Abstract
The Vibrio fischeri LuxR protein is the founding member of a family of acyl-homoserine lactone-responsive quorum-sensing transcription factors. Previous genetic evidence indicates that in the presence of its quorum-sensing signal, N-(3-oxohexanoyl) homoserine lactone (3OC6-HSL), LuxR binds to lux box DNA within the promoter region of the luxI gene and activates transcription of the luxICDABEG luminescence operon. We have purified LuxR from recombinant Escherichia coli. Purified LuxR binds specifically and with high affinity to DNA containing a lux box. This binding requires addition of 3OC6-HSL to the assay reactions, presumably forming a LuxR-3OC6-HSL complex. When bound to the lux box at the luxI promoter in vitro, LuxR-3OC6-HSL enables E. coli RNA polymerase to initiate transcription from the luxI promoter. Unlike the well-characterized LuxR homolog TraR in complex with its signal (3-oxo-octanoyl-HSL), the LuxR-30C6-HSL complex can be reversibly inactivated by dilution, suggesting that 3OC6-HSL in the complex is not tightly bound and is in equilibrium with the bulk solvent. Thus, although LuxR and TraR both bind 3-oxoacyl-HSLs, the binding is qualitatively different. The differences have implications for the ways in which these proteins respond to decreases in signal concentrations or rapid drops in population density.
Acyl-homoserine lactone (acyl-HSL) quorum-sensing systems occur in dozens of Proteobacteria and control different specific sets of genes in different species. In general, acyl-HSL quorum-sensing systems involve members of the LuxR family of transcription factors as signal receptors (16, 21). LuxR is the Vibrio fischeri N-(3-oxohexanoyl)-HSL (3OC6-HSL) receptor, functioning as a transcriptional activator of the luminescence genes in this marine bacterium (27). 3OC6-HSL is a diffusible signal required for LuxR activation of the luxICDABEG operon (8, 19, 29).
There is a substantial literature on LuxR, but our knowledge suffers from the fact that full-length native LuxR has not been purified and studied in vitro. The LuxR polypeptide contains two domains (3, 4, 18). 3OC6-HSL is thought to interact with LuxR through a signal-binding region involving residues 81 to 129 in the N-terminal domain (about two-thirds of the polypeptide). The interaction between LuxR and 3OC6-HSL is specific in that 3OC6-HSL analogs show limited or no LuxR-mediated activation of the lux operon (25). The C-terminal one-third of LuxR is a DNA-binding domain, which contains a helix-turn-helix motif (residues 200 to 224) (4). Specific amino acids between residues 193 and 220 in the C terminus are critical for DNA binding, while others in this region are critical for lux operon activation but not for DNA binding (10, 31). This two-domain structure seems to be conserved among LuxR family members, and it is supported by the crystal structure of the LuxR homolog TraR, a quorum-sensing signal receptor from Agrobacterium tumefaciens (32, 37).
The DNA sequences required for LuxR activation at the luxI promoter have also been studied in vivo. There is a 20-bp inverted repeat termed the lux box, centered 42.5 bp upstream of the luxI transcription start site, that is required for LuxR activation of luxI transcription (5, 11, 20).
Purification of full-length LuxR has proven difficult (20, 28). Recently several other members of the LuxR family have been purified (22, 23, 33, 38). The crystal structure for TraR in complex with its cognate signal, 3OC8-HSL, and its target lux box-like sequence indicates that functional TraR is a dimer and that the signal is entirely buried within its binding pocket (32, 37). The purification of functional TraR required the presence of 3OC8-HSL during the growth of recombinant TraR-producing bacteria to ensure stability in vivo. In subsequent purification steps, the incorporated acyl-HSL copurified with TraR and could be removed only with extensive dialysis (38). The acyl-HSL requirement for TraR stability in vivo, the tenacious binding of the acyl-HSL to TraR, and the discovery that the acyl-HSL is completely buried within the acyl-HSL binding region of TraR have led to the hypothesis (39) that the acyl-HSL is required for proper folding of nascent TraR, thereby protecting mature TraR from proteolytic digestion within the bacterial cell, and that acyl-HSL binding to TraR in vivo is essentially irreversible. According to this hypothesis, during the transition exiting the quorum-satisfied state, the decrease in population density will lead to a decline in the expression of quorum-controlled genes as previously formed TraR-AI functional complexes are eventually proteolyzed. Furthermore, in the nonquorum state, acyl-HSL-responsive TraR will not be available in a cell in the absence of acyl-HSL.
We have developed a LuxR purification procedure based on insights about TraR. Like TraR, the acyl-HSL signal is required during bacterial growth to obtain active LuxR. However, unlike purified TraR, the activity of purified LuxR is dependent on added 3OC6-HSL, indicating that this signal is not tightly bound. This suggests that LuxR binding of acyl-HSL has a significantly different character than acyl-HSL binding to TraR.
MATERIALS AND METHODS
Bacterial strains and growth media.
E. coli JM107 (36) is F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14 Δ(lac-proAB) thi gyrA96 endA1 hsdR17 relA1 glnV44. E. coli BL21(DE3) (30) is ompT gal dcm lon hsdSB and is lysogenized with λDE3 carrying the phage T7 RNA polymerase gene under the control of the lacUV5 promoter and lacI. The culture medium was Luria broth (LB) supplemented where indicated with ampicillin (300 μg/ml), 25 μM 3OC6-HSL (N-[β-ketocaproyl]-l-homoserine lactone; Sigma, St. Louis Mo.), and 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG; Sigma).
Plasmid construction.
A plasmid encoding a LuxR derivative with a six-histidine (H6) C-terminal tag (pMLU117) was constructed as follows. Plasmid pKE705 (10) carries the V. fischeri MJ1 luxR gene coding region under the control of the artificial tac promoter-Shine-Dalgarno region. pKE705 was modified by deletion of an unnecessary SphI restriction site in the nonfunctional tet gene by partial digestion with SphI followed by blunting with T4 DNA polymerase, creating pMU102. The artificial Shine-Dalgarno region upstream of luxR in pMU102 was replaced with the native luxR Shine-Dalgarno region by replacing the approximately 220-bp EcoRI-XbaI luxR′ fragment in pMU102 with a PCR-generated luxR′ fragment that had its native Shine-Dalgarno region and 24 bp of upstream DNA preceded by a synthetic EcoRI site formed from the PCR primer (5′-AAGAATTCAAAACTTGCGACAAACAATAGGTAAGG). The 3′ end contained the XbaI site from pMU102. The resulting plasmid, pMLU89, contains luxR under the control of the tac promoter and the native luxR translation initiation region. Finally, the plasmid coding for LuxR with a six-histidine tag was constructed by replacing the 3′-SphI-XhoI fragment of luxR in pMLU89 with a PCR fragment generated from an internal luxR primer and the synthetic XhoI end primer (5′-AAACTCGAGTTAATGATGATGATGATGATGATTTTTAAAGTATGGGCAATC). The resulting plasmid, pMLU117, was used to produce sufficient C-terminal His-tagged LuxR for N-terminal amino acid sequence analysis.
The plasmid used for overexpression of native LuxR (pMLU115) was constructed as follows. The luxR-coding region was amplified by PCR with pMU102 as the template. The reverse primer included the downstream BamHI site; the upstream primer was 5′-AAGCCATGGGTATGAAAAACATAAATGCCGACGAC-3′ and included a synthetic NcoI site centered on the luxR ATG start codon determined below. The approximately 770-bp NcoI-BamHI fragment was cloned into NcoI-BamHI-digested pET-3d (Novagen, Madison, Wis.) from which its unnecessary HindIII site had been deleted by deletion of a small EcoRI-EcoRV-EcoRV vector fragment. In the resulting plasmid, pMLU115, luxR expression is controlled by the phage T7 promoter.
Determination of the luxR translation start codon.
E. coli JM107 containing the LuxR-C-terminal His tag vector pMLU117 was grown in 25 ml of LB at 37°C. IPTG was added at an optical density at 600 nm (OD600) of 0.5, and incubation was continued for an additional 3 h. Cells were harvested by centrifugation, suspended, and lysed in the presence of 8 M urea, and the denatured LuxR-His6 peptide was purified by affinity chromatography on a His-Trap column (Pharmacia, Piscataway, N.J.) according to the manufacturer's denaturing purification protocol. The resulting LuxR-His6 preparation was further purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The band at 28 kDa was excised from the gel and transferred to a Problot polyvinylidene difluoride membrane (Bio-Rad, Hercules, Calif.) according to the manufacturer's instructions. The N-terminal amino acid sequence was determined at the University of Iowa Molecular Analysis Facility.
Purification of LuxR.
The purification protocol is based on that used for the A. tumefaciens LuxR homolog TraR (38). Colonies of the LuxR-overexpressing strain BL21(DE3)[pMLU115] freshly grown overnight on LB-ampicillin plates at 37°C were used to inoculate 1 liter of LB-ampicillin prewarmed at 37°C. The culture was grown at 37°C to an OD600 of 0.5 and was then quickly chilled in an ice bath to 28°C. IPTG and 3OC6-HSL were added, together with additional ampicillin (200 μg/ml), and culture growth was continued at 28°C for 4 h. Cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C and were frozen overnight at −80°C. All subsequent steps were performed at 0 to 4°C. The cells were suspended in 16 ml of A buffer (50 mM Tris-HCl [pH 7.0 at 22°C], 100 mM KCl, 50 mM NaCl, 2 mM EDTA, 2 mM dithiothreitol, 10% glycerol, 0.5% Tween 20) containing 25 μM 3OC6-HSL and were lysed by sonication. The lysate was centrifuged at 20,000 × g for 10 min, and the resulting supernatant fluid was centrifuged at 100,000 × g for 1.5 h. The cleared 100,000 × g cell extract was chromatographed on a 5-ml HiTrap heparin column (Amersham Pharmacia Biotech, Piscataway, N.J.) equilibrated with A buffer plus 25 μM 3OC6-HSL. The bound proteins were eluted using a 25-ml NaCl step gradient from 0.15 to 1 M. Fractions containing LuxR were pooled and dialyzed against 500 ml of A buffer containing 25 μM 3OC6-HSL and 20% (wt/vol) polyethylene glycol (molecular weight, 8,000) for 4 h to simultaneously lower the NaCl concentration and concentrate the sample twofold. After dialysis the sample was diluted 1:1 with a KCl- and NaCl-free version of A buffer plus 25 μM 3OC6-HSL to obtain a final salt concentration of 75 mM and was immediately loaded onto a tandemly arranged pair of ion-exchange columns comprising a 5-ml HiTrap Q Sepharose Fast Flow column and a 5-ml HiTrap SP Sepharose Fast Flow column (Amersham Pharmacia Biotech). Both columns had been equilibrated with B buffer (like buffer A, except that 50 mM KCl and 25 mM NaCl were used) plus 25 μM 3OC6-HSL. After the tandem columns were washed with B buffer, the Q column was removed, and the proteins bound to the SP column were eluted with a 100-ml linear NaCl gradient of 0.075 to 0.5 M. Fractions containing >95% pure LuxR (as judged by SDS-PAGE) were pooled and stored at −80°C at final concentrations of 10 to 50 μM monomer.
Gel shift experiments.
The gel shift protocol was based on the methods of Fried and Crothers (14) and Garner and Revzin (17). The DNA probes were derived from a 400-bp PCR product containing the luxI-luxR regulatory region. This PCR product was labeled at both ends using [γ32-P]ATP plus T4 nucleotide kinase and was subsequently cleaved with MwoI, resulting in a 160-bp fragment and a 240-bp fragment (see Fig. 1A). Protein-DNA binding reaction mixtures contained approximately 1 fmol each of the two DNA probes in a final volume of 20 μl of DNA binding buffer (20 mM Tris-HCl [pH 7.4 at 22°C], 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 100 μg of bovine serum albumin/ml, and 5% glycerol). Purified LuxR and 3OC6-HSL were added as indicated, and the reaction mixtures were incubated at 26°C for 20 min. One microliter of loading dye (0.1% xylene cyanol in 50% glycerol) was added, and the reaction mixtures were loaded onto a native 5% Tris-glycine-EDTA gel at 4°C. Following electrophoresis at 10 V/cm at 4°C, the probes were detected and quantitated using a Typhoon model 8600 phosphorimager with ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
FIG. 1.
3OC6-HSL-dependent DNA binding of purified LuxR. (A) The two DNA fragments comprising the luxI-luxR promoter region used as probes in the gel shift assay. The location of the 20-bp lux box sequence is indicated by the open rectangle. The vertical dashed line indicates the MwoI restriction site used to produce the two probes from a 400-bp PCR product. The −10 regions, along with the transcription start sites (+1) and Shine-Dalgarno regions (SD) for the divergent luxR and luxI promoters, are indicated. (B) Gel shift assay of LuxR binding to luxI promoter DNA. All lanes contain approximately 1 fmol of an equimolar mixture of the two probes and the indicated concentration of LuxR either with or without 6 μM 3OC6-HSL as indicated.
DNase I protection assays.
DNase I footprinting was based on previously described methods (26). The DNA fragment was the same as the probe used in gel shift experiments (Fig. 1A), except that the full-length 400-bp PCR fragment was first cloned as an EcoRI-BamHI fragment on a plasmid. The fragment was isolated by digesting the plasmid at the luxI end with BamHI, labeling with [γ32-P]ATP and T4 DNA kinase, and digesting with EcoRI. The end-labeled 400-bp fragment was purified after sizing by 5% PAGE. Approximately 0.01 pmol of the 32P-labeled DNA probe, along with the indicated amounts of LuxR protein or E. coli RNA polymerase (σ70 saturated; Epicentre, Madison, Wis.), was incubated in 120 μl of the gel shift DNA binding buffer, either with or without 6 μM 3OC6-HSL, for 20 min at 25°C to allow protein-DNA complexes to form. Six microliters (150 mU) of RQ1 DNase I (in 25 mM Tris-HCl [pH 7.4 at 22°C]-30 mM MgCl2,-30 mM CaCl; Promega, Madison, Wis.) was then added, and incubation was continued for an additional 30 s. The reactions were stopped by addition of 25 μl of footprint stop solution (3 M ammonium acetate, 0.25 M EDTA, 15 μg of sheared calf thymus DNA/ml), and DNA was precipitated with 370 μl of ethanol. DNase I digestion products were separated on 5% denaturing polyacrylamide gels and were visualized and quantitated using phosphorimager technology.
In vitro transcription assays.
Runoff assays were performed as previously described (35) by using purified E. coli RNA polymerase (σ70 saturated; Epicentre) and purified LuxR. The linear DNA template was a purified 612-bp restriction fragment carrying the divergent luxI-luxR promoters. Transcription from the luxI promoter on this fragment produces a 300-base runoff transcript. The predicted luxR promoter transcript is 170 bases in length. In early experiments, gels contained two molecular size standards to identify the 300-base luxI runoff transcript; in later experiments, gels run under identical conditions gave identical band patterns in the luxI template lanes, and thus the molecular size standards were no longer included. In brief, DNA template (1 nM) in transcription buffer (10 mM Tris-HCl [pH 7.4 at 22°C], 50 mM KCl, 8 mM MgCl2) containing 10 μM 3OC6-HSL was preincubated either without or with LuxR at the indicated concentrations for 5 min at 25°C. RNA polymerase was then added (10 nM), and incubation was continued for 20 min. Nucleotide triphosphates containing [α32-P]ATP were added in the presence of 50 μg of heparin/ml, and a single round of transcription was allowed to proceed for 15 min. Reactions were stopped by addition of footprint stop solution (see above), and the transcripts were separated on a 5% denaturing polyacrylamide gel. Bands were visualized and quantitated by phophorimager technology.
RESULTS
LuxR purification.
The luxR gene from V. fischeri MJ1 was previously reported to encode a 250-amino-acid polypeptide (20). However, an in-frame AUG two codons upstream of the reported start codon could also serve as a translation start site for luxR. A carboxy-terminal histidine-tagged derivative of LuxR, overexpressed in E. coli by using the native luxR Shine-Dalgarno region, was purified by nickel-affinity chromatography and subjected to N-terminal sequence analysis of the first 12 amino acids. More than 90% of the LuxR-His6 polypeptide initiated at the upstream AUG codon, indicating that native LuxR is encoded by the longer 252-codon open reading frame.
LuxR purification was based on a method used for purifying the LuxR homologue TraR from A. tumefaciens (38). The luxR coding region specifying the longer, 252-residue protein was placed downstream of the T7 promoter and Shine-Dalgarno region of plasmid pET-3d. E. coli BL21(DE3) carrying the resulting plasmid pMLU115 was grown in LB, and luxR expression was induced by IPTG in the presence or absence of the V. fischeri signal molecule 3OC6-HSL (at 25 μM). A gel mobility shift assay (see below) demonstrated that clarified cell extracts of cultures grown in the presence of 3OC6-HSL specifically bound DNA containing a lux box sequence in a 30C6-HSL-dependent manner. Extracts of cultures grown in the absence of 30C6-HSL did not show specific lux box binding, and the 3OC6-HSL could not be replaced with octanoyl-HSL (data not shown). LuxR was purified from the clarified extract of cells grown in the presence of 3OC6-HSL as described in Materials and Methods. SDS-PAGE analysis of the pooled fractions and Western blot analysis using a LuxR-specific antiserum indicated a single polypeptide species (>95% purity) with an Mr of 28,000; matrix-assisted laser desorption ionization—time-of-flight (MALDI-TOF) analysis of the purified protein indicated a monomeric molecular weight of 28,698 (data not shown). Both values are in agreement with the calculated LuxR molecular weight of 28,744 for the 252-amino-acid LuxR monomer.
LuxR binding to DNA depends on added 3OC6-HSL.
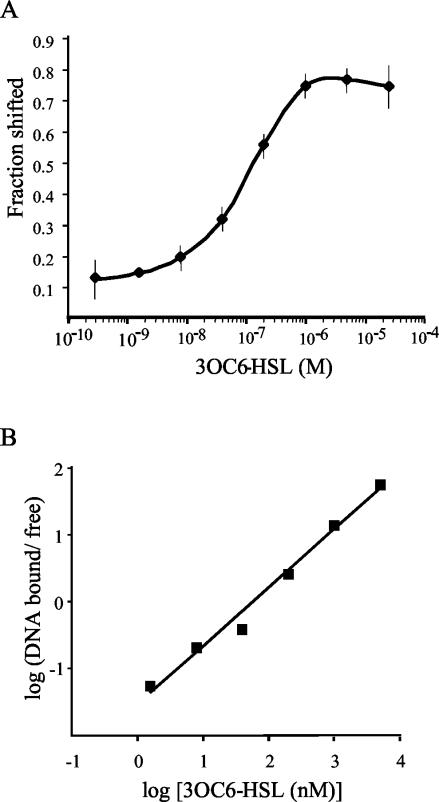
LuxR DNA binding activity was tested in a mobility shift assay using two DNA probes, only one of which contained a lux box (Fig. 1A). In the absence of 3OC6-HSL, LuxR did not bind to either probe at the highest LuxR concentration tested (3.5 nM) (Fig. 1B). In the presence of 10 μM 3OC6-HSL, 3.5 nM LuxR was sufficient to shift nearly all of the lux box probe without affecting the mobility of the nonspecific probe (Fig. 1B; compare lanes 1 to 4). Titration of LuxR in the presence of 10 μM 3OC6-HSL indicated that, under these conditions, 0.2 nM LuxR polypeptide was sufficient to shift 50% of the specific probe (Fig. 1B). In similar experiments, titration of 3OC6-HSL in the presence of 3.5 nM LuxR showed that 100 nM 3OC6-HSL was required to shift 50% of the specific probe (Fig. 2A). A Hill plot of the data in Fig. 2A yielded a Hill coefficient of 0.9, indicating that the stimulation by 3OC6-HSL of LuxR-DNA complex formation is a noncooperative process (Fig. 2B).
FIG. 2.
Estimation of LuxR binding affinity for 3OC6-HSL. (A) Gel shift reactions using increasing concentrations of 3OC6-HSL in the presence of 3.5 nM LuxR were run. The fractional amount of the specific probe shifted (y axis) was determined at each concentration of 3OC6-HSL (x axis) and averaged. Error bars, standard errors of the means. (B) Hill plot of the data in Fig. 2A.
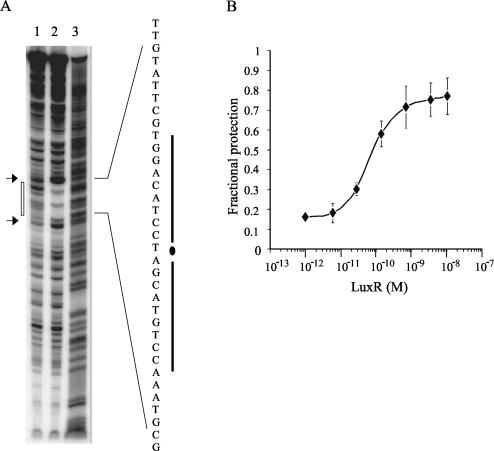
DNase I footprinting was used to test whether LuxR binds specifically to the lux box. In the presence of 10 μM 3OC6-HSL, LuxR protected a 28-bp region that included the 20-bp lux box (Fig. 3A). The presence of two unprotected bases within this region suggests that, like TraR (36), LuxR may bind to only one face of the DNA. Additionally, LuxR binding resulted in enhanced DNase I digestion approximately 8 bp upstream and downstream of the lux box. DNase I protection assays can also be used as an alternative method to measure binding to DNA under equilibrium conditions (2). We used LuxR concentrations varying over 4 orders of magnitude in the DNase I protection assay to measure the LuxR-lux box DNA dissociation constant (Fig. 3B). Under our experimental conditions, 0.1 nM LuxR was sufficient to give a fractional protection of 0.5, in good agreement with the apparent affinity indicated by the gel shift experiments. Because the footprint experiments indicate that the luxI promoter fragment contains only a single LuxR binding site, it is not surprising that a Hill plot of the data in Fig. 3B indicated noncooperative binding of this DNA fragment by LuxR (Hill coefficient, 0.85 [data not shown]).
FIG. 3.
DNase I protection of the lux box by LuxR. (A) A 32P-labeled probe derived from the luxI-luxR promoter region was preincubated in the presence of 10 μM 3OC6-HSL either without (lane 1) or with (lane 2) 10 nM LuxR and was then partially digested with DNase I, as described in Materials and Methods. Lane 3, Maxam-Gilbert A+G sequencing ladder of the probe. The open vertical box on the left delineates the region protected by LuxR in lane 2. The sequence of this protected region is given on the right, including the sequence comprising the lux box (indicated by heavy vertical lines). Arrows indicate two positions of enhanced DNase I digestion in lane 2. (B) Estimation of LuxR-DNA affinity by DNase I protection assays. A series of DNase I footprinting reactions using increasing concentrations of LuxR in the presence of 10 μM 3OC6-HSL was run. The fractional protection (y axis) of several bands within the LuxR-protected region was determined at each concentration of LuxR (x axis) and averaged. Error bars, standard errors of the means.
LuxR stabilizes RNA polymerase binding to the luxI promoter.
A previous study concluded that E. coli RNA polymerase-σ70 alone could not bind to the luxI promoter in vitro. However, in the presence of a C-terminal DNA-binding domain fragment of LuxR (LuxRΔN), which by itself had no specific lux box binding activity, both proteins formed a DNase I-resistant complex encompassing the lux box-luxI promoter region (28). We tested whether full-length LuxR can similarly mediate RNA polymerase binding to the luxI promoter in vitro. In agreement with the previous study, E. coli RNA polymerase alone failed to protect the luxI promoter region from DNase I attack (Fig. 4; compare lanes 1 and 4). When RNA polymerase was added in the presence of LuxR and exogenous 3OC6-HSL, the LuxR-protected region was extended through the luxI promoter region to approximately bp +20 (Fig. 4; compare lanes 2, 3, and 4).
FIG. 4.
LuxR-3OC6-HSL stimulation of RNA polymerase binding to the luxI promoter. All lanes contain 6 μM 3OC6-HSL and the same end-labeled probe used in the footprint shown in Fig. 2. Where indicated, LuxR is present at 3.5 nM and E. coli RNA polymerase (RNAP) is present at 3.3 nM. Elements of the luxI promoter are indicated to the right of the gel.
LuxR stimulates transcription from the luxI promoter in vitro.
The truncated LuxRΔN peptide had also been shown to be necessary and sufficient to activate transcription of the luxI promoter by E. coli RNA polymerase in vitro (29). However, high levels of the LuxRΔN peptide (>2,500 nM) were required to mediate this activation, presumably because it lacked the 3OC6-HSL binding region and was therefore insensitive to the facilitating effects of the signal. We showed above that 3OC6-HSL-dependent binding to the lux box by full-length purified LuxR occurs at nanomolar concentrations. Therefore, we used a runoff transcription assay to test whether low levels of full-length LuxR can stimulate transcription of the luxI promoter by E. coli RNA polymerase in vitro. In the absence of LuxR, RNA polymerase failed to produce a transcript from the luxI promoter (Fig. 5), even at the highest concentrations tested (64 nM [data not shown]). Addition of LuxR at levels as low as 5 nM activated transcription from the luxI promoter; quantitation of the 300-base luxI transcript indicated that maximum transcript levels occurred in the presence of 20 nM LuxR.
FIG. 5.
LuxR-3OC6-HSL stimulation of transcription from the luxI promoter. A 612-bp double-stranded linear DNA fragment carrying the luxI-luxR promoter region was used as a template in an in vitro transcription runoff assay. Reaction mixtures contained 1 nM DNA template, 10 nM E. coli RNA polymerase, [α32-P]ATP-labeled deoxynucleoside triphosphates, and 10 μM 3OC6-HSL, either with (lanes 1 to 6) or without (lane 7) purified LuxR, as indicated. Open arrows indicate calculated positions of the 395- and 130-base molecular size standards. Filled arrow points to the 300-base luxI runoff transcript. Because no significant transcription is seen in the absence of LuxR (lane 7), the LuxR-dependent transcripts above the 300-base luxI band are likely due to transcription that initiated at the luxI promoter but was extended beyond the template 3′ end by a loop-around mechanism described previously (24).
Although the luxR promoter is also present on the template DNA (12), a 170-base transcript predicted from the previously determined start site for this promoter is not seen (Fig. 5). Previous studies indicated that the catabolite repressor protein Crp and its effector cyclic AMP activate luxR transcription (6, 7). Another study identified the V. fischeri LitR protein as a positive transcriptional regulator of luxR (13). It is likely that expression from the luxR promoter in vitro requires CRP-cyclic AMP, LitR, or both.
DISCUSSION
The current model for quorum sensing in V. fischeri (recently reviewed in reference 15) holds that the association of 3OC6-HSL with LuxR is a key step enabling the activator to bind lux box DNA at the luxICDABEG operon and thereby facilitate the binding and activation of RNA polymerase at the luxI promoter. The role of 3OC6-HSL in potentiating these functions of LuxR has not been determined. In the case of A. tumefaciens, TraR is quickly degraded when synthesized in vivo in the absence of its cognate effector 3OC8-HSL. The presence of 3OC8-HSL during synthesis of the apoactivator promotes formation of a much more protease-resistant TraR dimer containing two molecules of 3OC8-HSL that, in an X-ray crystallographic structure of the complex, were shown to be buried in a closed pocket inside each TraR monomer (32, 37, 39). These two molecules of effector were tightly bound to TraR, forming an effectively irreversible holo-TraR complex that retained 3OC8-HSL throughout the purification procedure. Isolation of apo-TraR required extensive dialysis of the TraR-3OC8-HSL complex in the presence of 3% Tween 20 (38). The tight binding of 3OC8-HSL to TraR led to the conclusion that dissociation of 3OC8-HSL from the complex is unlikely to play a significant role in deactivation of the quorum-controlled genes in A. tumefaciens (39).
Our results indicate that cultures of the E. coli LuxR overexpression strain must also be grown in the presence of the cognate acyl-HSL (in this case 3OC6-HSL) in order to obtain extracts that have lux box-specific DNA binding activity. Addition of this effector to DNA binding reactions cannot rescue this activity from extracts of 3OC6-HSL-free cultures (data not shown). In contrast to what has been reported for TraR, the results presented here show that the specific DNA binding activity of extracts from 3OC6-HSL-supplemented cultures, as well as the activity of purified LuxR isolated from them, can easily and reversibly be rendered 3OC6-HSL dependent by simple dilution into 3OC6-HSL-free buffer. We conclude that formation of the LuxR-3OC6-HSL complex is reversible. We have determined an effective equilibrium constant for formation of this complex to be approximately 100 nM under the conditions of our assays. This value is consistent with previous in vivo studies that determined the effective response to 3OC6-HSL of lux operon reporters in E. coli (9), and it is also within the range of 3OC6-HSL concentrations thought to occur during quorum-sensing-mediated gene regulation in V. fischeri (1, 19). We have not been able to demonstrate whether the switch between the active and inactive forms of LuxR upon dilution or addition of 3OC6-HSL is due to a change in the oligomeric state of LuxR or whether it involves a conformational change in a single oligomeric species. The in vitro transcription assays highlight an additional distinction between TraR and LuxR. The former requires supercoiled promoter DNA for efficient transcription (38), while the latter can utilize a linear DNA template. These contrasting requirements may reflect different mechanisms of activation by LuxR and TraR.
The differences between LuxR and TraR are quite interesting and may point to ways in which these acyl-HSL binding proteins are used differently in different bacteria. The fact that LuxR immediately loses its ability to bind to target DNA when the acyl-HSL signal is diluted suggests that the V. fischeri quorum-sensing system can rapidly adjust to decreases in population density, whereas A. tumefaciens may respond more slowly, requiring degradation of active TraR. Members of the LuxR family of proteins show a significant but minimal sequence identity. The identity in the acyl-HSL binding regions of LuxR and TraR is less than 30% (34). Although many of the residues in this region of TraR that interact directly with the signal are conserved in LuxR, it is clear that other, nonconserved residues must somehow define important differences between these family members, not only in acyl-HSL specificity but also in how these systems might respond to changes in population densities. The purification of LuxR may now allow crystallization of this founding member of the family in order to identify the structural basis of this diversity.
Acknowledgments
This work was supported by a grant from the W. M. Keck Foundation. C.P.L. was supported by a U.S. Public Health Training Grant (T32-AI07511).
REFERENCES
- 1.Boettcher, K. J., and E. G. Ruby. 1995. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J. Bacteriol. 177:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenowitz, M., D. F. Senear, and R. E. Kingston. 1998. DNase I footprint analysis of protein-DNA binding., p. 12.4.1-12.4.16. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. John Wiley and Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 3.Choi, S. H., and E. P. Greenberg. 1991. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc. Natl. Acad. Sci. USA 88:11115-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, S. H., and E. P. Greenberg. 1992. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J. Bacteriol. 174:4064-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devine, J. H., G. S. Shadel, and T. O. Baldwin. 1989. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC 7744. Proc. Natl. Acad. Sci. USA 86:5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunlap, P. V., and E. P. Greenberg. 1985. Control of Vibrio fischeri luminescence gene expression in Escherichia coli by cyclic AMP and cyclic AMP receptor protein. J. Bacteriol. 164:45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunlap, P. V., and E. P. Greenberg. 1988. Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-LuxR protein regulatory circuit. J. Bacteriol. 170:4040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 9.Egland, K. A., and E. P. Greenberg. 2000. Conversion of the Vibrio fischeri transcriptional activator, LuxR, to a repressor. J. Bacteriol. 182:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egland, K. A., and E. P. Greenberg. 2001. Quorum sensing in Vibrio fischeri: analysis of the LuxR DNA binding region by alanine-scanning mutagenesis. J. Bacteriol. 183:382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egland, K. A., and E. P. Greenberg. 1999. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol. Microbiol. 31:1197-1204. [DOI] [PubMed] [Google Scholar]
- 12.Engebrecht, J., and M. Silverman. 1987. Nucleotide sequence of the regulatory locus controlling expression of bacterial genes for bioluminescence. Nucleic Acids Res. 15:10455-10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fidopiastis, P. M., C. M. Miyamoto, M. G. Jobling, E. A. Meighen, and E. G. Ruby. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131-143. [DOI] [PubMed] [Google Scholar]
- 14.Fried, M., and D. M. Crothers. 1981. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 9:6505-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 16.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 17.Garner, M. M., and A. Revzin. 1981. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 9:3047-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanzelka, B. L., and E. P. Greenberg. 1995. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 177:815-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, H. B., and E. P. Greenberg. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan, H. B., and E. P. Greenberg. 1987. Overproduction and purification of the luxR gene product: transcriptional activator of the Vibrio fischeri luminescence system. Proc. Natl. Acad. Sci. USA 84:6639-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 22.Minogue, T. D., M. Wehland-von Trebra, F. Bernhard, and S. B. von Bodman. 2002. The autoregulatory role of EsaR, a quorum-sensing regulator in Pantoea stewartii ssp. stewartii: evidence for a repressor function. Mol. Microbiol. 44:1625-1635. [DOI] [PubMed] [Google Scholar]
- 23.Nasser, W., M. L. Bouillant, G. Salmond, and S. Reverchon. 1998. Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol. Microbiol. 29:1391-1405. [DOI] [PubMed] [Google Scholar]
- 24.Oostra, B. A., A. C. Arnberg, G. Ab, and M. Gruber. 1981. Terminal strand-switching of E. coli RNA polymerase transcribing a truncated DNA fragment. Biochim. Biophys. Acta 655:446-448. [DOI] [PubMed] [Google Scholar]
- 25.Schaefer, A. L., B. L. Hanzelka, A. Eberhard, and E. P. Greenberg. 1996. Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J. Bacteriol. 178:2897-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz, A., and D. J. Galas. 1979. The interaction of RNA polymerase and Lac repressor with the lac control region. Nucleic Acids Res. 6:111-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shadel, G. S., and T. O. Baldwin. 1991. The Vibrio fischeri LuxR protein is capable of bidirectional stimulation of transcription and both positive and negative regulation of the luxR gene. J. Bacteriol. 173:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevens, A. M., K. M. Dolan, and E. P. Greenberg. 1994. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA 91:12619-12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens, A. M., and E. P. Greenberg. 1997. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J. Bacteriol. 179:557-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 31.Trott, A. E., and A. M. Stevens. 2001. Amino acid residues in LuxR critical for its mechanism of transcriptional activation during quorum sensing in Vibrio fischeri. J. Bacteriol. 183:387-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vannini, A., C. Volpari, C. Gargioli, E. Muraglia, R. Cortese, R. De Francesco, P. Neddermann, and S. D. Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welch, M., D. E. Todd, N. A. Whitehead, S. J. McGowan, B. W. Bycroft, and G. P. Salmond. 2000. N-Acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 19:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 35.Wu, W. F., M. L. Urbanowski, and G. V. Stauffer. 1995. Characterization of a second MetR-binding site in the metE metR regulatory region of Salmonella typhimurium. J. Bacteriol. 177:1834-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, R. G., T. Pappas, J. L. Brace, P. C. Miller, T. Oulmassov, J. M. Molyneaux, J. C. Anderson, J. K. Bashkin, S. C. Winans, and A. Joachimiak. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971-974. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 96:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu, J., and S. C. Winans. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA 98:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]