Y. Xylose-containing common structural unit in N-linked oligosaccharides of laccase from sycamore cells. Biochemistry. 1986;25:388–395. [Google Scholar]
- Tomiya N, Awaya J, Kurono M, Endo S, Arata Y, Takahashi N. Analyses of N-linked oligosaccharides using a two-dimensional mapping technique. Anal Biochem. 1988;171:73–90. doi: 10.1016/0003-2697(88)90126-1. [DOI] [PubMed] [Google Scholar]
- Tomiya N, Kurono M, Ishihara H, Tejima S, Endo S, Arata Y, Takahashi N. Structural analysis of N-linked oligosaccharides by a combination of glycopeptidase, exoglycosidases, and high-performance liquid chromatography. Anal Biochem. 1987;163:489–499. doi: 10.1016/0003-2697(87)90253-3. [DOI] [PubMed] [Google Scholar]
- Vitale A, Chrispeels MJ. Transient N-acetylglucosamine in the biosynthesis of phytohemagglutinin: attachment in the Golgi apparatus and removal in protein bodies. J Cell Biol. 1984;99:133–140. doi: 10.1083/jcb.99.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schaewen A, Sturm A, O'Neill J, Chrispeels MJ. Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 1993;102:1109–1118. doi: 10.1104/pp.102.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablackis E, Huang J, Müller B, Darvill AG, Albersheim P. Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 1995;107:1129–1138. doi: 10.1104/pp.107.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablackis E, York WS, Pauly M, Hantus S, Reiter WD, Chapple CCS, Albersheim P, Darvill A. Substitution of l-fucose by l-galactose in cell walls of Arabidopsis mur1. Science. 1996;272:1808–1810. doi: 10.1126/science.272.5269.1808. [DOI] [PubMed] [Google Scholar]