Abstract
Photosystem II (PSII), a large multisubunit pigment–protein complex localized in the thylakoid membrane of cyanobacteria and chloroplasts, mediates light-driven evolution of oxygen from water. Recently, a high-resolution X-ray structure of the mature PSII complex has become available. Two PSII polypeptides, D1 and CP43, provide many of the ligands to an inorganic Mn4Ca center that is essential for water oxidation. Because of its unusual redox chemistry, PSII often undergoes degradation followed by stepwise assembly. Psb27, a small luminal polypeptide, functions as an important accessory factor in this elaborate assembly pathway. However, the structural location of Psb27 within PSII assembly intermediates has remained elusive. Here we report that Psb27 binds to CP43 in such assembly intermediates. We treated purified genetically tagged PSII assembly intermediate complexes from the cyanobacterium Synechocystis 6803 with chemical cross-linkers to examine intermolecular interactions between Psb27 and various PSII proteins. First, the water-soluble 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) was used to cross-link proteins with complementary charged groups in close association to one another. In the His27△ctpAPSII preparation, a 58-kDa cross-linked species containing Psb27 and CP43 was identified. This species was not formed in the HT3△ctpA△psb27PSII complex in which Psb27 was absent. Second, the homobifunctional thiol-cleavable cross-linker 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP) was used to reversibly cross-link Psb27 to CP43 in His27△ctpAPSII preparations, which allowed the use of liquid chromatography/tandem MS to map the cross-linking sites as Psb27K63↔CP43D321 (trypsin) and CP43K215↔Psb27D58AGGLK63↔CP43D321 (chymotrypsin), respectively. Our data suggest that Psb27 acts as an important regulatory protein during PSII assembly through specific interactions with the luminal domain of CP43.
Keywords: mass spectrometry, photosynthesis, protein structure
Photosystem II (PSII) is an integral membrane protein complex with more than 20 subunit proteins and numerous cofactors (1). In the thylakoid membranes of cyanobacteria, algae, and higher plants, PSII mediates light-induced oxidation of water to molecular oxygen and reduction of plastoquinones. These reactions are critically important for the conversion of solar energy to biochemical energy used by the vast majority of life on Earth. An X-ray structural model of mature, photoactive PSII from a cyanobacterium has recently been described at a 1.9-Å resolution (1), and a catalytic inorganic Mn4Ca metal cluster is known to be essential for the oxygen evolution reaction in PSII.
An unusual property of PSII is that it constantly undergoes assembly and disassembly because of the unavoidable damage that results from its normal chemistry under extreme redox conditions. Thus, at any given instant, in addition to the active PSII complexes, thylakoid membranes contain various PSII subassembly complexes that are incapable of oxygen evolution. These transient protein complexes, representing various stages in the elaborate process of PSII biogenesis and repair, have been especially difficult to capture and characterize because of their low abundance, structural heterogeneity, and thermodynamic instability.
Among multiple PSII assembly intermediates, Psb27-containing PSII complexes are of particular interest because Psb27 is involved in the repair cycle of PSII (2) and assembly of the Mn4Ca cluster (3). Psb27, an 11-kDa thylakoid lumen-localized protein, was first identified in a PSII preparation from the cyanobacterium Synechocystis sp. PCC 6803 (4). Kashino et al. (5) later determined the presence of this protein in a polyhistidine-tagged PSII preparation from the same organism. Furthermore, an enriched level of Psb27 was observed in purified PSII preparations from a ctpA deletion mutant (6) in a His-tagged CP47-containing background strain (7) in which the D1 protein is present in its precursor (pD1) form. A psb27 deletion mutation in Synechocystis 6803 results in impaired PSII repair activity after high-light photoinhibition (3). Similarly, in the vascular plant Arabidopsis, a psb27 deletion mutant exhibits delayed recovery after photoinhibition treatment (8). Interestingly, in a monomeric form of PSII, purified from Thermosynechococcus elongatus (2), Psb27 is present, whereas other luminal extrinsic proteins, such as PsbO, PsbU, and PsbV, are absent. In addition, such a PSII subcomplex does not have an Mn cluster and cannot oxidize water. On the other hand, the psb27 deletion strain of Synechocystis still assembles PSII centers that can evolve oxygen. Altogether, these findings have suggested that Psb27 is an important regulatory protein that allows optimal assembly of catalytically competent PSII complexes.
Three-dimensional solution structures of Psb27 were recently determined by using NMR spectroscopy (9, 10). The structural arrangement of Psb27 within the PS II assembly complex, however, remains unclear. In this article, we present experimental evidence that, in certain PSII assembly intermediate complexes, Psb27 binds to CP43, a chlorophyll protein that is known to provide ligands to the Mn cluster in mature PSII (1). Our studies involved cross-linking of Psb27 to other PSII proteins by the water-soluble carbodiimide 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and the thiol-cleavable cross-linker 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP) followed by identification of specific cross-linked species by MS.
Results
We have recently described the composition of a PSII preparation from a Synechocystis 6803 △ctpA mutant that was isolated by using an engineered His tag on the Psb27 protein (11). Cross-linking of proteins by the water-soluble protein cross-linking agent EDC is a widely used tool to study protein–protein interactions and low-resolution structure determination because it is specific for groups that interact by complementary charges that are in van der Waals’ contact with each other (12). It was successfully used to determine close proximities of PsbO and CP47 proteins of PSII (13, 14), and these results have subsequently been validated by crystallographic studies of PSII (15).
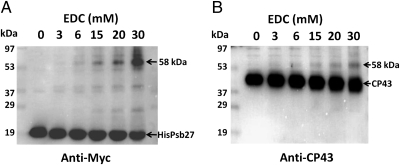
Treatment of a His27△ctpAPSII preparation with EDC resulted in various cross-linked species (Fig. 1). In the absence of EDC, HisPsb27 and CP43 migrated at 18 kDa (Fig. 1A) and 40 kDa (Fig. 1B), respectively. However, treatment with EDC resulted in the formation of a 58-kDa cross-linked product that was recognized by both anti-Myc antibodies (Fig. 1A) and anti-CP43 antisera (Fig. 1B). Moreover, the relative amounts of this 58-kDa band increased with increasing EDC concentrations, indicating that the formation of this cross-linked species depended on EDC. As is evident from the second lane of each blot in Fig. 1, without EDC treatment (0 mM EDC), no such cross-linked product was recognized by either anti-Myc or anti-CP43 antibodies. In addition to the 58-kDa band, two other weaker cross-reacting bands, migrating at 27 kDa and 39 kDa, respectively, were also observed (Fig. 1A). The identities of these bands remain unknown. Notably, neither of these two bands was recognized by the anti-CP43 antibodies (Fig. 1B). As evident in Fig. 1, EDC treatment did not result in cross-linking of the majority of the Psb27 and CP43 proteins in the sample, and only limited amounts of protein were cross-linked to the 58-kDa product. The primary reason is that we used relatively low concentrations of EDC to minimize formation of intercomplex, cross-linked products. In addition, with increasing concentrations of EDC, the CP43 band exhibited higher mobility (Fig. 1B), which could be the result of intramolecular cross-linking in CP43. Altogether, these results indicate that the 58-kDa cross-linked product consists of both Psb27 and CP43. The apparent molecular mass of the cross-linked complex is consistent with CP43 and the His/Myc-tagged Psb27 being present at a 1:1 stoichiometry.
Fig. 1.
EDC cross-linking of Psb27 and CP43. Concentration-dependent production of cross-linked species at 58 kDa is shown. His27△ctpAPSII complex was treated with EDC at various concentrations, and immunoblots were probed with anti-Myc antibodies that recognize the Myc epitope on the tagged Psb27 protein (A) or anti-CP43 antibodies (B). EDC concentrations are shown at the top of each blot. The locations of molecular mass standards are shown on the left. Cross-linked products and the parent polypeptides are identified on the right.
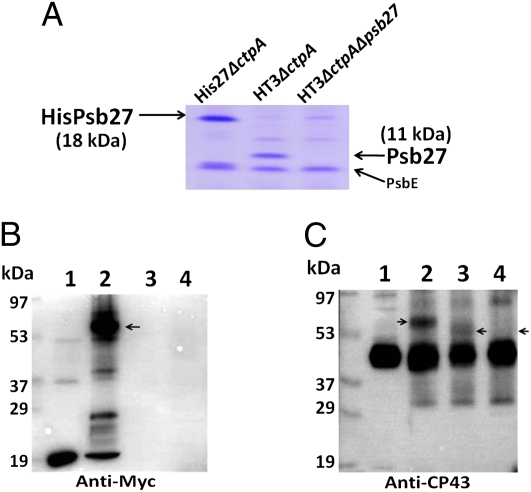
To more conclusively establish the identities of the cross-linked proteins, we examined the EDC cross-linked products by using different antibodies on three different PSII preparations from the His27△ctpA, HT3△ctpA, and HT3△ctpA△psb27 strains (Materials and Methods). As shown in Fig. 2A, the His/Myc-tagged Psb27 in His27△ctpAPSII migrated more slowly than did untagged Psb27 in HT3△ctpAPSII. As expected, neither form of this protein was detected in HT3△ctpA△psb27PSII, which was isolated from a psb27 deletion strain.
Fig. 2.
EDC cross-linking in three types of purified PSII complexes. (A) SDS/PAGE analysis showing slower migration of His/Myc-tagged Psb27 (18 kDa, lane 1) than native Psb27 (11 kDa, lane 2) after Coomassie Brilliant Blue (R-250) staining. Lane 1: His27△ctpAPSII; lane 2: HT3△ctpAPSII; lane 3: HT3△ctpA△psb27PSII. Two micrograms of chlorophyll a-containing samples were loaded in each lane. (B) Detection of cross-linked species. Lane 1: His27△ctpAPSII; lane 2: His27△ctpAPSII + EDC; lane 3: HT3△ctpAPSII + EDC; lane 4: HT3△ctpA△psb27PSII + EDC. EDC (30 mM) cross-linking reaction was performed for 30 min in the dark at 23 °C. Immunoblot was probed with anti-Myc antibodies. (C) Same as in B except probed with anti-CP43 antibodies. Arrows indicate the increased mobility of CP43–Psb27 cross-linked complex (lane 3) compared with His/Myc-tagged Psb27–CP43 cross-linked complex (lane 2) and the absence of any cross-linked product in lane 4 because of the absence of Psb27 in HT3△ctpA△psb27PSII.
The results shown in Fig. 2B are similar to those in Fig. 1A; as expected, the anti-Myc antibodies did not cross-react with any protein in the HT3△ctpAPSII (lane 3) and HT3△ctpA△psb27PSII (lane 4) samples. In Fig. 2C, the CP43–Psb27 cross-linked species exhibited a higher mobility (lane 3) compared with CP43–HisPsb27 (lane 2). As described above, this observation is consistent with the presence of a His/Myc tag on Psb27 in His27△ctpAPSII and its absence in HT3△ctpAPSII. We also determined that EDC treatment did not result in cross-linked products between Psb27 and either CP47 or D1, two other major proteins in PSII (Figs. S1 and S2).
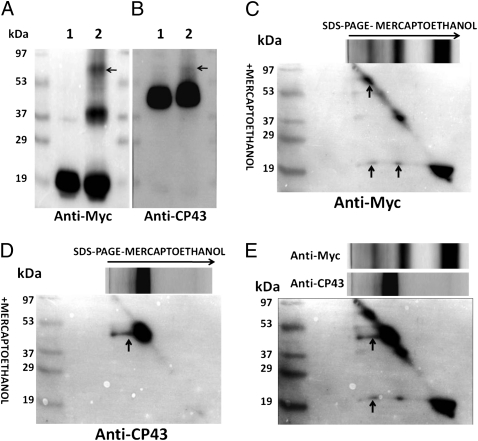
To further probe the interactions between Psb27 and CP43, we used DTSSP, a homobifunctional cleavable succinimide ester that reacts with the ε-amino groups of lysine residues and with the N termini of proteins (16). Fig. 3 A and B show the separation pattern of control and a DTSSP-treated sample probed with anti-Myc and anti-CP43 antibodies, respectively. Cross-linking led to two additional bands with apparent molecular masses of 39 kDa and 58 kDa (Fig. 3A, lane 2), which are similar to the two cross-linked species identified by EDC cross-linking (Fig. 1). Consequently, observation of the 58-kDa cross-linked species with both cross-linkers further demonstrates that Psb27 is closely associated with CP43 in these PSII assembly intermediates.
Fig. 3.
DTSSP cross-linking in His27△ctpAPSII preparation. (A) Lane 1: His27△ctpAPSII; lane 2: His27△ctpAPSII + DTSSP. The cross-linking reaction was performed by using 10 mM DTSSP for 30 min in the dark at 23 °C. Immunoblot was probed with anti-Myc antibodies. The location of the 58-kDa CP43–Psb27 cross-linked species is indicated with an arrow. (B) Same as in A except probed with anti-CP43 antibodies. (C) Results of 2D diagonal electrophoresis of His27△ctpAPSII complex treated with DTSSP and probed with anti-Myc antibodies. During SDS/PAGE, β-mercaptoethanol was omitted in the first dimension and was included in the second dimension. (D) Same as in C except probed with anti-CP43 antibodies. (E) Overlay of C and D. Protein spots that represent tagged Psb27 and CP43 are found as a vertical series below the 58-kDa cross-linking complex, indicating intermolecular cross-links between these two components.
The His27△ctpAPSII sample treated with DTSSP was next analyzed by 2D diagonal electrophoresis. Proteins that were not modified by chemical reduction migrated in a diagonal line because an identical gel system was used in both the first and second dimensions except for the presence of a reducing agent in the second dimension. When cleavage of two or more cross-linked polypeptides by DTT treatment occurred before fractionation in the second dimension, each component that was formerly cross-linked by DTSSP migrated independently in the gel, and the released proteins were observed along a vertical line below the cross-linked product (Fig. 3 C and D). Proteins that reacted with the anti-Myc and anti-CP43 antibodies formed two spots vertically below that of a cross-linked complex with an apparent molecular mass of 58 kDa, indicating that CP43 and Psb27 had been cross-linked by DTSSP (Fig. 3E), as seen after an overlay of Fig. 3 C and D. The similar apparent molecular masses also suggest that only Psb27 and CP43 are present in this cross-linked complex. Below the 39-kDa cross-linked complex, Psb27 was detected as a spot (Fig. 3C). This 39-kDa complex may represent a complex of Psb27 (18 kDa) with one or more PSII subunits, the identities of which remain unknown.
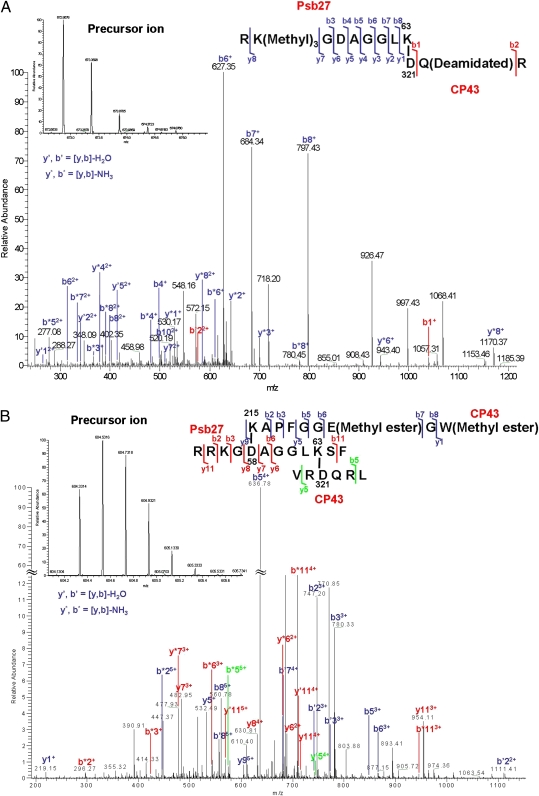
To gain insight into the accurate location of Psb27 within PSII assembly intermediates, we performed in-gel digestion with trypsin of the protein(s) corresponding to the 58-kDa band and analyzed the product by liquid chromatography/tandem MS. With the MassMatrix (version 2.3.8) search engine, we could identify 21% and 37% of the amino acid sequences of CP43 and Psb27, respectively. All product ion spectra representing cross-linked peptides were evaluated, and we determined that the K63 residues in Psb27 and the D321 residue in CP43 are cross-linked. Fig. 4A shows the product ion spectrum of cross-linked peptides RKGDAGGLK63 from Psb27 and D321QR from CP43. Interestingly, we also found an unusual posttranslational modification, lysine methylation, on K56 of Psb27. The role of such a modification remains unclear. Nevertheless, the series of product ions confirmed the sequence of the peptides and established the cross-linking site.
Fig. 4.
Product ion (tandem MS) spectra obtained for cross-linked peptides. (A) Product ion (tandem MS) spectra of the cross-linked peptides RKGDAGGLK63 (Psb27) and D321QR (CP43). The fragmentation pattern indicates that cross-links are formed specifically between K63 of Psb27 peptide 55–63 and D321 of CP43 peptide 321–323. Shown is the product ion map of cross-link fragment after collision-induced dissociation. The abundant b′, y′ and b*, y* ions are labeled. Ions showing cross-linking were m/z = 672.8678 (2+) (Inset). (B) Product ion (tandem MS) spectra of the intermolecular cross-links after chymotrypsin digestion. Peptide 54–65 of Psb27 protein was sandwich–cross-linked to peptide 215–223 and 319–324 of CP43, respectively. Cross-links were formed specifically between K215 of CP43 peptide 215–223 and D58 of Psb27 peptide 54–65 and between K63 of Psb27 peptide 54–65 and D321 of CP43 peptide 319–324, respectively. Ions showing cross-linking were of m/z = 604.3314 (5+) (Inset). The abundant b′, y′ and b*, y* ions are labeled.
These two cross-linked sites were also identified in an independent experiment in which the 58-kDa band was digested with chymotrypsin (Fig. 4B). In addition, we determined that D58 of Psb27 was also cross-linked to K215 in a large luminal extrinsic loop in CP43 (1). Thus, we were able to map the cross-linking sites as Psb27K63↔CP43D321 (trypsin) and CP43K215↔Psb27D58AGGLK63↔CP43D321 (chymotrypsin), respectively.
Discussion
In the recent 1.9-Å structural model of PSII, the locations of three luminal extrinsic proteins, PsbO, PsbU and PsbV have been well defined. However, two other extrinsic proteins, PsbQ (17) and Psb27, are absent in this structure. The crystal structure of PSII, however, only represents a “snapshot” of PSII during its life cycle. Thus, studies on the PSII assembly intermediates should provide further insights on the structural dynamics of PSII, a multisubunit protein complex. Previous studies have shown that Psb27 is involved in PSII repair cycles (2) and in at least two consecutive PSII assembly intermediates, His27△ctpAPSII and His27PSII (11). Our current study unequivocally shows that Psb27 is closely associated with the luminal domain of CP43.
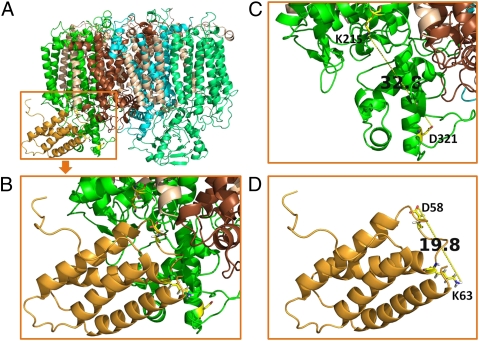
Based on these experimental data, we propose a schematic model of Psb27 within a PSII [Protein Data Bank (PDB) ID 3ARC] structural monomer (Fig. 5). In this model, Psb27 is located in the cavity formed by loop C and loop E of CP43 and the loop between the first and second transmembrane helices of the D1 protein (loop A). In this interaction, D58 and K63 of Psb27 form salt bridges with the K215 of loop C and D321 of loop E in CP43, respectively. Adopting this orientation, the highly conserved helices 3 and 4 (9) of Psb27 come into close contact with the loop E area, which is facing away from the pseudosymmetrical axis of D1–D2. This orientation takes into account any serious structural conflicts between Psb27 and loop A of the D1 protein. A spatial conflict, however, may still occur between the H2–H3 loop of Psb27 (9) and the N-terminal 10 aa residues of PsbO (PDB ID 3ARC). An implication of such a spatial conflict is that the binding affinity of PsbO is reduced significantly in the presence of Psb27 in the PSII assembly intermediates, consistent with the experimental observation that PsbO is absent in all △ctpA PSII preparations (6, 11). The possible reasons are (i) the unprocessed pD1 tail may impose potential steric clashes with the PsbO interface that normally interacts with CP43 and (ii) Psb27 decreases the binding affinity of PsbO to CP43 by blocking the PsbO N-terminal interaction domain, as discussed here. Decreased levels of PsbO were indeed observed for His27PSII (11), where pD1 had been processed to D1 and the steric clash with the pD1 tail had been removed. According to the in silico analysis carried out by Fagerlund and Eaton-Rye (18), the best structural model has Psb27 docked beneath CP43 and D1 in a position that the bulk of PsbO would usually occupy.
Fig. 5.
A schematic model of binding of Psb27 to the luminal domain of CP43 (PDB IDs 3ARC and 2KMF). (A) Front view showing Psb27 (bright orange) binding to loop C and loop E of CP43 (green). D1, brown; D2, cyan; CP47, light green; small subunit peptides, wheat. (B) A magnified view of the model shown in A. CP43K215, Psb27D58, Psb27K63, and CP43D321 are represented as sticks. (C) Distance between K215 and D321 of CP43 in the X-ray structural model (PDB ID 3ARC). (D) Distance between D58 and K63 of Psb27 in the NMR model (PDB ID 2KMF, model 1). All images were prepared with PyMOL software (30).
The present study questions whether luminal domains of proteins such as CP43, CP47, and D1 in PSII assembly intermediates adopt conformations similar to those in fully functional PSII. PSII assembly is a dynamic process with sequential binding of multiple extrinsic proteins and concomitant multistepped Mn-cluster photoassembly. Experimental data gathered on cyanobacteria and from higher plants for the past 20 y have strongly supported a significant conformational rearrangement between nonfunctional PSII and the functional PSII (19–21). It has also been also suggested that the binding affinity of CP43 to other intrinsic PSII components (e.g., D1) is even reduced when the Mn cluster is not assembled (22).
Our experiment with the zero-length cross-linker establishes that the distance between D58 and K63 ranges between 11.3 Å and 19.8 Å, as in the NMR structure (PDB ID 2KMF, models 1 and 10, respectively) (Fig. 5D). The significant difference between this distance and that of the 32.8-Å span between K215 and D321 in CP43 in the 1.9-Å structural model of PSII (Fig. 5C) indicates that, in the PSII assembly intermediates, the luminal domain of CP43 may adopt a conformation that is significantly different from that in the fully functional PSII. In the Psb27-containing PSII assembly intermediates, the distance between K215 and D321 on CP43 is in the range of 11.3–19.8 Å. In other words, the luminal domains of CP43 may adopt a more compact and condensed state in the presence of Psb27 than that in the fully functional and active PSII. This conformational feature may be characterized by blocking the premature binding of extrinsic proteins, thus preventing Mn-cluster assembly, as was observed experimentally (2, 6). It is conceivable that, upon removal of Psb27 during subsequent PSII assembly steps, more stretched luminal domains are formed before Mn-cluster photoactivation. The subsequent association of the extrinsic proteins PsbO, PsbU, PsbV, and PsbQ presumably further stabilizes such domains.
The close association of Psb27 and CP43 does not necessarily exclude the possibility that Psb27 may interact with the CP47 protein and (or) the D1 protein on the luminal side. In Arabidopsis, the LPA19 protein, a homolog of Psb27, interacts with the soluble C terminus of both pD1 and mature D1, as determined with yeast two-hybrid analyses (23). In our study, however, no cross-linked product of Psb27 and pD1 could be detected (Fig. S2).
Each protein subunit in PSII is synthesized separately in the cell, and assembly occurs under appropriate biological conditions. Formation of each interface in the PSII protein complex should be thermodynamically driven by many factors, including hydrophobic interactions, electrostatic-charge interactions, and new hydrogen bonding. The EDC cross-linking results suggest that regions of complementary charge exist between these two PSII assembly intermediate subunits, CP43 and Psb27. Thus, we can hypothesize that association and dissociation between CP43 and Psb27 may depend on, or be triggered by, changing ionic strength or pH. From the crystal structures, the large extrinsic loops E of CP43 and that of CP47 appear to provide the most binding sites for the extrinsic proteins PsbV, PsbU, and PsbO (24). Our results provide a refined picture for the structural accommodation of transiently bound Psb27 in the PSII structure. We suggest that CP43, in addition to its roles as a reaction center light-harvesting antenna and an enabler in Mn-cluster coordination (25), serves as a binding protein for Psb27 during PSII assembly. Considering the presence of Psb27 and the absence of the Mn cluster, we propose a gatekeeper role for Psb27 in the process of Mn-cluster assembly.
Materials and Methods
Mutant Construction.
Generation of His27△ctpA and HT3△ctpA mutants was previously reported (6, 11). HT3 mutant, a strain with C-terminally His6-tagged CP47 protein, was a generous gift from Terry Bricker (Louisiana State University, Baton Rouge, LA) (7). The HT3△ctpA△psb27 mutant was generated by transforming the HT3△ctpA strain with the △psb27 construct reported in ref. 3. Purification of tagged PSII complexes was performed essentially as described in ref. 5. Protein electrophoresis and immunodetection were performed as in refs. 26 and 27, unless otherwise indicated.
EDC Cross-Linking.
PSII preparations were resuspended at 0.2 mg/mL chlorophyll a in 25% glycerol, 10 mM MgCl2, 5 mM CaCl2, and 50 mM MES buffer (pH 6.5) and then incubated with a range of EDC concentrations (0–30 mM) for 30 min in the dark at 23 °C. The treated samples were desalted, and buffer was exchanged with a Zeba column (Thermo Scientific) according to the manufacturer's protocol.
DTSSP Cross-Linking and 2D Diagonal Electrophoresis.
PSII samples were resuspended at 0.2 mg/mL chlorophyll a in 25% glycerol, 10 mM MgCl2, 5 mM CaCl2, and 50 mM Hepes buffer (pH 7.5) and then incubated with 10 mM DTSSP for 30 min in the dark at 23 °C. The cross-linking reaction was terminated by adding stop solution [Tris⋅Cl (pH 7.5)] to a final concentration of 50 mM for 15 min followed by Zeba column desalting and buffer exchange.
The proteins cross-linked by DTSSP were resolved by 2D diagonal electrophoresis followed by immunodetection. In the first dimension, the treated samples were fractionated by SDS/PAGE in the absence of any reducing agent. Immediately after the first-dimension electrophoresis, the gel was cut to strips and equilibrated for 30 min at 30 °C in 125 mM Tris⋅HCl (pH 6.8) and 50 mM DTT. The equilibrated first-dimension gel strip was then layered onto a second-dimension SDS/PAGE slab gel.
Proteolytic Digestion and Peptide Clean-Up.
The gel slices containing cross-linked protein samples corresponding to 58 kDa were subjected to overnight in-gel digestion by trypsin or chymotrypsin with a substrate-to-enzyme ratio of 50:1 at 37 °C for trypsin and 30 °C for chymotrypsin. The peptide mixtures were extracted and cleaned up with C18 NuTips (Glygen) wetted in 80% acetonitrile and 0.1% formic acid and preequilibrated in 0.1% formic acid. Next, the tips were washed with 0.1% formic acid, and the bound peptides were eluted with 80% acetonitrile and 0.1% formic acid. The eluents were vacuum-dried, resuspended in 20 μL of 0.1% formic acid, and then subjected to liquid chromatography/tandem MS analysis.
MS Analysis.
Peptides were analyzed on an LTQ Orbitrap XL (Thermo Fisher). Samples were loaded and eluted with an autosampler and an Ultra 1D+ ultra performance liquid chromatograph (Eksigent). The 75-μm diameter nanospray column was pulled (Sutter Instruments) and packed with Magic C18AQ reverse-phase media (Michrom Bioresources). Columns were mounted in a PicoView nanospray source (New Objective) and eluted with a 200-min gradient at 260 nL/min. The gradient was set from 2–60% solvent B (acetonitrile and 0.1% formic acid) over 180 min, ramped to 80% solvent B in 10 min, and reequilibrated at 100% solvent A (water and 0.1% formic acid) for 10 min. One full MS acquisition was followed by six collision-induced dissociation scans. The Orbitrap parameters were as follows: spray voltage, 2.0 kV; capillary temperature, 200 °C; tandem MS selection threshold, 1,000 counts; activation q, 0.25; activation time, 30 ms. Tandem MS data were centroided during acquisition.
Database Search.
Thermo RAW files were converted to MGF files with MSQuant (version 2.0) with default settings (28). The National Center for Biotechnology Information nonredundant database (version 20100915; restricted to bacteria) was searched with Mascot 2.2.06 (Matrix Science) with the following settings: enzyme, trypsin or chymotrypsin; MS tolerance, 10 ppm; tandem MS tolerance, 0.6 Da; maximum number of missed cleavages, 3; peptide charge of 1+, 2+, and 3+; variable modification, oxidation of M.
Cross-linked peptides were identified with the MassMatrix database search engine (29) (www.massmatrix.net). The database search settings were as follows: enzyme, trypsin or chymotrypsin; variable modification, carbamylation of NQ, deamination of NQ, methyl ester of C-term, DE, methylation of R-, K-, and N-term, oxidation of M; precursor ion tolerance, 10 ppm; product ion tolerance, 0.8 Da; maximum number of PTM per peptide, 2; minimum peptide length, 6 aa; maximum peptide length, 40 aa; minimum pp score, 5.0; maximum number of match per spectrum, 1; maximum number of combinations per match, 1; cross-link, EDC; cross-link mode, exploratory; cross-link sites cleavability, noncleavable by enzyme; maximum number of cross-links per peptide, 2. All identifications of cross-linked peptides were manually confirmed by interpretation of product ion spectra.
Theoretical product ion (tandem MS) spectra were generated (Protein Prospector MS-Product, http://prospector.ucsf.edu) for cross-linked peptides that were identified in the MassMatrix search, and a table of product ions was made for each cross-linked peptide. The product ion spectra were manually compared with each table, and the best match was identified.
Supplementary Material
Acknowledgments
We thank Dr. Terry M. Bricker for the HT3 strain and other members of the laboratories of H.B.P. and M.L.G. for collegial discussions. Funding of this research is provided by National Science Foundation Grant MCB0745611 (to H.B.P.) and National Institutes of Health National Center for Research Resources Grant 2P41RR000954 (to M.L.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111597108/-/DCSupplemental.
References
- 1.Umena Y, Kawakami K, Shen JR, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 2.Nowaczyk MM, et al. Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell. 2006;18:3121–3131. doi: 10.1105/tpc.106.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roose JL, Pakrasi HB. The Psb27 protein facilitates manganese cluster assembly in photosystem II. J Biol Chem. 2008;283:4044–4050. doi: 10.1074/jbc.M708960200. [DOI] [PubMed] [Google Scholar]
- 4.Ikeuchi M, Inoue Y, Vermaas W. Characterization of photosystem II subunits from the cyanobacterium Synechocystis sp. PCC6803. In: Mathis P, editor. Photosynthesis: From Light to Biosphere. Vol III. Dordrecht, The Netherlands: Kluwer; 1995. pp. 297–300. [Google Scholar]
- 5.Kashino Y, et al. Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry. 2002;41:8004–8012. doi: 10.1021/bi026012+. [DOI] [PubMed] [Google Scholar]
- 6.Roose JL, Pakrasi HB. Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J Biol Chem. 2004;279:45417–45422. doi: 10.1074/jbc.M408458200. [DOI] [PubMed] [Google Scholar]
- 7.Bricker TM, Morvant J, Masri N, Sutton HM, Frankel LK. Isolation of a highly active photosystem II preparation from Synechocystis 6803 using a histidine-tagged mutant of CP 47. Biochim Biophys Acta. 1998;1409:50–57. doi: 10.1016/s0005-2728(98)00148-0. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, et al. A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol Biol. 2006;61:567–575. doi: 10.1007/s11103-006-0031-x. [DOI] [PubMed] [Google Scholar]
- 9.Cormann KU, et al. Structure of Psb27 in solution: Implications for transient binding to photosystem II during biogenesis and repair. Biochemistry. 2009;48:8768–8770. doi: 10.1021/bi9012726. [DOI] [PubMed] [Google Scholar]
- 10.Mabbitt PD, et al. Solution structure of Psb27 from cyanobacterial photosystem II. Biochemistry. 2009;48:8771–8773. doi: 10.1021/bi901309c. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Roose JL, Cameron JC, Pakrasi HB. A genetically tagged Psb27 protein allows purification of two consecutive photosystem II (PSII) assembly intermediates in Synechocystis 6803, a cyanobacterium. J Biol Chem. 2011;286:24865–24871. doi: 10.1074/jbc.M111.246231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hackett CS, Strittmatter P. Covalent cross-linking of the active sites of vesicle-bound cytochrome b5 and NADH-cytochrome b5 reductase. J Biol Chem. 1984;259:3275–3282. [PubMed] [Google Scholar]
- 13.Bricker TM, Odom WR, Queirolo CB. Close association of the 33 kDa extrinsic protein with the apoprotein of CPa1 in photosystem II. FEBS Lett. 1988;231:111–117. [Google Scholar]
- 14.Seidler A. Intermolecular and intramolecular interactions of the 33-kDa protein in photosystem II. Eur J Biochem. 1996;242:485–490. doi: 10.1111/j.1432-1033.1996.0485r.x. [DOI] [PubMed] [Google Scholar]
- 15.Zouni A, et al. Crystal structure of Photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
- 16.Schweizer E, Angst W, Lutz HU. Glycoprotein topology on intact human red blood cells reevaluated by cross-linking following amino group supplementation. Biochemistry. 1982;21:6807–6818. doi: 10.1021/bi00269a029. [DOI] [PubMed] [Google Scholar]
- 17.Roose JL, Kashino Y, Pakrasi HB. The PsbQ protein defines cyanobacterial Photosystem II complexes with highest activity and stability. Proc Natl Acad Sci USA. 2007;104:2548–2553. doi: 10.1073/pnas.0609337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fagerlund RD, Eaton-Rye JJ. The lipoproteins of cyanobacterial photosystem II. J Photochem Photobiol B. 2011;104:191–203. doi: 10.1016/j.jphotobiol.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Johnson GN, Rutherford AW, Krieger A. A change in the midpoint potential of the quinone QA in Photosystem II associated with photoactivation of oxygen evolution. Biochim Biophys Acta. 1995;1229:202–207. [Google Scholar]
- 20.Stewart DH, Brudvig GW. Cytochrome b559 of photosystem II. Biochim Biophys Acta. 1998;1367:63–87. doi: 10.1016/s0005-2728(98)00139-x. [DOI] [PubMed] [Google Scholar]
- 21.Leuschner C, Bricker TM. Interaction of the 33 kDa extrinsic protein with photosystem II: Rebinding of the 33 kDa extrinsic protein to photosystem II membranes which contain four, two, or zero manganese per photosystem II reaction center. Biochemistry. 1996;35:4551–4557. doi: 10.1021/bi9522615. [DOI] [PubMed] [Google Scholar]
- 22.Burnap RL. D1 protein processing and Mn cluster assembly in light of the emerging Photosystem II structure. Phys Chem Chem Phys. 2004;6:4803–4809. [Google Scholar]
- 23.Wei L, et al. LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis. J Biol Chem. 2010;285:21391–21398. doi: 10.1074/jbc.M110.105064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guskov A, et al. Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- 25.Debus RJ. Protein ligation of the photosynthetic oxygen-evolving center. Coord Chem Rev. 2008;252:244–258. doi: 10.1016/j.ccr.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashino Y, et al. Low-molecular-mass polypeptide components of a photosystem II preparation from the thermophilic cyanobacterium Thermosynechococcus vulcanus. Plant Cell Physiol. 2002;43:1366–1373. doi: 10.1093/pcp/pcf168. [DOI] [PubMed] [Google Scholar]
- 27.Kashino Y, Koike H, Satoh K. An improved sodium dodecyl sulfate-polyacrylamide gel electrophoresis system for the analysis of membrane protein complexes. Electrophoresis. 2001;22:1004–1007. doi: 10.1002/1522-2683()22:6<1004::AID-ELPS1004>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 28.Mortensen P, et al. MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J Proteome Res. 2010;9:393–403. doi: 10.1021/pr900721e. [DOI] [PubMed] [Google Scholar]
- 29.Xu H, Freitas MA. MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.