Abstract
We describe approaches for distances between pairs of two-dimensional surfaces (embedded in three-dimensional space) that use local structures and global information contained in interstructure geometric relationships. We present algorithms to automatically determine these distances as well as geometric correspondences. This approach is motivated by the aspiration of students of natural science to understand the continuity of form that unites the diversity of life. At present, scientists using physical traits to study evolutionary relationships among living and extinct animals analyze data extracted from carefully defined anatomical correspondence points (landmarks). Identifying and recording these landmarks is time consuming and can be done accurately only by trained morphologists. This necessity renders these studies inaccessible to nonmorphologists and causes phenomics to lag behind genomics in elucidating evolutionary patterns. Unlike other algorithms presented for morphological correspondences, our approach does not require any preliminary marking of special features or landmarks by the user. It also differs from other seminal work in computational geometry in that our algorithms are polynomial in nature and thus faster, making pairwise comparisons feasible for significantly larger numbers of digitized surfaces. We illustrate our approach using three datasets representing teeth and different bones of primates and humans, and show that it leads to highly accurate results.
Keywords: homology, Mobius transformations, morphometrics, Procrustes
To document and understand physical and biological phenomena (e.g., geological sedimentation, chemical reactions, ontogenetic development, speciation, evolutionary adaptation, etc.), it is important to quantify the similarity or dissimilarity of objects affected or produced by the phenomena under study. The grain size or elasticity of rocks, geographic distances between populations, or hormone levels and body masses of individuals—these can be readily measured, and the resulting numerical values can be used to compute similarities/distances that help build understanding. Other properties like genetic makeup or gross anatomical structure cannot be quantified by a single number; determining how to measure and compare these is more involved (1–4). Representing the structure of a gene (through sequencing) or quantification of an anatomical structure (through the digitization of its surface geometry) leads to more complex numerical representations. Even though such representations are not measurements allowing direct comparison among samples of genes or anatomical structures, they form an essential initial step for such quantitative comparisons. The one-dimensional, sequential arrangement of genomes and the discrete variation (four nucleotide base types) for each of thousands of available correspondence points help reduce the computational complexity of determining the most likely alignment between genomes; alignment procedures are now increasingly automated (5). The resulting, rapidly generated and massive datasets, analyzed with increasing sophistication and flexible in-depth exploration due to advances in computing technology, have led to spectacular progress. For instance, phylogenetics has begun to unravel mysteries of large-scale evolutionary relationships experienced as extraordinarily difficult by morphologists (6).
Analyses of massive developmental and genetic datasets outpace those on morphological data. The comparative study of gross anatomical structures has lagged behind mainly because it is harder to determine corresponding parts on different samples, a prerequisite for measurement. The difficulty stems from the higher dimension (two for surfaces vs. one for genomes), the continuous rather than discrete nature of anatomical objects,* and from large degrees of shape variations.
In standard morphologists’ practice, correspondences are first assessed visually; then, some (10–100, at most) feature points can usually be defined as equivalent and/or identified as landmarks. Just as comparisons of tens of thousands of nucleotide base positions are used to determine similarity among genomes, the coordinates of these dozens of feature points (or measurements they define) are used to evaluate patterns of shape variation and similarity/difference (7). However, as stated in 1936 by G. G. Simpson, the paleontologist chaperon of the Modern Synthesis in the study of evolution, the “difficulty in acquiring personal knowledge” (ref. 8, p. 3) of morphological evidence limits our understanding of the evolutionary significance of morphological diversity; this remains true today. New techniques for generating and analyzing digital representations have led to major advances (see, e.g., refs. 9–11), but they typically still require determinations of anatomical landmarks by observers whose skill of identifying anatomical correspondences takes many years of training.
Several groups have sought to determine automatic correspondence among morphological structures. Existing successful methods typically introduce an effective dimensional reduction, using, e.g., 2D outlines and/or images (12) or, in one of the few studies attempting automatic biological correspondences in 3D as a method for evolutionary morphologists, “automatically detected crest lines” (13) on surfaces obtained by computed tomography (CT) scans to register modern human skulls to each other (14) or to pre-Neanderthal, Homo heidelbergensis skulls (15); another example is Wiley et al. (9). Studies using 2D outlines or images sacrifice a lot of the original geometric information available in the 3D objects on which they are based; such specifically limited representations cannot easily be incorporated into other studies. More generally and most importantly, none of these methods are independent of user input. When outlines or standard 2D views are used, precise observations of the 3D anatomical structures are required by the trained technician who creates the outlines or 2D views (16, 17). Several methods for 3D alignment use iterative closest point (ICP) algorithms (18), which require observer input to fix an initial guess (then further improved via local optimization); ICP-generated correspondences can also have large distortions and discontinuities of shape. In refs. 19 and 20, surfaces are matched by using the Gromov–Hausdorff distances between them, and applications to several shape analysis problems are given. However, Gromov–Hausdorff distances are hard to compute and have to be approximated; the gradient descent optimization used in practice does not guarantee convergence to a global (rather than local) minimum.
Determination of correspondences or similarities among 3D digitizations of general anatomic surfaces that is both (i) fully automated and (ii) computationally fast (to handle the large datasets that are becoming increasingly available as imaging technologies become more widespread and efficient; ref. 21) is still elusive. Our aim here is to fill this gap by fully automating the determination of correspondences among gross anatomical structures. Success in this pursuit will help bring to phenomic studies the rate, objectivity, and exhaustiveness of genomic studies. Large-scale initiatives to phenotype model species after systematically knocking out each gene (22), as well as analysis of computational simulations of organogenesis (23), stand to greatly benefit from automating the determination of correspondence among, and measurement of, morphological structures.
In this paper, we describe several distances between surfaces that can be used for such fully automated anatomic correspondences, and we test their relevance for biologically meaningful tasks on several anatomical dataset examples (high-resolution digitizations of bones and teeth). The paper is organized as follows. Section 1 gives the mathematical background for our algorithms: conformal geometry and optimal mass transportation (also known as Earth Mover’s distance). In section 2, we use these ingredients to define distances or measures of dissimilarity, including a generalization to surfaces of the Procrustes distance. Section 3 presents the results obtained by our algorithms for three different morphological datasets and an application.
No technical advance stands on its own; this paper is no exception. Conformal geometry is a powerful mathematical tool (permitting the reduction of the study of surfaces embedded in 3D space to 2D problems) that has been useful in many computational problems; ref. 24 provides an introduction to both theory and algorithms, with many applications, including the use of conformal images of anatomical structures, combined with user-prescribed landmarks and/or special features, for registration purposes, seeking “optimal” correspondence between pairs of surfaces (25). Earth Mover’s distances (26) and continuous optimal mass transportation (27) have been used in image registration and for more general image analysis and parameterization (28); in ref. 29, (quadratic) mass transportation is used to relax the notion of Gromov–Hausdorff distance. Procrustes distances for discrete point sets are familiar to morphologists and other researchers working on shape analysis (7, 11). The mathematical and algorithmic contribution of our work is the combination in which we use and generalize these ingredients to construct appropriate distance metrics, paired with efficient, fully automatic algorithms not requiring user guidance. They open the door to new applications requiring a large number of distance computations.
1. Mathematical Components
Conformal Geometry.
A mapping φ from one two-dimensional (smooth) surface  to another,
to another, 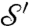 , defines for every point
, defines for every point 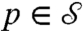 a corresponding point
a corresponding point 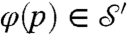 . If the mapping is smooth itself, it maps a smooth curve Γ on
. If the mapping is smooth itself, it maps a smooth curve Γ on  to a corresponding smooth curve Γ′ on
to a corresponding smooth curve Γ′ on 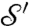 , which is called the image of Γ. Two curves Γ1 and Γ2 on
, which is called the image of Γ. Two curves Γ1 and Γ2 on 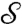 that intersect in a point s are mapped to curves
that intersect in a point s are mapped to curves 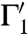 ,
, 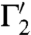 that intersect as well, in s′ = φ(s). Consider the two (straight) lines ℓ1 and ℓ2 that are tangent to the curves Γ1 and Γ2 at their intersection point s; the angle between Γ1 and Γ2 at s is then taken to mean the angle between the two lines ℓ1 and ℓ2; similarly, the angle between the curves
that intersect as well, in s′ = φ(s). Consider the two (straight) lines ℓ1 and ℓ2 that are tangent to the curves Γ1 and Γ2 at their intersection point s; the angle between Γ1 and Γ2 at s is then taken to mean the angle between the two lines ℓ1 and ℓ2; similarly, the angle between the curves 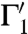 and
and 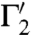 [at s′ = φ(s)] is the angle between their tangent lines at s′. The mapping φ is called conformal if for any two smooth curves Γ1 and Γ2 on
[at s′ = φ(s)] is the angle between their tangent lines at s′. The mapping φ is called conformal if for any two smooth curves Γ1 and Γ2 on 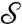 , the angle between their images
, the angle between their images 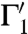 and
and 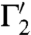 is the same as that between Γ1 and Γ2 at the corresponding intersection point.
is the same as that between Γ1 and Γ2 at the corresponding intersection point.
Riemann’s uniformization theorem (24) guarantees that every (reasonable) 2D surface 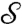 in our standard 3D space that is a disk-type surface (i.e., that has a boundary but no holes) can be mapped conformally to the 2D unit disk D = {z∣z = x + iy,|z| ≤ 1}, with the boundary of the disk corresponding to the boundary of
in our standard 3D space that is a disk-type surface (i.e., that has a boundary but no holes) can be mapped conformally to the 2D unit disk D = {z∣z = x + iy,|z| ≤ 1}, with the boundary of the disk corresponding to the boundary of 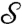 .† This mapping is called “conformally flattening”.‡ This flattening process is accompanied by area distortion; the conformal factor f(x,y) on the disk, varying from point to point, indicates the area distortion factor produced by the operation.
.† This mapping is called “conformally flattening”.‡ This flattening process is accompanied by area distortion; the conformal factor f(x,y) on the disk, varying from point to point, indicates the area distortion factor produced by the operation.
One important practical implication of this theorem is that the family of conformal maps between two surfaces can be characterized naturally via the flattened representations of the surfaces: If γ is a conformal mapping from 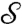 to
to 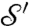 , and φ (φ′) is a flattening (i.e., a conformal map to the disk D) of
, and φ (φ′) is a flattening (i.e., a conformal map to the disk D) of 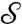 (
(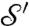 ), then the family of all possible conformal mappings from
), then the family of all possible conformal mappings from 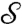 to
to 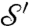 is given by γ = φ′-1∘m∘φ, where m ranges over all the conformal bijective self-mappings of the unit disk D. We shall call such m disk-preserving Möbius transformations; they constitute a group, the disk-preserving Möbius transformation group
is given by γ = φ′-1∘m∘φ, where m ranges over all the conformal bijective self-mappings of the unit disk D. We shall call such m disk-preserving Möbius transformations; they constitute a group, the disk-preserving Möbius transformation group 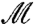 . Each m in
. Each m in 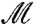 is characterized by three parameters and is given by the closed-form formula
is characterized by three parameters and is given by the closed-form formula 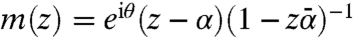 , where θ∈[0,2π), |α| < 1. For our applications, it is important that the flattening process (starting from a triangulated digitized version of
, where θ∈[0,2π), |α| < 1. For our applications, it is important that the flattening process (starting from a triangulated digitized version of 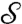 ) and more importantly the disk-preserving Möbius transformations can be computed fast and with high accuracy; for more details, see refs. 30 and 31.§ Note that the flattening map of a surface
) and more importantly the disk-preserving Möbius transformations can be computed fast and with high accuracy; for more details, see refs. 30 and 31.§ Note that the flattening map of a surface 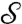 is not unique; one can choose any arbitrary point of
is not unique; one can choose any arbitrary point of 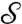 to be mapped to the origin of the disk D, and any direction through this point to become the “x axis.” The transition from choosing one (center, direction) pair to another is simply a disk-preserving Möbius transformation. It is convenient to equip the disk D with its hyperbolic measure dη(x,y) = [1 - (x2 + y2)]-2dxdy, invariant with respect to Möbius transformations; correspondingly, we set f(x,y) = [1 - (x2 + y2)]2f(x,y), so that fdη = fdxdy.
to be mapped to the origin of the disk D, and any direction through this point to become the “x axis.” The transition from choosing one (center, direction) pair to another is simply a disk-preserving Möbius transformation. It is convenient to equip the disk D with its hyperbolic measure dη(x,y) = [1 - (x2 + y2)]-2dxdy, invariant with respect to Möbius transformations; correspondingly, we set f(x,y) = [1 - (x2 + y2)]2f(x,y), so that fdη = fdxdy.
Optimal Mass Transportation.
An integrable function μ is a (normalized) mass distribution on a domain D if μ(u)≥0 is well defined for each u∈D, and ∫Dμ(u)du = 1. If τ is a differentiable bijection from D to itself, the mass distribution μ′ = τ∗μ on D defined by μ(u) = μ′(τ[u])Jτ(u) (where Jτ is the Jacobian of the map τ) is the transportation (or push-forward) of μ by τ in the sense that, for any arbitrary (nonpathological) function F on D, ∫DF(u)μ′(u)du = ∫DF(τ[u])μ(u)du. The total transportation effort is given by 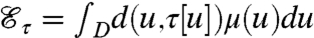 , where d(u,v) denotes the distance between two points u and v in D.
, where d(u,v) denotes the distance between two points u and v in D.
If two mass distributions μ and ν on D are given, then the optimal mass-transportation distance between μ and ν (in the sense of Monge; see ref. 32, p. 4) is the infimum of the transportation effort 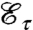 , taken over all the measurable bijections τ from D to D for which ν equals the transportation of μ by τ. This set of bijections is hard to search; the determination of an optimal mass-transportation scheme becomes more tractable if the mass “at u” need not all end up at the same end point. One then considers measures π on D × D with marginals μ and ν (which means that, for all continuous functions F, G on D, ∫D×DF(u)dπ(u,v) = ∫DF(u)μ(u)du and ∫D×DG(v)dπ(u,v) = ∫DG(v)ν(v)dv); the optimal mass transportation in this more general Kantorovitch formulation is the infimum over all such measures π of Eπ = ∫D×Dd(u,v)dπ(u,v). A comprehensive treatment of optimal mass transport is in ref. 32.
, taken over all the measurable bijections τ from D to D for which ν equals the transportation of μ by τ. This set of bijections is hard to search; the determination of an optimal mass-transportation scheme becomes more tractable if the mass “at u” need not all end up at the same end point. One then considers measures π on D × D with marginals μ and ν (which means that, for all continuous functions F, G on D, ∫D×DF(u)dπ(u,v) = ∫DF(u)μ(u)du and ∫D×DG(v)dπ(u,v) = ∫DG(v)ν(v)dv); the optimal mass transportation in this more general Kantorovitch formulation is the infimum over all such measures π of Eπ = ∫D×Dd(u,v)dπ(u,v). A comprehensive treatment of optimal mass transport is in ref. 32.
2. New Distances Between Two-Dimensional Surfaces
Conformal Wasserstein Distance (cW).
One can use optimal mass transport to compare conformal factors f and f′ obtained by conformally flattening two surfaces, 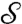 and
and 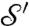 . If m is a disk-preserving Möbius transformation, then f and m∗f = f∘m-1 are both equally valid conformal factors for
. If m is a disk-preserving Möbius transformation, then f and m∗f = f∘m-1 are both equally valid conformal factors for 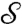 . A standard approach to take this equivalence into account is to “quotient” over
. A standard approach to take this equivalence into account is to “quotient” over 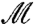 , which leads to the conformal Wasserstein distance:
, which leads to the conformal Wasserstein distance:
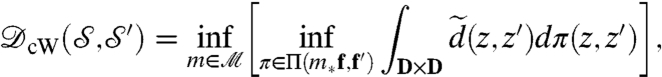 |
[1] |
where 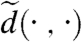 is the (conformally invariant) hyperbolic distance ¶ in D;
is the (conformally invariant) hyperbolic distance ¶ in D; 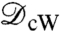 satisfies then all the properties of a metric (33). In particular,
satisfies then all the properties of a metric (33). In particular, 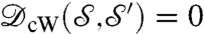 if
if 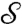 and
and 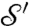 are isometric. However, computing this metric requires solving a Kantorovitch mass-transportation problem for every candidate m; even though the whole procedure has polynomial runtime complexity, it is too heavy to be used in practice for large datasets.
are isometric. However, computing this metric requires solving a Kantorovitch mass-transportation problem for every candidate m; even though the whole procedure has polynomial runtime complexity, it is too heavy to be used in practice for large datasets.
Conformal Wasserstein Neighborhood Dissimilarity Distance (cWn).
We propose another natural way to use Kantorovich’s optimal mass transport to compare surfaces 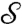 and
and 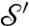 . Instead of determining the most efficient way to transport “mass” f from z to z′, we can quantify how dissimilar the “landscapes” are, defined by f and f′ near z, respectively, z′, and replace the distance
. Instead of determining the most efficient way to transport “mass” f from z to z′, we can quantify how dissimilar the “landscapes” are, defined by f and f′ near z, respectively, z′, and replace the distance 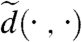 by a measure of neighborhood dissimilarity. The neighborhood N(0,R) around 0 is given by N(0,R) = {z; |z| < R}; neighborhoods around other points are obtained by letting the disk-preserving Möbius transformations act on N(0,R): For any m in
by a measure of neighborhood dissimilarity. The neighborhood N(0,R) around 0 is given by N(0,R) = {z; |z| < R}; neighborhoods around other points are obtained by letting the disk-preserving Möbius transformations act on N(0,R): For any m in 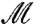 such that z = m(0), N(z,R) is the image of N(0,R) under the mapping m. Next we define the dissimilarity between f at z and f′ at z′:
such that z = m(0), N(z,R) is the image of N(0,R) under the mapping m. Next we define the dissimilarity between f at z and f′ at z′:
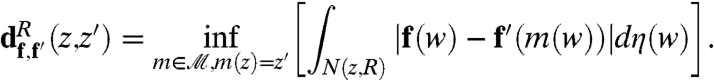 |
It is straightforward to check that, for all m,m′ in 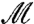 ,
, 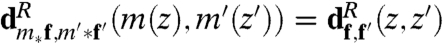 . We now use optimal transport and define the conformal Wasserstein neighborhood dissimilarity distance between f and f′:
. We now use optimal transport and define the conformal Wasserstein neighborhood dissimilarity distance between f and f′:
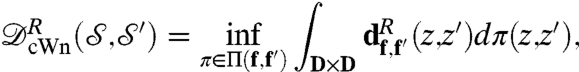 |
[2] |
where the superscript recalls that this definition depends on the choice of the parameter R. For a proof that this formula defines a true distance between (generic) surfaces 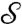 and
and 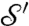 , and further mathematical properties, see refs. 33 and 34. One practical difference with
, and further mathematical properties, see refs. 33 and 34. One practical difference with 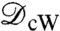 is that [2] requires solving only one Kantorovitch mass-transportation problem once the special dissimilarity cost is computed, resulting in a simpler optimization problem. To implement the computation of these distances, we discretize the integrals and the optimization searches, picking collections of discrete points on the surfaces; the minimizing measure π in the definition of
is that [2] requires solving only one Kantorovitch mass-transportation problem once the special dissimilarity cost is computed, resulting in a simpler optimization problem. To implement the computation of these distances, we discretize the integrals and the optimization searches, picking collections of discrete points on the surfaces; the minimizing measure π in the definition of 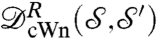 can then be used to define a correspondence between points of
can then be used to define a correspondence between points of 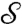 and
and 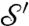 .
.
Continuous Procrustes Distance Between Surfaces.
Both cW and cWn are intrinsic: They use only information “visible” from within each surface, such as geodesic distances between pairs of points; consequently, they do not distinguish a surface from any of its isometric embeddings in 3D. The continuous Procrustes (cP) distance (35) described in this section uses some extrinsic information as well; it fails to distinguish two surfaces only if one is obtained by applying to the other a rigid motion (which is a very special isometry).
The (standard) Procrustes distance is defined between discrete sets of points 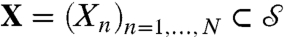 and
and 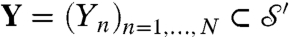 by minimizing over all rigid motions:
by minimizing over all rigid motions: 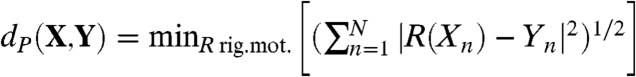 , where |·| denotes the standard Euclidean norm ∥. Often X,Y are sets of landmarks on two surfaces, and dP(X,Y) is interpreted as a distance between these surfaces. This practice has several drawbacks: (i) dP(X,Y) depends on the (subjective) choice of X,Y, which makes it a not necessarily “well-defined” or easily reproducible proxy for a surface distance; (ii) the (relatively) small number of N landmarks on each surface disregards a wealth of geometric data; and (iii) identifying and recording the xn, yn is time consuming and requires expertise.
, where |·| denotes the standard Euclidean norm ∥. Often X,Y are sets of landmarks on two surfaces, and dP(X,Y) is interpreted as a distance between these surfaces. This practice has several drawbacks: (i) dP(X,Y) depends on the (subjective) choice of X,Y, which makes it a not necessarily “well-defined” or easily reproducible proxy for a surface distance; (ii) the (relatively) small number of N landmarks on each surface disregards a wealth of geometric data; and (iii) identifying and recording the xn, yn is time consuming and requires expertise.
We eliminate all these drawbacks by a landmark-free approach, introducing the continuous Procrustes distance. Instead of relying on experts to identify “corresponding” discrete subsets of 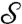 and
and 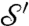 , we consider a family of continuous maps
, we consider a family of continuous maps 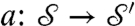 between the surfaces and rely on optimization to identify the “best” a. The earlier exact correspondence of one point Yn to one point Xn, and the (tacit) assumption that X (Y) collectively represent all the noteworthy aspects of
between the surfaces and rely on optimization to identify the “best” a. The earlier exact correspondence of one point Yn to one point Xn, and the (tacit) assumption that X (Y) collectively represent all the noteworthy aspects of 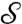
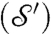 in a balanced way, are recast as requiring that the “correspondence map” be area-preserving (35)—that is, for every (measurable) subset Ω of
in a balanced way, are recast as requiring that the “correspondence map” be area-preserving (35)—that is, for every (measurable) subset Ω of 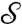 ,
, 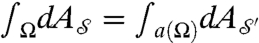 , where
, where 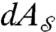 and
and 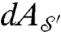 are the area elements on the surfaces induced by their embeddings in
are the area elements on the surfaces induced by their embeddings in 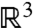 . We denote
. We denote 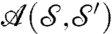 the set of all these area-preserving diffeomorphisms. For each a in
the set of all these area-preserving diffeomorphisms. For each a in 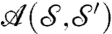 , we set
, we set 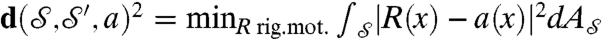 ; the continuous Procrustes distance between
; the continuous Procrustes distance between 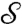 and
and 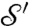 is then
is then
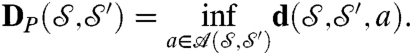 |
[3] |
Formula 3 defines a metric distance on the space of surfaces (up to rigid motions, for congruent surfaces the distance is 0) (35). Minimizing over rigid motions is easy; there exist closed-form formulas, as in the discrete case. But the second set over which to minimize, 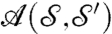 , is an unwieldy, formally infinite-dimensional manifold, hard to explore.** For “reasonable” surfaces (e.g., surfaces with uniformly bounded curvature), transformations a close to optimal are close to conformal (35). This crucial insight allows limiting the search to the much smaller space of maps obtained by small deformations of conformal maps. Concretely, we compose a conformal map (represented as a Möbius transformation)
, is an unwieldy, formally infinite-dimensional manifold, hard to explore.** For “reasonable” surfaces (e.g., surfaces with uniformly bounded curvature), transformations a close to optimal are close to conformal (35). This crucial insight allows limiting the search to the much smaller space of maps obtained by small deformations of conformal maps. Concretely, we compose a conformal map (represented as a Möbius transformation) 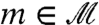 with maps χ and ϱ, where ϱ is a smooth map that roughly aligns high-density peaks, and χ is a special deformation (following ref. 37) using local diffusion to make χ∘ϱ∘m area preserving (up to approximation error). For each choice of peaks p, p′ in the conformal factors of
with maps χ and ϱ, where ϱ is a smooth map that roughly aligns high-density peaks, and χ is a special deformation (following ref. 37) using local diffusion to make χ∘ϱ∘m area preserving (up to approximation error). For each choice of peaks p, p′ in the conformal factors of 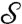 ,
, 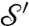 , the algorithm (i) runs through the one-parameter family of m that map p to p′; (ii) constructs a map ϱ that aligns the other peaks, as best possible; and (iii) computes
, the algorithm (i) runs through the one-parameter family of m that map p to p′; (ii) constructs a map ϱ that aligns the other peaks, as best possible; and (iii) computes 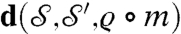 . Repeat for all choices of p, p′; the ϱ∘m that minimizes d is then deformed to be area preserving, producing the map a = χ∘ϱ∘m;
. Repeat for all choices of p, p′; the ϱ∘m that minimizes d is then deformed to be area preserving, producing the map a = χ∘ϱ∘m; 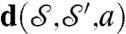 and a are our approximate
and a are our approximate 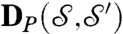 and correspondence map, respectively (more in SI Appendix, Materials).
and correspondence map, respectively (more in SI Appendix, Materials).
3. Application to Anatomical Datasets
To test our approach, we used three independent datasets, representing three different regions of the skeletal anatomy, of humans, other primates, and their close relatives. Digitized surfaces were obtained from high-resolution X-ray computed tomography (μCT) scans (see SI Appendix, Materials) of (A) 116 second mandibular molars of prosimian primates and nonprimate close relatives, (B) 57 proximal first metatarsals of prosimian primates, New and Old World monkeys, and (C) 45 distal radii of apes and humans. For every pair of surfaces, the output of our algorithms consists of (i) a correspondence map for the whole surface (i.e., not just a few points), and (ii) a nonnegative number giving their dissimilarity (where zero means they are isometric or congruent). Typical running times for a pair of surfaces were approximately 20 s for cP and approximately 5 min for cWn. To evaluate the performance of the algorithms, we compared the outcomes to those determined independently by morphologists. Using the same set of digitized surfaces, geometric morphometricians collected landmarks on each, in the conventional fashion (7), choosing them to reflect correspondences considered biologically and evolutionarily meaningful (see SI Appendix, Materials). These landmarks determine “discrete” Procrustes distances for every two surfaces (see section 2), here called observer-determined landmarks Procrustes (ODLP) distances. For each of the three distances, we obtain thus a (symmetric) matrix.
Comparing the Distance (Dissimilarity) Matrices.
We compare cWn and cP with ODLP matrices in two different ways. Sets of distances are far from independent, and it is traditional to assess the relationship between distance matrices by a Mantel correlation analysis (38): First correlate the entries in the two square arrays, and then compute the fraction, among all possible relabelings of the rows/columns for one of them, that leads to a larger correlation coefficient; this Mantel significance is a stronger indicator than the correlation coefficient itself. Table 1 gives the results for our datasets.
Table 1.
Results of Mantel correlation analysis for cP and cWN versus ODLP distances
| Obs. 1/cP | Obs. 2/cP | Obs. 1/cWn | Obs. 2/cWn | |||||
| Dataset |
r |
P |
r |
P |
r |
P |
r |
P |
| Teeth | 0.690 | 0.0001 | not applicable | 0.373 | 0.0001 | not applicable | ||
| First metatarsal | 0.640 | 0.0001 | 0.620 | 0.0001 | 0.365 | 0.0001 | 0.392 | 0.0001 |
| Radius | 0.240 | 0.0001 | not applicable | 0.075 | 0.166 | not applicable | ||
In all cases, the Mantel significance between ODLP and cP distances is higher than that between ODLP and cWn, indicating that distances computed using cP match those determined by morphometricians better than those using cWn.
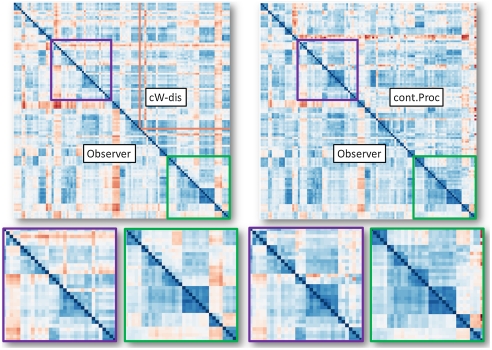
Fig. 1 illustrates the relationship of cP, cWn, and ODLP distances in a different way. In each of the two square matrices (corresponding to cP and cWn, each vs. ODLP), the color of each pixel indicates the value of the entry (using a red-blue colormap, with deep blue representing 0, and saturated red the largest value); upper right triangular halves correspond to cP or cWn; (identical) lower left halves to ODLP. The same ordering of samples is used in the three cases, with samples ordered so that nearby samples typically have smaller distances. This type of display is especially good to compare the structure of two distance matrices for small distances, often the most reliable.†† Note the better symmetry along the diagonal for ODLP/cP comparison on the left: In this comparison, as in the previous one, cP outperforms cWn.
Fig. 1.
For small distances, the structures of the matrices with cP, cWn distances and distances based on observer landmarks (ODLP) are very similar, with cP (on the right) the most similar to ODLP. The dataset illustrated here is dataset (A).
Comparing Scores in Taxonomic Classification.
Accurately placed ODL usually result in smaller ODLP distances between specimens representing individuals of the same species/genus than between individuals of different species/genera. To assess whether this result holds as well for the algorithmic cP and cWn distances, we run three taxonomic classification analyses on each dataset, one using ODLP distances, and two using cP and cWn distances,‡‡ with a “leave one out” procedure: Each specimen (treated as unknown) is assigned to the taxonomic group of its nearest neighbor among the remainder of the specimens in the dataset (treated as known). Table 2 lists success rates (in percentage) for three different classification queries for the three datasets. For each dataset and for each query, N is the number of objects and “No.” is the number of groups. Classifications based on the cP distances are similar in accuracy to those based on the ODLP distances, outperforming the cWn distances for all three of our anatomic datasets.
Table 2.
Success rates (percentage) of leave-one-out classification, based on the cP, cWn, and ODLP distances
| Dataset | Teeth | First metatarsal | Radius | |||||||||||||
| Classification |
No. |
N | cP |
Obs. 1 |
cWn |
No. |
N | Obs. 1 |
cP |
Obs. 2 |
cWn |
No. |
N | cP |
Obs. 1 |
cWn |
| Genera | 24 | 99 | 90.9 | 91.9 | 68 | 13 | 59 | 76.3 | 79.9 | 88.1 | 50.8 | 4 | 45 | 84.4 | 77.8 | 68.9 |
| Family | 17 | 106 | 92.5 | 94.3 | 75.1 | 9 | 61 | 83.6 | 91.8 | 93.4 | 68.9 | not applicable | ||||
| Above family | 5 | 116 | 94.8 | 95.7 | 83.3 | 2 | 61 | 100 | 100 | 100 | 98.4 | not applicable | ||||
Note: A similar classification based on topographic variables is less accurate; for the 99 teeth belonging to 24 genera, only 54 (54%) were classified correctly with a classification based on the four topographic variables, energy, shearing quotient, relief index, and orientation patch count (details in SI Appendix, Materials).
Comparing the Correspondence Maps.
Morphometric analyses are based on the identification of corresponding landmark points on each of 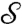 and
and 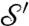 ; the cP algorithm constructs a correspondence map a from
; the cP algorithm constructs a correspondence map a from 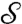 to
to 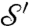 . (The correspondence induced by cWn is less smooth and will not be considered here.) For each landmark point L on
. (The correspondence induced by cWn is less smooth and will not be considered here.) For each landmark point L on 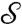 , we can compare the location on
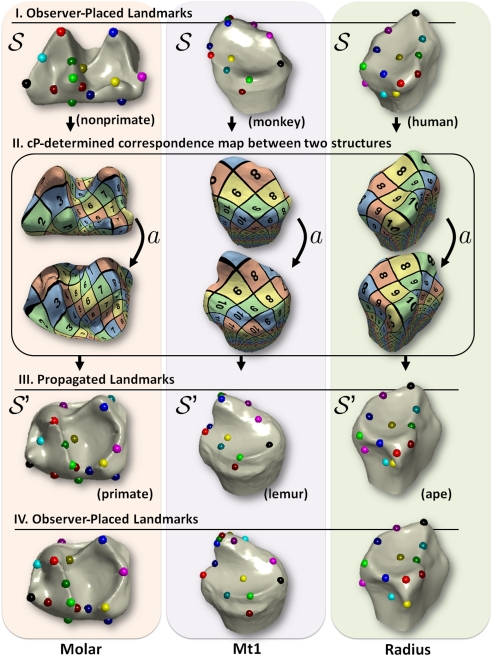
, we can compare the location on 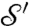 of its images a(L) with the location of the corresponding landmark points L′. Fig. 2 shows that the “propagated” landmarks a(L) typically turn out to be very close to those of the observer-determined landmarks L′ (more in SI Appendix, Materials).
of its images a(L) with the location of the corresponding landmark points L′. Fig. 2 shows that the “propagated” landmarks a(L) typically turn out to be very close to those of the observer-determined landmarks L′ (more in SI Appendix, Materials).
Fig. 2.
Observer-placed landmarks can be propagated from structure 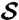 (I) using cP-determined correspondence maps (II) to another specimen
(I) using cP-determined correspondence maps (II) to another specimen 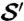 (III). The similarity between propagated landmarks in III and observer placed landmarks in IV on
(III). The similarity between propagated landmarks in III and observer placed landmarks in IV on 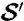 shows the success of the method and makes explicit the geometric basis for the observer determinations.
shows the success of the method and makes explicit the geometric basis for the observer determinations.
An Application.
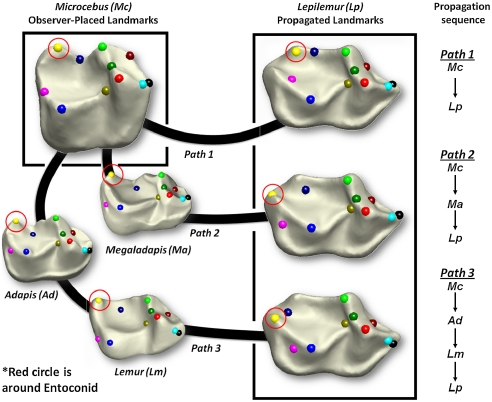
These comparisons show our algorithms capture biologically informative shape variation. But scientists are interested in more than overall shape! We illustrate how correspondence maps could be used to analyze more specific features. In comparative morphological and phylogenetic studies, anatomical identification of certain features (e.g., particular cusps on teeth) is controversial in some cases; an example of this is the distolingual corner of sportive lemur (Lepilemur) lower molars in Dataset (A) (2, 41), illustrated in Fig. 3.
Fig. 3.
Observer-placed landmarks on a tooth of Microcebus are propagated using cP-determined correspondence maps to a tooth of Lepilemur. Path 1 is direct, paths 2 and 3 have intermediate steps, representing stepwise propagation between teeth of other taxa.
In such controversial cases, transformational homology (42) hypotheses are usually supported by a specific comparative sample or inferred morphocline (2, 43, 44). Lepilemur is thought by some researchers to lack a cusp known as an entoconid (Fig. 3) but to have a hypertrophied metastylid cusp that “takes the place” of the entoconid (2) in other taxa. Yet, in comparing a Lepilemur tooth to a more “standard” primate tooth, like that of Microcebus, both seem to have the same basic cusps; alternatives to the viewpoint of ref. 2 have therefore also been argued in the literature (41). However, another lemur, Megaladapis (now extinct), arguably a closer relative of Lepilemur than Microcebus, has an entoconid that is very small and a metastylid that is rather large, thus providing an evolutionary argument supporting the original hypothesis. (For more details, see SI Appendix, Materials.) Such arguments can now be made more precise. We can propagate (as in Fig. 2) landmarks from the Microcebus to the Lepilemur molar; this direct propagation matches the entoconid cusp of Microcebus with the controversial cusp of Lepilemur (Fig. 3, path 1), supporting ref. 41. In contrast, when we propagate landmarks in different steps, either from Microcebus to Megaladapis and then to Lepilemur (Fig. 3, path 2), or through the extinct Adapis and extant Lemur (Fig. 3, path 3), the Lepilemur metastylid takes the place of the Microcebus entoconid, supporting ref. 2. Automatic propagation of landmarks via mathematical algorithms recenters the controversy on the (different) discussion of which propagation channel is most suitable.
Summary and Conclusion.
New distances between 2D surfaces, with fast numerical implementations, were shown to lead to fast, landmark-free algorithms that map anatomical surfaces automatically to other instances of anatomically equivalent surfaces, in a way that mimics accurately the detailed feature-point correspondences recognized qualitatively by scientists, and that preserves information on taxonomic structure as well as observer-determined landmark distances. Moreover, the correspondence maps thus generated can incorporate, in their tracking of point features, evolutionary relationships inferred to link different taxa together.
Our approach makes morphology more accessible to nonspecialists and allows the documentation of anatomical variation and quantitative traits with previously unmatched comprehensiveness and objectivity. More frequent, rapid, objective, and comprehensive construction of morphological datasets will allow the study of morphological diversity’s evolutionary significance to be better synchronized with studies incorporating genetic and developmental information, leading to a better understanding of anatomical form and its genetic basis, as well as the evolutionary processes that have contributed to their diversity among living things on Earth.
Supplementary Material
Acknowledgments.
Access was provided by curators and staff at the American Museum of Natural History (to dental specimens to be molded, cast, and µCT-scanned); D. Krause and J. Groenke of Stony Brook University, Department of Anatomical Sciences (to facilities of the Vertebrate Paleontology Fossil Preparation Lab); and C. Rubin and S. Judex of Stony Brook University, Department of Biomedical Engineering and the Center for Biotechnology (to µCT scanners, for digital imaging of tooth casts). A National Science Foundation (NSF) Doctoral Dissertation Improvement Grant through Physical Anthropology, the Evolving Earth Foundation, and the American Society of Physical Anthropologists provided funding (D.M.B.). Y.L., J.P., T.F., and I.D. were supported by NSF and Air Force Office of Scientific Research grants.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 18189.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112822108/-/DCSupplemental.
*These differences may seem innocuous but they lead to an exponential increase in the size of the “search spaces” to be explored by comparison algorithms.
†We shall restrict ourselves to this case here, although our approach is more general; see ref. 30).
‡The uniformization theorem holds for more general surfaces as well. For instance, surfaces without holes, handles, or boundaries can be mapped conformally to a sphere; if one point is removed from such a surface, it can be mapped conformally to the full plane. Surfaces with holes or handles can still be conformally flattened to a piece of the plane.
§If the digitization of the surface is given as a point cloud, standard fast algorithms can be used to determine an appropriate (e.g., Delauney) triangulation.
¶This distance is the geodesic distance on D induced by the hyperbolic Riemann metric tensor dη on D. The geodesic from the origin to any point z in D is the straight line connecting them, and 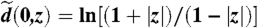 .
.
∥It is interesting to note that in ref. 36, a Kantorovich version of dP was introduced, and its equivalence to the Gromov–Wasserstein distance (when the shapes are endowed with Euclidean distances) was proved.
**This manifold can be viewed as the continuous analog to the exponentially large group of permutations.
††In some modern data analysis methods, such as diffusion-based or graph-Laplacian-based methods, only the small distances are retained, to be used in spectral methods that “knit” the larger scale distances of the dataset together more reliably.
‡‡We do not claim this method as an alternative for automatic species or genus identification. Reliable automatic species recognition uses, in addition, auditory, chemical, nongeometric morphological, and other data, analyzed by a range of methods; see, e.g., refs. 17, 39, and 40 and references therein. Comparison of taxonomic classification based on human-expert generated vs. algorithm-computed distances is meant only as a quantitative evaluation based on biology rather than mathematics.
References
- 1.Sargis EJ, Boyer DM, Bloch JI, Silcox MT. Evolution of pedal grasping in primates. J Hum Evol. 2007;53:103–107. doi: 10.1016/j.jhevol.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz JH, Tattersall I. Evolutionary Relationships of Living Lemurs and Lorises (Mammalia, Primates) and Their Potential Affinities with European Eocene Adapidae. New York: Am Museum Natural History; 1985. [Google Scholar]
- 3.MacLeod N. Generalizing and extending the eigenshape method of shape visualization and analysis. Paleobiology. 1999;25:107–138. [Google Scholar]
- 4.Bookstein FL. In: Morphometrics and Computed Homology: An Old Theme Revisited. Automated Taxon Identification in Systematics. MacLeod N, editor. London: CRC; 2007. pp. 69–82. [Google Scholar]
- 5.Liu K, Raghavan S, Nelesen S, Linder CR, Warnow T. Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science. 2009;324:1561–1564. doi: 10.1126/science.1171243. [DOI] [PubMed] [Google Scholar]
- 6.Murphy WJ, et al. Resolution of the early placental mammal radiation using bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 7.Mitteroecker P, Gunz P. Advances in geometric morphometrics. Evol Biol. 2009;36:235–247. [Google Scholar]
- 8.Simpson GG. Studies of the earliest mammalian dentitions. Dent Cosmos. 1936;78:791–800. 940–953. [Google Scholar]
- 9.Wiley DF, et al. Evolutionary morphing. Proc IEEE Visualization. 2005;8 [Google Scholar]
- 10.Polly PD, MacLeod N. Locomotion in fossil carnivora: An application of eigensurface analysis for morphometric comparison of 3D surfaces. Palaeontol Electron. 2008;11.2.8A:1–13. [Google Scholar]
- 11.Zelditch ML, Swiderski DL, Sheets DH, Fink WL. Geometric Morphometrics for Biologists. San Diego: Elsevier Academic; 2004. [Google Scholar]
- 12.Lohmann GP. Eigenshape analysis of microfossils: A general morphometric method for describing changes in shape. Math Geol. 1983;15:659–672. [Google Scholar]
- 13.Thirion JP, Gourdon A. The 3D marching lines algorithm. Graph Model Im Proc. 1996;58:503–509. [Google Scholar]
- 14.Subsol G, Thirion JP, Ayache N. A general scheme for automatically building 3D morphometric anatomical atlases: Application to a skull atlas. Med Image Anal. 1998;2:37–60. doi: 10.1016/s1361-8415(01)80027-x. [DOI] [PubMed] [Google Scholar]
- 15.Subsol G, Mafart B, Silvestre A, de Lumley MA. 3d image processing for the study of the evolution of the shape of the human skull: presentation of the tools and preliminary results. In: Mafart B, Delingette H, Subsol G, editors. Three-Dimensional Imaging in Paleoanthropology and Prehistoric Archaeology. Vol. 1049. Oxford: Archeopress; 2002. pp. 37–45. (British Archeological Reports International Series). [Google Scholar]
- 16.Gaston KJ, O’Neill MA. Automated species identification—why not? Philos Trans R Soc Lond B Biol Sci. 2004;359:655–667. doi: 10.1098/rstb.2003.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weeks PJD, O’Neill MA, Gaston KJ, Gauld ID. Species-identification of wasps using principal component associative memories. Image Vision Comput. 1999;17:861–866. [Google Scholar]
- 18.Besl PJ, McKay ND. A method for registration of 3-D shapes. IEEE Trans Pattern Anal Mach Intell. 1992;14:239–256. [Google Scholar]
- 19.Mémoli F, Guillermo Sapiro. A theoretical and computational framework for isometry invariant recognition of point cloud data. Found Comput Math. 2005;5:313–347. [Google Scholar]
- 20.Bronstein AM, Bronstein MM, Kimmel R. Generalized multidimensional scaling: A framework for isometry-invariant partial surface matching. Proc Natl Acad Sci USA. 2006;103:1168–1172. doi: 10.1073/pnas.0508601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt EJ, et al. Micro-computed tomography-based phenotypic approaches in embryology: Procedural artifacts on assessments of embryonic craniofacial growth and development. BMC Dev Biol. 2010;10:18. doi: 10.1186/1471-213X-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott A. Mouse project to find each gene’s role. Nature. 2010;465:410. doi: 10.1038/465410a. [DOI] [PubMed] [Google Scholar]
- 23.Salazar-Ciudad I, Jernvall J. A computational model of teeth and the developmental origins of morphological variation. Nature. 2010;464:583–586. doi: 10.1038/nature08838. [DOI] [PubMed] [Google Scholar]
- 24.Gu X, Yau ST. Computational Conformal Geometry. Boston: Intl Press; 2008. [Google Scholar]
- 25.Gu X, Wang Y, Chan TF, Thompson PM, Yau ST. Genus zero surface conformal mapping and its application to brain surface mapping. IEEE Trans Med Imaging. 2004;23:949–958. doi: 10.1109/TMI.2004.831226. [DOI] [PubMed] [Google Scholar]
- 26.Rubner Y, Tomasi C, Guibas LJ. The Earth mover’s distance as a metric for image retrieval. Int J Comput Vis. 2000;40:99–121. [Google Scholar]
- 27.Haker S, Zhu L, Tannenbaum A, Angenent S. Optimal mass transport for registration and warping. Int J Comput Vis. 2004;60:225–240. [Google Scholar]
- 28.Dominitz A, Tannenbaum A. Texture mapping via optimal mass transport. IEEE Trans Vis Comput Graph. 2010;16:419–433. doi: 10.1109/TVCG.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mémoli F. On the use of Gromov-Hausdorff distances for shape comparison. In: Botsch M, Parajola P, Chen B, Zwicker M, editors. Proc Point Based Graphics; Boston: AK Peters; 2007. pp. 81–90. [Google Scholar]
- 30.Lipman Y, Puente J, Daubechies I. Conformal Wasserstein distances: Comparing disk and sphere-type surfaces in polynomial time II: Computational aspects. Math Comput. 2011 arXiv:1103.4681. [Google Scholar]
- 31.Lipman Y, Funkhouser T. Mobius voting for surface correspondence. ACM Trans Graph. 2009;28:72.1–12. Proc SIGGRAPH. [Google Scholar]
- 32.Villani C. Graduate Studies in Mathematics. Vol. 58. New Providence, RI: Am Math Soc; 2003. Topics in optimal transportation. [Google Scholar]
- 33.Lipman Y, Daubechies I. Conformal wasserstein distances: Comparing surfaces in polynomial time. Adv Math. 2011;227:1047–1077. [Google Scholar]
- 34.Lipman Y, Daubechies I. Surface comparison with mass transportation. 2010. arXiv:0912.3488v2.
- 35.Lipman Y, Al-Aifari R, Daubechies I. The continuous Procrustes distance between two surfaces. Communication on Pure and Applied Mathematics. 2011 arXiv:1106:4588. [Google Scholar]
- 36.Mémoli F. Gromov–Hausdorff distances in Euclidean spaces. 2008. Computer Vision and Pattern Recognition Workshop (IEEE Computer Society); 10.1109/CVPRW.2008.4563074.
- 37.Moser J. On the volume elements on a manifold. Trans Am Math Soc. 1965;120:286–294. [Google Scholar]
- 38.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 39.MacLeod N, editor. Automated Taxon Identification in Systematics: Theory, Approaches, and Applications. London: CRC; 2007. p. 339. [Google Scholar]
- 40.MacLeod N. Understanding morphology in systematic contexts: 3D specimen ordination and 3D specimen recognition. In: Wheeler Q, editor. The New Taxonomy. London: CRC; 2008. pp. 143–210. [Google Scholar]
- 41.Swindler. Primate Dentition: An Introduction to the Teeth of Non-Human Primates. Cambridge, UK: Cambridge Univ Press; 2002. p. 312. [Google Scholar]
- 42.Patterson C. Morphological characters and homology. Systematics Association Special Volume No 21. 1982:21–74. [Google Scholar]
- 43.Osborn HF. Evolution of Mammalian Molar Teeth: To and From the Tritubercular Type. New York: Macmillan; 1907. [Google Scholar]
- 44.Van Valen L. Homology and causes. J Morphol. 1982;173:305–312. doi: 10.1002/jmor.1051730307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.