Abstract
The binding of bacteria and platelets may play a central role in the pathogenesis of infective endocarditis. Platelet binding by Streptococcus gordonii strain M99 is predominantly mediated by the 286-kDa cell wall-anchored protein GspB. This unusually large protein lacks a typical amino-terminal signal peptide and is translocated from the cytoplasm via a dedicated transport system. A 14-kb segment just downstream of gspB encodes SecA2 and SecY2, two components of the GspB-specific transport system. The downstream segment also encodes several putative glycosyl transferases that may be responsible for the posttranslational modification of GspB. In this study, we compared the abilities of M99 and two GspB− mutant strains to bind various lectins. GspB was found to have affinity for lectins that bind N-acetylglucosamine. We also examined variant forms of GspB that lack a carboxy-terminal cell wall-anchoring domain and thus are free of covalent linkage to cell wall peptidoglycan. Like native GspB, these truncated proteins appear to be heavily glycosylated, as evidenced by migration during sodium dodecyl sulfate-polyacrylamide gel electrophoresis with an apparent molecular mass >100 kDa in excess of the predicted mass, negligible staining with conventional protein stains, and reactivity with hydrazide following periodate oxidation. Furthermore, analysis of the carbohydrate associated with the GspB variants by high-pH anion-exchange chromatography revealed the presence of ∼70 to 100 monosaccharide residues per GspB polypeptide (primarily N-acetylglucosamine and glucose). Analysis of GspB in protoplasts of secA2 or secY2 mutant strains, which do not export GspB, indicates that GspB is glycosylated in the cytoplasm of these strains. The combined data suggest that the native GspB is a glycoprotein and that it may be glycosylated prior to export.
The binding of bacteria and platelets is thought to play a central role in the pathogenesis of infective endocarditis. Platelets on the surfaces of damaged cardiac valves may provide an attachment site for bacteria circulating in the bloodstream, thereby initiating infection (6, 8, 9, 13). The subsequent deposition of platelets onto the infected valve surface may also be facilitated by bacterium-platelet binding (5, 21), leading to the formation of the hallmark lesion of this disease, the macroscopic endovascular vegetation.
Numerous mechanisms for the attachment of bacteria to platelets have been proposed. Our own studies indicate that platelet binding by Streptococcus gordonii strain M99 is predominantly mediated by the cell surface protein GspB (2). This 280-kDa protein has a cell wall-anchoring domain characteristic of many gram-positive bacterial surface proteins (an LPXTG motif, a hydrophobic domain, and a charged tail [10, 18, 19]). However, several features of GspB are unusual. First, it does not have a typical amino-terminal signal for export. Instead, GspB is predicted to have a 90-amino-acid signal peptide, which is approximately three times longer than signals for export mediated by the general protein secretion (Sec) system in gram-positive bacteria (25, 27). Second, GspB migrates during sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with an apparent molecular mass that is much greater than predicted. Third, a majority of the protein is comprised of ∼190 semiconserved repeats of the peptide motif SASESASTSASV. This feature of an extended region of serine-rich repeats has recently been noted for several other streptococcal surface proteins, including the S. gordonii DL1 sialic acid-binding adhesin Hsa (23) and the Streptococcus parasanguis fimbria-associated protein Fap1 (28).
GspB is also unusual because a 13.6-kb region downstream of the 9.2-kb gspB structural gene (Fig. 1) is required for its expression. This region encodes SecA2 and SecY2, which are homologues of two highly conserved components of the general Sec system. Mutation of either secA2 or secY2 results in the accumulation of GspB in the cytoplasm, indicating that they are required for the export of GspB. However, mutation of these genes has no apparent effect on the transport of other proteins, which suggests that SecA2 and SecY2 selectively mediate the export of GspB (2). Although the export of GspB by a dedicated export pathway is consistent with the fact that GspB does not have a typical N-terminal signal peptide, the specific features of the GspB sequence or structure that are recognized by SecA2 and SecY2 for export are unknown.
FIG. 1.
gspB-sec locus of M99. GspB is a cell surface-anchored platelet binding adhesin. Gly, Nss, and Gtf are likely to function in carbohydrate metabolism: Gly is predicted to be a cytoplasmic glycosyl transferase (family 8); Nss is similar to nucleotide sugar synthetases; and Gtf is 46% similar to the Bacillus subtilis polyglycerol phosphate α-glucosyl transferase (S06048). SecA2 and SecY2 are similar to the SecA ATPases and the SecY transmembrane translocases of various organisms (components of the general secretory pathway), respectively, and are required specifically for the export of GspB. The proteins encoded by orf1 to orf4 show no similarity to any proteins of known function.
The segment downstream of gspB also encodes several proteins that are likely to function in carbohydrate metabolism, including two that have conserved domains characteristic of glycosyl transferases. Mutation of at least one of the putative glycosyl transferase genes (gtf) abolished GspB expression (2). We proposed that Gtf, along with the other transferases encoded within the gspB-sec operon, might be responsible for the glycosylation of GspB, which would account for the aberrant electrophoretic mobility of the protein. We now report that GspB is indeed a glycoprotein and that its glycosylation occurs independently of its export.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
The bacterial strains and plasmids used in this study are listed in Table 1. S. gordonii strains were grown in Todd-Hewitt Broth (THB; Difco Laboratories) at 37°C in a 5% CO2 environment. Biotinylated lectins were purchased from Vector Laboratories. Succinylated wheat germ agglutinin (WGA) agarose and N-acetylglucosamine were obtained from EY Laboratories. Streptavidin-conjugated horseradish peroxidase, orthophenylenediamine (OPD), sodium perborate-containing phosphate-citrate buffer, Dulbecco's phosphate-buffered saline (DPBS), isopropyl β-d-thiogalactopyranoside, tunicamycin, monensin, bacitracin, and cross-linked phosphorylase b were from Sigma.
TABLE 1.
Strains and plasmids used in the present study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strain | ||
| M99 | S. gordonii parental strain | 22 |
| PS321 | M99 Δ[nss-gtf] | 2 |
| PS426 | M99 secY2::pVA891 | 2 |
| PS436 | M99 gspB::pM995′Bint | 2 |
| PS463 | M99 gspB::pM99midBint; secretes GspB105 | 2 |
| PS465 | M99 gspB::pM993′Bint; secretes GspB274 | 2 |
| PS466 | PS463 secY2::pVA891 | 2 |
| PS469 | M99 secA2::pVA891 | 2 |
| PS497 | M99 gspB::pM99B194His6int; secretes GspB194 | This study |
| PS498 | PS426 gspB::pM99B194His6int | This study |
| PS526 | PS469 gspB::pM99B194His6int | This study |
| Plasmid | ||
| pBluescript | Ampr | Stratagene |
| pVA891 | erm (gram positive) cat tet ori (E. coli) | 12 |
| pEVP3 | cat (gram positive and E. coli) ori (E. coli) | 4 |
| pB194His6int | SpeI-NheI fragment of gspB + His6 in pEVP3 | This study |
| pM995′Bint | HindIII-HincII fragment of gspB in pVA891 | 2 |
| pM99midBint | SpeI fragment of gspB in pEVP3 | 2 |
| pM993′Bint | NheI-NsiI fragment of gspB in pEVP3 | 2 |
Whole-cell lectin binding assay.
S. gordonii strains were grown for 18 h in 3 ml of THB, washed twice, and then suspended in 3 ml of DPBS. Fifty microliters of the washed cell suspensions was added to each well of 96-well plates, and the bacteria were allowed to attach to the plastic surface for 2 to 3 h at room temperature (RT). After unattached bacteria were removed by aspiration, the cell monolayers were washed with 100 μl of DPBS. On examination by light microscopy, the bacterial-cell monolayers were confluent and remained attached to the wells throughout the assay. Biotinylated lectins were diluted to 1 (WGA), 2 (concanavalin A [ConA] and succinylated WGA), or 10 (all other lectins) μg ml−1 in 100 mM Tris-150 mM NaCl, pH 7.5 (TBS) containing 1× blocking reagent (Roche), 1 mM CaCl2, and 1 mM MgCl2. Fifty microliters of the lectin solutions was added to the immobilized bacteria, and the plates were incubated for 1 h at RT with gentle rocking. For studies that examined the ability of N-acetylglucosamine or glucose to inhibit the binding of selected lectins to S. gordonii, a specified amount of either monosaccharide was included in the lectin solution. The supernatant liquid was then removed by aspiration, and the wells were washed three times with 100 μl of TBS. Fifty microliters of streptavidin-conjugated horseradish peroxidase (0.1 μg ml−1 in TBS) was added to each well, and the plates were incubated for 45 min at RT. The wells were then washed twice with 100 μl of TBS. Two hundred microliters of a solution of 0.4-mg of OPD ml−1 in citrate-phosphate buffer was added to each well, and the contents were mixed by gently vortexing the plates. The absorbance at 450 nm (A450) was measured ∼20 min after the addition of the OPD substrate. The data are reported as the mean ± standard deviation of the A450 for the specified lectin minus the averaged value of wells that were treated identically except for the omission of the biotinylated lectin. Differences in lectin binding were compared by the unpaired t test, using the Welch modification.
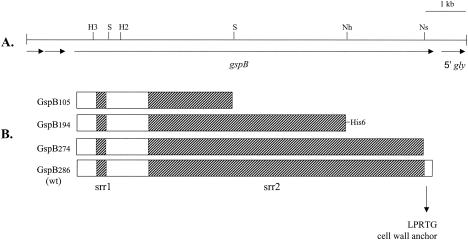
Construction of PS497, PS498, and PS526.
The S. gordonii strain PS497, which secretes a truncated, C-terminally His6-tagged version of GspB, was constructed as follows. A 3-kb SpeI-NheI DNA fragment spanning the central region of serine-rich repeat region 2 (srr2) (Fig. 2), and including codons 1059 to 2062 of 3072, was cloned in pBluescript pKS(−). The plasmid was digested with NotI, and the Klenow fragment of DNA polymerase was used to fill in the 5′ overhanging ends. The plasmid DNA was then digested with SacI and ligated to a double-stranded DNA linker that had been made by annealing the 5′-end-phosphorylated primers 5′-CACCACCACCACCACCACTAAGGATCCGAGCT-3′ and 5′-CGGATCCTTAGTGGTGGTGGTGGTGGTG-3′. The linker has a 4-nucleotide SacI-compatible 3′ overhang adjacent to a BamHI restriction site (underlined). In-frame fusion of six histidine and one stop codons to codon 2062 of gspB (along with four codons derived from the pBluescript multicloning site) was confirmed by DNA sequence analysis. The gspB fragment was then excised with BamHI and ligated to pEVP3 that had been digested with the same restriction enzyme. The resulting plasmid, pB194His6int, was propagated in Escherichia coli strain DH5α and then introduced into M99 by natural transformation as described previously (2). To construct strains PS498 and PS526 (which express but do not secrete the C-terminally His6-tagged version of GspB), pB194His6int was used to transform the secY2 mutant strain PS426 or the secA2 mutant strain PS469, respectively. Integration of pB194His6int at the expected chromosomal site in each mutant strain was confirmed by Southern blot analysis.
FIG. 2.
Features of the serine-rich repeat protein GspB and the corresponding genetic locus. (A) Restriction map of the M99 chromosomal region spanning gspB. The locations of selected restriction sites in gspB are indicated. H3, HindIII; H2, HincII; Nh, NheI; Ns, NsiI; S, SpeI. (B) Diagram of GspB and truncated derivatives. The number associated with the GspB designation indicates the predicted molecular mass in kilodaltons. GspB194 was engineered with a C-terminal His6 tag. The hatched sections correspond to srr1 and srr2. wt, wild type.
Purification of truncated GspB derivatives.
The C-terminally truncated GspB derivatives GspB105, GspB194, and GspB274 (Fig. 2) were purified from the culture supernatants of S. gordonii strains PS463, PS497, and PS465, respectively. As a negative control, a sample was prepared from the GspB− mutant strain PS436 in parallel. The strains were grown for 18 h in 40 ml of THB, and the cultures were used to inoculate 400 ml of fresh medium. After incubation for 8 h at 37°C, the cells were removed by centrifugation at 14,000 × g for 20 min. Proteins were precipitated from the supernatant culture medium by the addition of ammonium sulfate to a final concentration of 50%. The precipitated proteins were recovered by centrifugation at 18,000 × g for 25 min and then solubilized in 7 ml of TBS. Any remaining cellular debris was removed by centrifugation at 3,200 × g for 15 min, followed by passage through 0.45-μm-pore-size filters (Millipore). Ammonium sulfate was removed by passing the filtered solutions over EconoPac 10DG desalting columns (Bio-Rad), and the desalted samples were added to 250 μl of succinylated-WGA agarose resin and tumbled for 15 h at 4°C. The solution of unbound proteins was removed, and the resin was washed three times with 3 ml of TBS. The bound GspB was eluted with 3 ml of 500 mM N-acetylglucosamine in 10 mM Tris-HCl, pH 8. The yield of GspB194 was 4 to 500 pmol (80 to 100 μg of the core polypeptide), as determined by a quantitative dot blot immunoassay using an anti-His6 monoclonal antibody.
Monosaccharide composition analysis.
Prior to monosaccharide analysis, free N-acetylglucosamine was separated from the eluted GspB derivatives (or the negative control sample prepared in parallel) by two rounds of desalting into distilled H2O. The desalted samples were then concentrated to 400 pmol ml−1 in YM-100 Centricon units (Millipore). The identification and quantification of monosaccharides bound to the purified GspB variants were accomplished by high-pH anion-exchange chromatography, combined with pulsed amperometric detection, at the Glycotechnology Resource Training Center at the University of California, San Diego. In brief, samples were hydrolyzed for 4 h at 100°C in 2 N trifluoroacetic acid. The hydrolysates were dried under vacuum, washed with methanol, dried again, and then dissolved in distilled H2O. The levels of fucose, glucose, N-acetylglucosamine, N-acetylgalactosamine, and galactose were determined by isocratic chromatography on a CarboPac PA10 column (Dionex) in 18 mM NaOH at a flow rate of 1 ml min−1. Rhamnose, mannose, and xylose were measured on a CarboPac MA1 column (Dionex) developed in 60 mM NaOH for 5 min, followed by a gradient elution of 60 to 660 mM NaOH over 30 min at a flow rate of 0.4 ml min−1.
Preparation of antiserum against recombinant GspB.
A 2.3-kb SpeI fragment of gspB that included the nonrepeat region between srr1 and srr2, along with the first 1.3 kb of srr2 (Fig. 2), was cloned in pBluescript pKS(−). The gspB fragment was then excised from the plasmid using BamHI and NotI and ligated to pET28b (Novagen) that had been digested with the same restriction enzymes. This resulted in the in-frame fusion of six histidine codons at both the 5′ and 3′ ends (i.e., N- and C-terminal His6 tags), which was confirmed by DNA sequence analysis of the plasmid DNA. Recombinant-protein expression was induced in the E. coli host strain BL21(DE3) with 0.4 mM isopropyl β-d-thiogalactopyranoside, and the recombinant GspB (rGspB) was purified using nickel affinity chromatography. The purified rGspB was used to immunize New Zealand White rabbits (Covance).
Analysis of secreted, cell wall, and protoplast proteins.
For analysis of secreted products, proteins were precipitated from culture supernatants by using trichloroacetic acid as described previously (1). Cell wall proteins were extracted from S. gordonii strains by mutanolysin treatment (14). The mutanolysin extraction buffer included raffinose (26% [wt/vol]) to maintain the integrity of the protoplasts. For analysis of cytoplasmic components, protoplasts (generated by digestion of the cell wall with mutanolysin) were lysed by suspension in SDS-PAGE sample buffer, followed by boiling them for 10 min. Proteins were separated by SDS-PAGE through 3 to 8% Tris-acetate gels (Invitrogen) under reducing conditions. The proteins were either stained with the SYPRO Ruby protein gel stain (Molecular Probes) or transferred to BioTrace NT nitrocellulose membranes using the XCell SureLock transfer apparatus (Invitrogen). For Western blot analysis, membranes were incubated for 1 h in a suspension of 1× blocking reagent (Roche) in DPBS. Anti-rGspB polyclonal rabbit serum was used at a dilution of 1:1,000, and anti-His6 mouse monoclonal antibody (Novagen) was used at a 1:2,000 dilution. Peroxidase-conjugated anti-rabbit or anti-mouse serum (Sigma) was used at a 1:25,000 dilution. The blots were developed with the SuperSignal West Pico chemiluminescent substrate (Pierce). Detection of carbohydrate on the blotted proteins was performed with the DIG-glycan detection kit as recommended by the manufacturer (Roche), except that a peroxidase-conjugated anti-digoxigenin antibody was used in place of the alkaline phosphatase-conjugated anti-digoxigenin.
Effect of inhibitors of glycosylation on GspB expression.
PS497 was grown for 18 h in THB and then diluted 1:10 in THB or THB containing glycosylation inhibitors at the concentrations used by Erickson and Herzberg (7) for treatment of protoplasted cells: 20 μg of tunicamycin (an inhibitor of N-linked glycosylation)/ml, 0.07 μg of monensin (an inhibitor of O-linked glycosylation)/ml, or 10 μg of bacitracin (a general inhibitor of glycosylation)/ml. The cultures were incubated for 8 h at 37°C and then centrifuged for 15 min at 3,000 × g. The supernatants were transferred to clean tubes, and proteins were precipitated using trichloroacetic acid as described above. The proteins were dissolved in SDS-PAGE sample buffer and then boiled for 10 min before being loaded into wells of 3 to 8% polyacrylamide gels. The proteins were transferred to nitrocellulose, and GspB194 was detected by Western blotting, using an anti-His6 monoclonal antibody.
Treatment of GspB with deglycosylating enzymes.
Purified GspB105 and GspB194 were treated with a mixture of glycosidases (ProZyme) as recommended by the manufacturer. In brief, 4 pmol (400 or 800 ng) of the GspB derivatives was incubated for 18 h at 37°C under denaturing conditions with a mixture of enzymes consisting of 5 U of peptide:N-glycosidase F (PNGase F) 0.005 U of sialidase A, 0.00125 U of endo-O-glycosidase, 0.003 U of β(1,4)galactosidase, and 0.04 U of glucosaminidase. As a control, 5 μg of the control glycoprotein fetuin was treated with the same mixture. The enzymatically treated GspB and fetuin samples were combined with SDS-PAGE sample buffer, heated for 10 min at 70°C and then loaded into wells of NuPAGE bis-Tris 4 to 12% gradient gels (Invitrogen). Following electrophoresis, the proteins were stained with SYPRO Ruby or subjected to Western blotting and examined for changes in electrophoretic mobility compared with the same glycoproteins that had not been treated with glycosidases.
RESULTS
GspB affects the ability of M99 to bind several lectins.
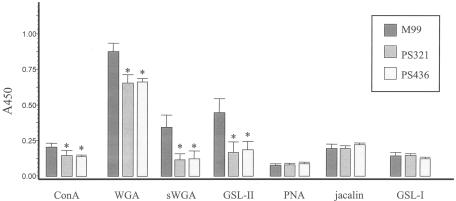
The ability of lectins to bind to defined carbohydrate structures has been useful for identifying carbohydrate moieties on eukaryotic glycoproteins. To characterize the carbohydrate associated with GspB, the affinities of M99 for a number of lectins were assessed. As negative controls, two strains that do not express GspB were also examined (PS321 and PS436). Of the 18 lectins analyzed (Table 2), only 7 showed detectable levels of binding to M99 (Fig. 3). Three of the seven lectins (WGA, succinylated WGA, and GSL-II) showed significantly greater binding (P < 0.0001) to M99 than the GspB− strains. The three lectins are known to bind N-acetylglucosamine (GlcNAc). Of note, GSL-II is unique among lectins in the ability to react only with terminal, nonreducing GlcNAc residues. Lower but still significant GspB-dependent binding of ConA was also detected (P < 0.05), which indicated that the GspB-linked carbohydrate may also contain glucose or mannose.
TABLE 2.
Lectins used in the current study
| Lectin | Abbreviation | Carbohydrate specificitya |
|---|---|---|
| Concanavalin Ab | ConA | α-Man > α-Glc |
| Wheat germ agglutininb | WGA | β-GlcNAc, SA |
| Succinylated WGA | sWGA | β-GlcNAc |
| Griffonia simplicifolia lectin I | GSL-I | α-Gal, α-GalNAc |
| G. simplicifolia lectin II | GSL-II | Terminal GlcNAc |
| Peanut agglutinin | PNA | Terminal β-galactosidase |
| Artocarpus integrifolia lectin | Jacalin | α-Gal, Galβ(1,3)GalNAc |
| Lens culinaris agglutinin | LCA | α-Man, α-Glc |
| Pisum sativum agglutinin | PSA | α-Man, α-Glc |
| Sophora japonica agglutinin | SJA | Terminal GalNAc > Gal |
| Soybean agglutinin | SBA | Terminal GalNAc > Gal |
| Vicia villosa agglutinin | VVA | Terminal GalNAc |
| Ulex europaeus agglutinin I | UEA I | α-Fuc |
| Dolicos biflorus agglutinin | DBA | Terminal α-GalNAc |
| Solanum tuberosum lectin | STL | β-GlcNAc oligomers |
| Lycopersicon esculentum lectin | LEL | GlcNAc |
| Datura stramonium lectin | DSL | β-GlcNAc oligomers |
| Erythrin cristagalli lectin | ECL | Galβ(1,4)GlcNAc, Gal |
Man, mannose; Glc, glucose; Gal, galactose; GalNAc, N-acetylgalactosamine; Fuc, fucose; SA, sialic acid.
Lectin is known to bind some bacterial-cell walls.
FIG. 3.
Lectin binding to S. gordonii strain M99 and the GspB− strains PS321 and PS436 (left to right). Biotinylated forms of the indicated lectins were assessed for binding to bacteria that were immobilized in 96-well plates. The asterisks indicate values that are significantly different from that of the parental strain, M99 (P < 0.05). The error bars indicate standard deviations.
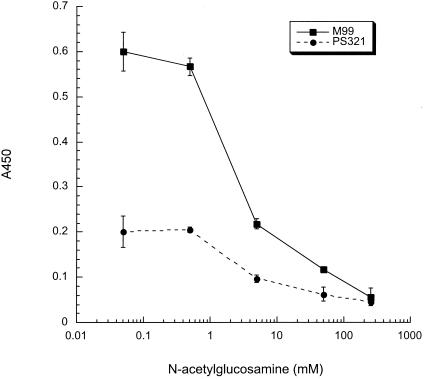
To confirm the specificity of lectin binding, we assessed whether these interactions could be inhibited by specific monosaccharides. The binding of succinylated WGA to M99 could be partially blocked by the addition of 5 mM GlcNAc, and it was completely inhibited by 250 mM GlcNAc (Fig. 4). In contrast, the binding of succinylated WGA to M99 was not affected by the presence of up to 250 mM glucose (data not shown). The binding of WGA and GSL-II to M99 was also specifically inhibited by GlcNAc (data not shown). The combined results further indicate that the binding of WGA, succinylated WGA, and GSL-II to M99 is mediated by GlcNAc residues on GspB and not by protein-protein or nonspecific (e.g., charge or hydrophobic) interactions.
FIG. 4.
Inhibition of succinylated-WGA binding to M99 by GlcNAc. The effect of GlcNAc on the binding of succinylated WGA to M99 and the GspB− strain PS321 was assessed by the whole-cell lectin binding assay.
Detection of carbohydrate on proteins extracted from the S. gordonii cell wall.
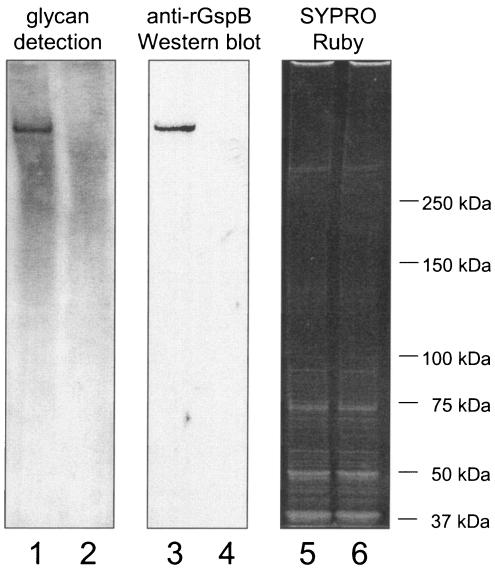
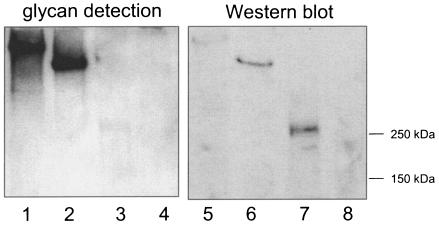
We next examined whether carbohydrate could be detected on GspB after the protein was extracted from the cell wall of M99. GspB is predicted to be covalently linked to the peptide cross-bridge of the cell wall peptidoglycan via a carboxy-terminal LPXTG motif and can be released from the bacterial-cell surface by treatment with the muralytic enzyme mutanolysin (a common method for removing LPXTG-linked proteins from streptococci) (14). Upon oxidation of the electrophoretically separated cell wall proteins with periodate, followed by reaction of any resultant aldehyde groups with digoxigenin-labeled hydrazide, a distinct glycoprotein band was detected among the M99 cell wall proteins that was absent from the cell wall proteins extracted from the gspB mutant strain PS436 (Fig. 5, lanes 1 and 2, respectively). The glycosylated protein migrated the same distance as GspB, as determined by Western blotting of samples run in parallel (lanes 3 and 4). These results indicate that GspB is indeed glycosylated. However, there was also detectable reactivity of the digoxigenin-labeled hydrazide with high-molecular-mass cell wall proteins from both strains. This suggested that some of the carbohydrate bound to GspB extracted from the cell wall of S. gordonii was likely derived from cell wall polysaccharides.
FIG. 5.
Detection of carbohydrate linked to GspB and other S. gordonii cell wall proteins. The proteins extracted from M99 (lanes 1, 3, and 5) or the gspB mutant strain PS436 (lanes 2, 4, and 6) were separated by electrophoresis through 3 to 8% polyacrylamide gradient gels. The proteins were then transferred to nitrocellulose and either examined for the presence of carbohydrate by using the DIG-glycan detection kit (lanes 1 and 2) or probed with a polyclonal anti-rGspB serum (lanes 3 and 4). To confirm that comparable amounts of total protein were loaded in each lane, gels run in parallel were stained with SYPRO Ruby (lanes 5 and 6).
Truncated derivatives of GspB are biochemically similar to native GspB.
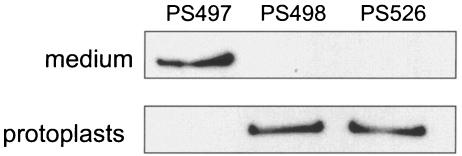
In order to characterize any GspB-linked carbohydrate that does not originate from peptidoglycan, we examined GspB derivatives that lack the C-terminal anchoring domain and are thus freely secreted by S. gordonii into the culture medium. Two of these derivatives, GspB105 and GspB274, have been described in a prior report (2). The third GspB derivative used in this study (GspB194) has a predicted mass of 194 kDa and was engineered with a C-terminal His6 tag to aid in tracking it during purification. Like GspB105 and GspB274, GspB194 is freely secreted by S. gordonii into the culture medium (Fig. 6, lane 1). This export is dependent on the accessory Sec components SecY2 and SecA2, as mutagenesis of the respective sec genes results in retention of GspB194 in the protoplasts (lanes 2 and 3).
FIG. 6.
Dependence of GspB194 export on components of the accessory Sec system. GspB194 was detected using an anti-His6 monoclonal antibody. The upper lanes contain proteins precipitated from 160 μl of the culture medium; the lower lanes were loaded with protoplasts of bacteria in 120 μl of culture. PS498 and PS526 are derivatives of strain PS497 that have mutations in secY2 and secA2, respectively.
We then examined whether the truncated forms of GspB could bind the same lectins as did the native, cell wall-associated GspB. All three variants could be purified by affinity chromatography using either WGA, succinylated WGA, or GSL-II and could be eluted from the affinity matrices with GlcNAc. However, the efficiency of purification (overall yield and purity) was highest when succinylated WGA was used (data not shown). The succinylated-WGA-purified GspB variants were therefore selected for further characterization.
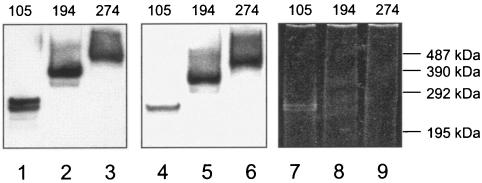
The three purified GspB derivatives were initially examined by SDS-PAGE. Each of the variants was readily detected by Western blot analysis, using an anti-rGspB serum (Fig. 7, lanes 1 to 3). Notably, each of the variants was found to migrate with a molecular mass ∼150 kDa greater than predicted. The purified proteins were subsequently examined for the presence of carbohydrate, using digoxigenin-labeled hydrazide as described above. Each of the GspB variants showed a strong positive reaction (lanes 4 to 6), whereas no reactivity was seen with the negative control protein (creatinase) (data not shown). The proteins were also examined for the ability to be stained with conventional protein stains. Like native GspB, GspB274 and GspB194 were refractory to staining with Coomassie or silver, which is a noted characteristic of highly anionic glycoproteins (1). Similarly, neither protein bound the fluorescent stain SYPRO Ruby (Fig. 7, lanes 8 and 9). The more truncated GspB derivative GspB105, although still resistant to staining with Coomassie, was detectable with SYPRO Ruby (Fig. 7, lane 7). The combined results suggest that the secreted GspB variants are glycosylated in a manner similar to that of the full-length native, cell wall-associated GspB.
FIG. 7.
SDS-PAGE analyses of truncated derivatives of GspB. The proteins were purified from S. gordonii culture supernatants. The predicted masses (in kilodaltons) of the GspB variants are indicated along the top of the gel. The proteins in lanes 1 to 3 (1 pmol of GspB per lane) underwent Western blot analysis using a polyclonal anti-rGspB serum. The proteins in lanes 4 to 6 (1 pmol per lane) were analyzed for the presence of carbohydrate, using the DIG-glycan detection kit. The proteins in lanes 7 to 9 (4 pmol per lane) were stained with SYPRO Ruby. Note that the proteins in lanes 8 and 9 are refractory to staining with SYPRO Ruby and are thus not readily apparent here. The molecular mass standards correspond to cross-linked multimers of phosphorylase b. Lanes 1, 4, and 7, GspB105; lanes 2, 5, and 8, GspB194; lanes 3, 6, and 9, GspB274.
Monosaccharide analysis of purified GspB variants.
Because the truncated forms of GspB expressed by S. gordonii appeared to be glycosylated similarly to the native protein, the monosaccharide compositions of purified GspB105 and GspB194 were determined. GspB274 was not analyzed, since the recovery and solubility of the protein upon desalting into water for monosaccharide analysis were very low. Eight monosaccharides known to be present in eukaryotic and some prokaryotic glycoproteins (Table 3) were used as standards for the identification and quantification of monosaccharides released from the GspB variants following acid hydrolysis. To control for carbohydrates not originating from GspB, we also analyzed material that was prepared in parallel from the gspB mutant strain PS436.
TABLE 3.
Monosaccharide analysis of carbohydrates associated with GspB105 and GspB194
| Carbohydrate | Contenta
|
Content/nmol of GspBb
|
||
|---|---|---|---|---|
| GspB105 | GspB194 | GspB105 | GspB194 | |
| GlcNAc | 1.42 | 2.99 | 14 | 30 |
| Glc | 4.05 | 7.26 | 41 | 73 |
| GalNAc | 0.49 | 0.17 | 5 | 2 |
| Gal | 0.77 | 0 | 8 | 0 |
| Rhamnose | 0 | 0 | 0 | 0 |
| Fucose | 0.09 | 0 | Trace (<1) | 0 |
| Mannose | 0 | 0 | 0 | 0 |
| Xylose | 0 | 0 | 0 | 0 |
Monosaccharides bound to ∼100 pmol of the purified GspB glycoproteins, with the amount detected in the negative control sample prepared in parallel subtracted.
Calculated by dividing the measured amount (in nanomoles) by the amount of each GspB variant analyzed (0.1 nmol).
Analysis of the monosaccharides released from GspB105 indicated the presence of GlcNAc and glucose at levels well above those measured in the negative control sample. These monosaccharides were present in a ratio of 14:41 residues per GspB105 polypeptide (Table 3). Minor amounts of N-acetylgalactosamine and galactose were also detected. Rhamnose, mannose, and xylose were not detected above background levels, although a trace amount of fucose was present. The total carbohydrate (68 nmol per nmol of GspB105) corresponds to ∼10% (wt/wt) carbohydrate.
The carbohydrate content of GspB194 was also examined. The same two major monosaccharides, GlcNAc and glucose, were present in a ratio of 30:73 residues per GspB194 polypeptide (Table 3). A minor amount of N-acetylgalactosamine was detected, whereas galactose was not. The total carbohydrate associated with GspB194 (105 nmol per nmol of GspB194) corresponds to ∼9% (wt/wt) of the total glycoprotein mass. The composite results suggest that the native GspB is likely to have a carbohydrate composition similar to that of the C-terminally truncated variants.
Linkage analysis.
Covalent attachment of carbohydrate to polypeptides can occur either via N linkage to asparagine (Asn) residues or via O linkage to serine (Ser) or threonine (Thr) residues. To characterize the type of linkages on GspB, the lectin-purified GspB variants were treated with a cocktail of deglycosylating enzymes known to remove carbohydrate from eukaryotic glycoproteins. At least one of these enzymes (PNGase F) has also been shown to deglycosylate prokaryotic glycoproteins (7, 15). Treatment with the glycosidases produced no detectable change in the electrophoretic mobility of the GspB variants (data not shown). In addition, growth of S. gordonii in the presence of tunicamycin (an inhibitor of N-linked glycosylation), monensin (an inhibitor of O-linked glycosylation), or bacitracin (a general inhibitor of glycosylation) had no effect on GspB expression or electrophoretic mobility (not shown). The combined results suggest that the carbohydrate linkages of GspB are different from the typical linkages found on eukaryotic glycoproteins.
GspB is glycosylated in the protoplasts of secA2 and secY2 mutant strains.
Since the glycosyl transferases encoded in the gspB-sec operon do not have amino-terminal export signals, these proteins are predicted to be localized to the cytoplasm. We therefore sought to determine whether GspB could be glycosylated prior to, or independently of, export. For this analysis, we used secY2 mutant strains, which do not export GspB (2) (Fig. 6). The protoplast proteins were subjected to SDS-PAGE and examined for the presence of carbohydrate, using the digoxigenin-hydrazide labeling described above. A single carbohydrate-containing protein was detected in the strains expressing either full-length GspB (GspB286) or GspB194 (Fig. 8, lanes 1 and 2). A glycoprotein was less readily detected in the GspB105-expressing secY2 mutant strain PS466 (lane 3), whereas no glycoprotein was seen in protoplasts of the gspB null mutant strain PS436 (lane 4). The glycoproteins migrated the same distance as did GspB or the GspB variants, as determined by Western blotting of samples run in parallel and probed with anti-rGspB polyclonal antiserum (lanes 5 to 8). This indicated that the glycoprotein detected in these protoplasts was indeed GspB. Similar results were obtained if the protoplasts were sonicated and debris was removed prior to analysis (data not shown), indicating that the glycoproteins were in the soluble versus membrane-associated fraction of the protoplasts. Furthermore, similar results were also obtained when analyzing GspB expressed by secA2 mutant strains (data not shown). The combined results indicate that GspB can be glycosylated in the cytoplasm of S. gordonii independently of export.
FIG. 8.
Detection of glycosylated GspB in S. gordonii protoplasts. Each lane contained material from protoplasts of cells in a 120-μl culture volume. Lanes 1 to 4, proteins were examined for carbohydrate. Lanes 5 to 8, proteins were subjected to Western blot analysis using the anti-rGspB serum. Lanes 1 and 5, PS426 (GspB286 SecY2−); lanes 2 and 6, PS498 (GspB194 SecY2−); lanes 3 and 7, PS466 (GspB105 SecY2−); lanes 4 and 8, PS436 (GspB−).
DISCUSSION
Once thought to be a rarity, the glycosylation of proteins by prokaryotes has been reported with increasing frequency in the last 2 decades (reviewed in references 3, 16, 17, and 26). The list of glycoproteins has grown well beyond the original family of surface (S) layer proteins and includes a number of pilins, flagellins, and fimbrial proteins, as well as enzymes, toxins, and adhesins. Although the function of glycosylation has not been clarified in all cases, proposed and defined roles include altered antigenicity, control of enzymatic activity, resistance to proteolytic processing, altered protein conformation, and the direct mediation of adherence.
By far the most extensively characterized bacterial glycoproteins are the S-layer proteins (17). These abundant, lattice-forming, cell surface proteins may have a total carbohydrate content of 2 to 20% (wt/wt). There is typically one type of glycan chain per polypeptide, but these can vary in length due to different degrees of polymerization (from 20 to 50 units) of repeating units of two to six monosaccharides. The glycan chains may have a core of one to three sugars and are attached at two to six sites per polypeptide, predominantly via an O linkage. Composite analyses of other bacterial glycoproteins indicate that they are often very different from, and more varied than, eukaryotic glycoproteins (3, 16, 26). This includes differences not only in the types of constituent monosaccharides but also in the glycan chain length, the interlinking core region of the glycan chain, the configuration of the glycosidic linkage between the carbohydrate and the protein, and the acceptor amino acid sequence.
The results presented here indicate that the S. gordonii surface protein GspB is a new example of a bacterial glycoprotein adhesin. Truncated derivatives of GspB, which lack the carboxy-terminal peptidoglycan linkage domain, are heavily glycosylated and have a total carbohydrate content of 9 to 10% (wt/wt). Analyses of the GspB-linked carbohydrate by high-pH anion-exchange chromatography, along with lectin binding profiles, indicate a predominance of glucose and GlcNAc residues, as well as minor amounts of galactose and N-acetylgalactosamine. At least some of the GlcNAc residues appear to be located at the nonreducing termini of the carbohydrate chains, as determined by affinity for the lectin GSL-II, but the order of the remaining sugars remains to be determined.
A recent report of the monosaccharides associated with the GspB homologue Fap1, a fimbrial glycoprotein of S. parasanguis, indicates both similarities, and distinct differences in monosaccharide content compared with GspB. Fap1 also has a predominance of GlcNAc and glucose, with minor amounts of galactose, N-acetylgalactosamine, and rhamnose (20). However, Fap1 has a higher percentage of GlcNAc than glucose (GlcNAc, glucose, galactose, N-acetylgalactosamine, and rhamnose were present in a ratio of 39:29:5:1:1). A comparison of the glycosyl transferases that modify GspB with those that act on Fap1 may provide insights into the specificity of and control over carbohydrate chain length and composition.
The monosaccharides associated with GspB appear to be quite different from those linked to another streptococcal protein, the platelet aggregation-associated protein (PAAP) of Streptococcus sanguis. In one of the earliest demonstrations of covalent linkage of a carbohydrate to a bacterial protein, the polysaccharide bound to PAAP was found to be rich in rhamnose and N linked to Asn residues (7). This suggests that streptococci may have a variety of glycoproteins that interact with platelets, as well as other human cells or tissues.
A number of glycosidases have been used in the characterization of glycoproteins. The glycosidase PNGase F can remove most N-linked oligosaccharides from eukaryotic glycoproteins, and it released the N-linked carbohydrates from PAAP (7) and from two Borrelia burgdorferi glycoproteins (15). However, the enzyme had no affect on the electrophoretic mobility of GspB194 or GspB105. In addition, endo-O-glycosidase had no effect on the GspB variants. This enzyme is more specific than PNGase F in that it removes only the Galβ(1,3)GalNAc core present on most O-linked eukaryotic glycoproteins, and any modification of the core structure will block the action of the enzyme. The negative result obtained by treatment with endo-O-glycosidase thus does not rule out the possibility that the GspB-bound carbohydrate is O linked. As seen with other bacterial glycoproteins, however, the O linkages on GspB are likely to be different from those of eukaryotic glycoproteins. Determination of the precise monosaccharide and anomeric configuration of the covalent linkage to the Ser or Thr residues of GspB will require further analysis.
In the GspB polypeptide, there are 1,599 Ser and Thr residues that are potential candidates for sites of covalent attachment of glycan chains. However, not all of these residues may undergo modification. In a study of the Campylobacter jejuni flagellin glycoprotein, Thibault et al. found that just 19 of 107 Ser and Thr residues were modified by pseudaminic acid polymers (24). The carbohydrate was not attached to random Ser or Thr residues or to any specific consensus peptide sequence. Instead, attachment was limited to residues accessible at the surface of the folded protein. Determination of the length and structure (linear versus branched) of the GspB-linked carbohydrate chains, along with the number and distribution of attachment sites per GspB polypeptide, will require more detailed chemical and structural analyses.
For most bacterial glycoproteins, it is unknown whether glycosylation occurs in the cytoplasm as opposed to the periplasm (of gram-negative species) or cell wall (of gram-positive species). However in Haemophilus influenzae, the HMW1 adhesin has been shown to be glycosylated in the cytoplasm (11). The E. coli TibA and AIDA-I autotransporter adhesins may also undergo glycosylation in the cytoplasm (3). The studies presented here indicate that in S. gordonii, the export of GspB is not a prerequisite to glycosylation of this protein. This is consistent with the observation that the gspB-sec operon-encoded enzymes that are likely to be involved in carbohydrate metabolism (Gly, Nss, and Gtf) lack apparent signal peptides and are predicted to be localized to the cytoplasm of S. gordonii.
The extensive glycosylation of GspB may have multiple effects on the function of the protein. Although it is possible that the carbohydrate residues could be directly involved in adherence to platelets, preliminary evidence has shown that neither 500 mM GlcNAc nor 250 mM glucose inhibits the binding of M99 to platelets (unpublished results). However, this does not exclude the possibility that the platelet receptor for GspB might recognize a distinct oligosaccharide component of GspB. Since we have not been able to detect a nonglycosylated version of GspB expressed by S. gordonii, it has not been possible to confirm whether glycosylation is required for platelet binding. Alternatively, glycosylation may directly affect the conformation or stability of the GspB polypeptide. One further possibility is that glycosylation might be necessary for export mediated by the accessory secretory proteins SecA2 and SecY2. We are using a combination of genetic and biochemical analyses to explore these possibilities.
Acknowledgments
This work was supported by grant AI41513 from the National Institutes of Health and by the Department of Veterans Affairs.
We thank B. Hayes for his assistance with the monosaccharide analysis.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Bensing, B. A., and P. M. Sullam. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44:1081-1094. [DOI] [PubMed] [Google Scholar]
- 3.Benz, I., and M. A. Schmidt. 2002. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45:267-276. [DOI] [PubMed] [Google Scholar]
- 4.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 5.Durack, D. T. 1975. Experimental bacterial endocarditis. IV. Structure and evolution of very early lesions. J. Pathol. 115:81-89. [DOI] [PubMed] [Google Scholar]
- 6.Durack, D. T., and P. B. Beeson. 1972. Experimental bacterial endocarditis. I. Colonization of a sterile vegetation. Br. J. Exp. Pathol. 53:44-49. [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson, P. R., and M. C. Herzberg. 1993. Evidence for the covalent linkage of carbohydrate polymers to a glycoprotein from Streptococcus sanguis. J. Biol. Chem. 268:23780-23783. [PubMed] [Google Scholar]
- 8.Ferguson, D. J., A. A. McColm, D. M. Ryan, and P. Acred. 1986. Experimental staphylococcal endocarditis and aortitis. Morphology of the initial colonization. Virchows Arch. A 410:43-48. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson, D. J., A. A. McColm, T. J. Savage, D. M. Ryan, and P. Acred. 1986. A morphological study of experimental rabbit staphylococcal endocarditis and aortitis. I. Formation and effect of infected and uninfected vegetations on the aorta. Br. J. Exp. Pathol. 67:667-678. [PMC free article] [PubMed] [Google Scholar]
- 10.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 11.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St Geme III. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 12.Macrina, F. L., R. P. Evans, J. A. Tobian, D. L. Hartley, D. B. Clewell, and K. R. Jones. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145-150. [DOI] [PubMed] [Google Scholar]
- 13.McGowan, D. A., and R. Gillett. 1980. Scanning electron microscopic observations of the surface of the initial lesion in experimental streptococcal endocarditis in the rabbit. Br. J. Exp. Pathol. 61:164-171. [PMC free article] [PubMed] [Google Scholar]
- 14.McNab, R., and H. F. Jenkinson. 1998. Lipoproteins and other cell-surface associated proteins in streptococci. Methods Cell Sci. 20:209-216. [Google Scholar]
- 15.Sambri, V., C. Stefanelli, and R. Cevenini. 1992. Detection of glycoproteins in Borrelia burgdorferi. Arch. Microbiol. 157:205-208. [DOI] [PubMed] [Google Scholar]
- 16.Schaffer, C., M. Graninger, and P. Messner. 2001. Prokaryotic glycosylation. Proteomics 1:248-261. [DOI] [PubMed] [Google Scholar]
- 17.Schaffer, C., and P. Messner. 2001. Glycobiology of surface layer proteins. Biochimie 83:591-599. [DOI] [PubMed] [Google Scholar]
- 18.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 20.Stephenson, A. E., H. Wu, J. Novak, M. Tomana, K. Mintz, and P. Fives-Taylor. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43:147-157. [DOI] [PubMed] [Google Scholar]
- 21.Sullam, P. M., U. Frank, M. R. Yeaman, M. G. Tauber, A. S. Bayer, and H. F. Chambers. 1993. Effect of thrombocytopenia on the early course of streptococcal endocarditis. J. Infect. Dis. 168:910-914. [DOI] [PubMed] [Google Scholar]
- 22.Sullam, P. M., F. H. Valone, and J. Mills. 1987. Mechanisms of platelet aggregation by viridans group streptococci. Infect. Immun. 55:1743-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi, Y., K. Konishi, J. O. Cisar, and M. Yoshikawa. 2002. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect. Immun. 70:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thibault, P., S. M. Logan, J. F. Kelly, J. R. Brisson, C. P. Ewing, T. J. Trust, and P. Guerry. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276:34862-34870. [DOI] [PubMed] [Google Scholar]
- 25.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Upreti, R. K., M. Kumar, and V. Shankar. 2003. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3:363-379. [DOI] [PubMed] [Google Scholar]
- 27.von Heijne, G., and L. Abrahmsen. 1989. Species-specific variation in signal peptide design. Implications for protein secretion in foreign hosts. FEBS Lett. 244:439-446. [DOI] [PubMed] [Google Scholar]
- 28.Wu, H., and P. M. Fives-Taylor. 1999. Identification of dipeptide repeats and a cell wall sorting signal in the fimbriae-associated adhesin, Fap1, of Streptococcus parasanguis. Mol. Microbiol. 34:1070-1081. [DOI] [PubMed] [Google Scholar]