Abstract
Class D β-lactamases with carbapenemase activity are emerging as carbapenem-resistance determinants in Gram-negative bacterial pathogens, mostly Acinetobacter baumannii and Klebsiella pneumoniae. Carbapenemase activity is an unusual feature among class D β-lactamases, and the structural elements responsible for this activity remain unclear. Based on structural and molecular dynamics data, we previously hypothesized a potential role of the residues located in the short-loop connecting strands β5 and β6 (the β5–β6 loop) in conferring the carbapenemase activity of the OXA-48 enzyme. In this work, the narrow-spectrum OXA-10 class D β-lactamase, which is unable to hydrolyze carbapenems, was used as a model to investigate the possibility of evolving carbapenemase activity by replacement of the β5–β6 loop with those present in three different lineages of class D carbapenemases (OXA-23, OXA-24, and OXA-48). Biological assays and kinetic measurements showed that all three OXA-10–derived hybrids acquired significant carbapenemase activity. Structural analysis of the OXA-10loop24 and OXA-10loop48 hybrids revealed no significant changes in the molecular fold of the enzyme, except for the orientation of the substituted β5–β6 loops, which was reminiscent of that found in their parental enzymes. These results demonstrate the crucial role of the β5–β6 loop in the carbapenemase activity of class D β-lactamases, and provide previously unexplored insights into the mechanism by which these enzymes can evolve carbapenemase activity.
Keywords: antibiotic resistance, protein engineering, protein evolution, X-ray structure
Although antimicrobial chemotherapy represented one of the major advances of medicine in the last century, the efficacy of antibiotics has been relentlessly challenged by the emergence of resistant bacteria. Because of their efficacy and safety, β-lactams, still represent the most widely used antibiotics (1). Among these β-lactams, carbapenems currently have a critical importance as they are active against a wide range of Gram-negative pathogens, including multidrug-resistant strains, and are increasingly used to treat severe infections acquired in the hospital setting. Thus, the worldwide emergence of carbapenem-resistant bacteria, commonly associated with either multidrug resistance or extensively-drug resistant phenotypes, is considered one of the major challenges for the treatment of bacterial infections (2, 3).
β-Lactamases (which hydrolyze the amide bond of the β-lactam ring) play a key role in the development of acquired resistance to β-lactams in many bacterial pathogens. Two structurally and mechanistically unrelated families of β-lactamases are known: (i) the most numerous active serine enzymes (belonging to the molecular classes A, C, and D) that hydrolyze their substrates via the formation of an acyl-enzyme covalent complex, and (ii) the zinc-dependent metallo-β-lactamases (class B). Serine-β-lactamases, besides having an impressive sequence heterogeneity, share common catalytically important residues: the S*xxV motif (motif 1, S* being the catalytic serine), SxV (motif 2), and K-T/S-G motif (motif 3) (4).
The production of β-lactamases capable of hydrolyzing carbapenems (carbapenemases) is one of the major mechanisms of carbapenem resistance. Known carbapenemases include enzymes of molecular classes A, B, and D, each showing distinct structural and biochemical properties but with the common ability to inactivate carbapenem antibiotics (4).
The class D (OXA-type) carbapenemases play a major role in acquired carbapenem resistance in Acinetobacter baumannii and, to a lesser extent, in Klebsiella pneumoniae (5). Four major lineages of acquired class D carbapenemases are known—namely OXA-23, OXA-24, OXA-48, and OXA-58 (some including closely related allelic variants)—that are notably different from each other at the level of primary structure (34–63% amino acid sequence divergence).
As a result of recently obtained X-ray structural data, some attempts have been made to unravel the structure–function relationships of these clinically relevant enzymes (6, 7). The 3D structure of OXA-24 shows an unusual tunnel-like entrance of the active site, stabilized by the interaction between tyrosine 112 and methionine 223, which was retained as critical for the carbapenemase activity of this enzyme and was proposed to be the main factor to dictate the biochemical properties of OXA-24. The tunnel-like entrance of OXA-24 was also hypothesized to be essential for the correct orientation of carbapenem substrates and, therefore, their entrance in the active site (6). Although this study did provide a structural basis for the unusual substrate profile of OXA-24 (i.e., the lack of hydrolysis of oxacillin substrates with bulkier side chains because of steric hindrance, with the residues constituting the tunnel-like active site entrance), it did not provide a general mechanism for carbapenem hydrolysis by class D carbapenemases. More recently, the structure of OXA-48 was obtained, which surprisingly does not show the same tunnel-like entrance. Despite this lack of a tunnel-like entrance, the enzyme is currently (in terms of turnover rates) the most efficient class D carbapenemase (kcat for imipenem, 5 s−1, a value still rather low compared with that of the best substrates of OXA-type enzymes; that is, the steady-state kcat value for cloxacillin or oxacillin can be as high as 500–600 s−1) (8). Moreover, structural analysis, molecular dynamics, and molecular modeling studies pointed to the potential role of the residues located in the β5–β6 loop. Indeed, we previously proposed a mechanism for carbapenem hydrolysis that involves an enzyme-assisted rotation of hydroxyethyl moiety of the acyl-enzyme intermediate, a process in which the residues of the β5–β6 loop would be directly involved and which would allow the deacylation water molecule to reach the scissile bond and perform the nucleophilic attack, leading to the breakdown of the acyl-enzyme complex (7).
To provide experimental evidence for the suggested role of the β5–β6 loop in carbapenemase activity of class D enzymes, we substituted the β5–β6 loop of the noncarbapenemase enzyme OXA-10 with the structurally-equivalent loops of three class D carbapenemases (OXA-23, OXA-24, and OXA-48), and tested the β-lactamase profile of the hybrid proteins and their functional and structural features. Indeed, the hybrid OXA-10 proteins acted as functional carbapenemases, thus demonstrating the crucial role of the β5–β6 loop in the evolution of carbapenemase activity and modulation of substrate specificity among class D β-lactamases.
Results and Discussion
Structural Variability of the β5–β6 Loop in Class D Carbapenemases vs. Noncarbapenemases.
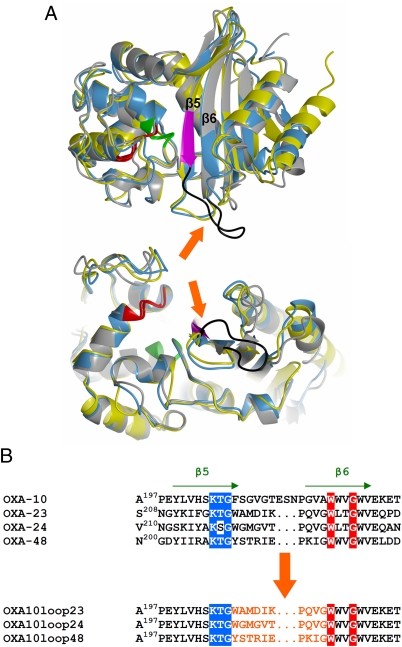
A thorough comparison of the structures of class D carbapenemases OXA-24 and OXA-48 with that of class D noncarbapenemase OXA-10 was done. Interestingly, a major difference was observed in the orientation and size of the β5–β6 loop, which appeared to be similar in OXA-24 and OXA-48 but different from that of OXA-10 (Fig. 1) and other class D β-lactamases whose structure has been obtained (9–13). This loop is close to the active site and connects two β-strands, one of which includes the catalytically relevant conserved K205TG residues (a conserved motif of serine β-lactamases, which delimits one side of the active site) (Fig. 1). The same loop also delimits one side of the active site in the carbapenemases OXA-24 and OXA-48, although it points outwards to the active site in OXA-10 (Fig. 1A). This structural heterogeneity could further support our previous hypothesis regarding the relevance of the residues in the β5–β6 loop in the enzyme catalytic properties, and more specifically in determining the fate of the acyl-enzyme complex in the case of carbapenems (i.e., inhibition or efficient hydrolysis) (7).
Fig. 1.
Comparison of the structural features of class D carbapenemases with the narrow-spectrum OXA-10 β-lactamase, showing the significant differences in orientation and length of the β5–β6 loop. (A) Side (Upper) and bottom (Lower) views of the superimposed structures of native OXA-10 (gray) (PDB code, 1FOF), OXA-24 (yellow) (PDB code, 2JC7), and OXA-48 (blue) (PDB code, 3HBR). The location of catalytically relevant conserved motifs is shown in green (SxxK motif, S being the catalytic serine 70 residue), red (SxV motif), and magenta (KTG motif). The β5–β6 loops, connecting the labeled β5 and β6 strands, are indicated by the arrows and, in the case of OXA-10, colored in black. (B) Amino acid sequence comparison of the β5–β6 loop region in OXA-10, OXA-23, OXA-24, and OXA-48. The relevance of the β5–β6 loop in the carbapenemase activity of OXA-23, OXA-24, and OXA-48 carbapenemases was probed by mutagenesis. The β5–β6 loop of these class D carbapenemases was grafted in the structurally equivalent position of the narrow-spectrum class D enzyme OXA-10, to obtain three OXA-10 loop variants named OXA-10loop23, OXA-10loop24, and OXA-10loop48, as shown.
Construction of OXA-10 Hybrids with β5–β6 Loops from Different Class D Carbapenemases by Rational Protein Design.
The relevance of the β5–β6 loop in the carbapenemase activity of class D carbapenemases was probed by mutagenesis, and OXA-10 was used as a protein model to evaluate the possibility to evolve carbapenemase activity in an enzyme lacking this property. OXA-10 variants differing in the nature of the residues located in the β5–β6 loop were designed using a structure-based rational approach in which the β5–β6 loop of OXA-10 was replaced with the corresponding structural element of class D carbapenemases OXA-24 and OXA-48 (Fig. 1B).
On the basis of sequence homology, and despite the fact that 3D structural data are not available for this enzyme, the same strategy was used to obtain an OXA-10 derivative carrying the β5–β6 loop of OXA-23, a class D carbapenemase closer to OXA-24 than OXA-48 (OXA-23 exhibits 59% and 35% identity with these enzymes, respectively). Interestingly, OXA-23 shows 4 amino acid substitutions in its β5–β6 loop compared with OXA-24 (Fig. 1B), and thus was used as a further probe for the impact of structure diversity in this element on the functional properties of the enzymes.
These OXA-10 laboratory variants were then cloned and expressed in Escherichia coli DH5α to investigate their potential contribution to β-lactam resistance and investigate their biochemical and structural properties. The authenticity of the purified OXA-10 derivatives has been assessed by sequencing of the plasmid constructs and by peptide mass fingerprint MALDI-TOF mass spectrometry, which confirmed the correct substitution of the β5–β6 loop residues (Table S1).
β5–β6 Loop Variants of OXA-10 Exhibit Carbapenem-Hydrolyzing Activity.
The contribution of OXA-10 loop variants to resistance to β-lactam antibiotics in an E. coli DH5α laboratory strain was investigated by measuring the minimal inhibitory concentrations (MICs) for several antibiotics (Table 1). The strains producing the enzyme variants exhibited high MIC values for penicillins, although somewhat lower than those of the strain producing the wild-type enzyme. These data indicate that the substitution of the β5–β6 loop in OXA-10 yields a functional enzyme, which can efficiently confer β-lactam resistance in vitro. The MIC values of carbapenems for the various strains were rather low for all mutants. However, it was already observed that the contribution of carbapenem-hydrolyzing enzymes in regards to the resistance to carbapenems in E. coli was very limited, and the fast permeation of carbapenems in this microorganism was demonstrated to be determinant in the resulting antibacterial activity of these compounds (14).
Table 1.
Antimicrobial susceptibility profile of E. coli DH5α producing OXA-10 and variants thereof
| MIC (μg/mL) | |||||
| β-Lactam(s) | OXA-10 | OXA-10loop23 | OXA-10loop24 | OXA-10loop48 | pBC-SK |
| Ampicillin | >256 | 64 | 128 | 128 | 4 |
| Piperacillin | 32 | 16 | 16 | 16 | 1 |
| Ticarcillin | >256 | 128 | 256 | >256 | 8 |
| Imipenem | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| Meropenem | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 |
| Ertapenem | <0.01 | 0.12 | <0.01 | 0.03 | <0.01 |
| Aztreonam | 0.5 | 0.12 | 0.12 | 0.12 | 0.25 |
MIC values for the strain carrying the empty vector (pBC-SK) are shown for comparison.
A more sensitive method was then devised to quickly obtain relevant information regarding the impact of the various substitutions on the OXA-10 catalytic properties, especially regarding carbapenems. The carbapenemase plate assay (see Materials and Methods for details) was based on the microbial titration of imipenem after incubation with crude extracts of the E. coli strains producing the wild-type OXA-10 and the various OXA-10 variants (a β-lactamase–negative strain was used as the negative control), using Bacillus subtilis as the indicator organism. This method yielded interesting results, as it clearly highlighted different behaviors between the wild-type OXA-10 and the loop variants (Table 2 and Fig. S1). Indeed, wild-type OXA-10 expectedly proved unable to significantly degrade imipenem after 4 h of incubation, as shown by the constant growth inhibition of B. subtilis. In contrast, all loop variants showed a complete degradation of the antibiotic within 2 to 3 h by the physiological amount of β-lactamase produced by the respective E. coli clones, as shown by the absence of growth inhibition of the indicator organism.
Table 2.
Results of the carbapenemase plate assay
| Inhibition zone diameter (mm) with crude extracts from E. coli DH5α strains producing: | |||||
| Time (h) | OXA-10 | OXA-10loop23 | OXA-10loop24 | OXA-10loop48 | Control* |
| 0 | 20 | 20 | 20 | 20 | 20 |
| 1 | 20 | 11 | 8 | 15 | 20 |
| 2 | 20 | No inhibition | No inhibition | 7 | 20 |
| 3 | 20 | No inhibition | No inhibition | No inhibition | 20 |
| 4 | 20 | No inhibition | No inhibition | No inhibition | 20 |
See Materials and Methods for details.
*The controls consisted of imipenem incubated with either buffer or a crude extract of E. coli DH5α carrying the empty plasmid vector (pBC-SK) and identical results were obtained with both samples.
These data indicated that the loop variants definitely acquired carbapenem-hydrolyzing activity. Interestingly, the same result was achieved independently of the nature and origin of the loop, indicating that OXA-10 was able to accommodate structural elements originating from rather close (e.g., OXA-48) but also very distant proteins (e.g., OXA-23 and OXA-24). To better analyze the differences in the biochemical properties of wild-type OXA-10 and the loop variants, two of these variants (containing the most distant heterologous loops; that is, from OXA-24 and OXA-48) were purified to near homogeneity and subjected to kinetic analysis.
Similar catalytic efficiencies were obtained with the loop variants and the wild-type OXA-10 for the hydrolysis of ampicillin, oxacillin, and cephalothin (Table 3), and only minor differences could be observed in the kcat and Km values (up to 5- or 10-fold variations, respectively). This finding confirms the excellent plasticity of OXA-10, as the enzyme is not only tolerant to the substitution of its β5–β6 loop but also largely maintains its original biochemical properties unaltered. This theory is further reinforced by the comparison of the kinetic data of the OXA-10 loop variants with that of the native carbapenemase. Indeed, OXA-24 is characterized by a very low turnover rate of oxacillin, and a very large Km value for this substrate (Table 3), but such a typical behavior is not observed with the OXA-10loop24 variant. These results thus indicate that the residues constituting the β5–β6 loop of OXA-10 are not relevant for the hydrolysis of ampicillin, oxacillin, or cephalothin. Most strikingly, kinetic analysis confirmed the acquisition of a significant carbapenem-hydrolyzing activity, essentially reflected by the impressive increase of the kcat value (up to 175-fold). An increase of the Km value is also observed in the loop variants, which determines a significant change in the behavior of OXA-10 toward carbapenems. Indeed, the wild-type enzyme is readily acylated (low Km and high kcat/Km values) but very slowly deacylated (low turnover rates), imipenem being a poor substrate (or inhibitor) of the enzyme. This process actually allowed for the direct structural observation of the acylated imipenem or meropenem complex structures of OXA-13, an enzyme very close to OXA-10 (12). In contrast, the loop variants now show kinetic parameters for the hydrolysis of imipenem that are closer to that measured with cephalothin. It is important to mention that the turnover rates of these laboratory loop variants are actually higher than those measured for the parent carbapenemases, being three- to ninefold greater than that of OXA-48 (the class D carbapenemase showing the highest turnover rate for imipenem) and OXA-24, respectively (Table 3) (6, 7). This finding indicates that these rationally designed mutants are actually more efficient than the natural enzymes, although it could not be excluded that the substitution of the loop might affect other factors, such as protein stability and folding.
Table 3.
Steady-state kinetic parameters for the hydrolysis of various β-lactam antibiotics of the purified OXA-10 β-lactamase and variants thereof
| Substrate | OXA-10 variant (parent enzyme) | kcat (s−1) | Km (μM) | kcat/Km (M−1·s−1) |
| Ampicillin | WT | 530 | 77 | 6.9 × 106 |
| Loop 24 (OXA-24) | 850 ± 40 (220) | 130 ± 20 (80) | 6.5 × 106 (2.8 × 106) | |
| Loop 48 (OXA-48) | 510 ± 20 (955) | 83 ± 12 (395) | 6.1 × 106 (2.4 × 106) | |
| Oxacillin | WT | 660 | 96 | 6.9 × 106 |
| Loop 24 (OXA-24) | 460 ± 12 (0.18) | 360 ± 30 (1,270) | 1.3 × 106 (1.4 × 102) | |
| Loop 48 (OXA-48) | 180 ± 3 (130) | 19 ± 2 (95)* | 9.5 × 106 (1.4 × 106) | |
| Cephalothin | WT | 2.9 ± 0.1 | 11 ± 1.4 | 2.6 × 105 |
| Loop 24 (OXA-24) | 10 ± 0.5 (–)† | 260 ± 30 (–)† | 3.8 × 104 (–)† | |
| Loop 48 (OXA-48) | 10 ± 0.3 (44) | 100 ± 10 (195) | 1.0 × 105 (2.3 × 105) | |
| Imipenem | WT | <0.1 | 0.04 ± 0.007* | N. D. |
| Loop 24 (OXA-24) | 9 ± 0.6 (1) | 60 ± 10 (0.58) | 1.5 × 105 (1.7 × 106) | |
| Loop 48 (OXA-48) | 14 ± 0.3 (4.8) | 45 ± 3 (13) | 3.1 × 105 (3.7 × 105) |
Data for ampicillin and oxacillin with wild-type OXA-10 are from Danel et al. (8). The kinetic parameters for the parent carbapenemases OXA-24 and OXA-48 are indicated in the parentheses (6, 7).
*Low Km values were determined as Ki using either 60 μM cephalothin or 180 μM ampicillin as the reporter substrates.
†Data not available.
Structural Properties of the OXA-10 Loop Variants.
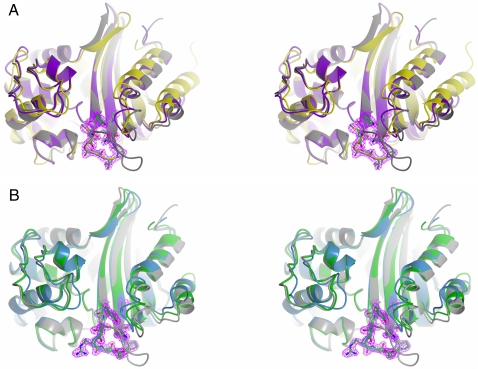
Once we determined that the substitution of the β5–β6 loop of OXA-10 could yield laboratory variants with significant carbapenem-hydrolyzing activity, two questions needed to be addressed from the structural standpoint: (i) What would be the conformation adopted by the grafted carbapenemase loop? (ii) What would be the impact of the replacement of the β5–β6 loop of OXA-10 on its overall fold and the active site properties?
Crystals of the purified loop variants (OXA-10loop24 and OXA-10loop48) were readily obtained, although both proteins crystallized in different conditions, likely reflecting the different nature and composition of the grafted loop. In addition, these two variants crystallized in different space groups and with significantly different cell dimensions and content (Table 4). Both OXA-10loop24 and OXA-10loop48 variants adopted the dimeric quaternary structure of wild-type OXA-10 (10, 11). Comparison of the overall fold of the OXA-10 loop variants with that of the wild-type enzyme did not reveal any significant differences, except in the region of the β5–β6 loop (Fig. 2). Minor differences were observed in the extremities of strands β5 and β6, reflecting the introduction of a shorter loop connecting these two structural elements. A comparison of the OXA-10 loop variants with OXA-24 (Fig. 2A) and OXA-48 (Fig. 2B) clearly indicates that the loops originating from carbapenemase enzymes and grafted in OXA-10 at structurally equivalent positions maintain the conformation presented on their respective wild-type carbapenemases, and are oriented inward toward the active site.
Table 4.
Data collection and refinement statistics for the crystal structures of OXA-10 loop variants
| Enzyme or atom | OXA-10loop24 | OXA-10loop48 |
| Enzyme | ||
| PDB code | 3QNB | 3QNC |
| X-ray source | ESRF ID23-2 | ID14-1 |
| Wavelength (Å) | 0.873 | 0.933 |
| Data coll. Temp. (K) | 100 | 100 |
| Space group | P32 | P212121 |
| Cell dimensions (Å) | a = b = 81.33; c = 151.92 | a = 49.21; b = 97.01; c = 125.87 |
| Subunits/asu | 4 | 2 |
| Matthews coeff. (Å3Da−1) | 2.66 | 2.74 |
| Solvent content (%) | 53.72 | 55.10 |
| Resolution limits (Å) | 33.43–1.95 (2.06–1.95) | 28.76–1.60 (1.68–1.60) |
| Reflections measured | 262,333 (37,459) | 554,619 (72,850) |
| Unique reflections | 81,931 (11,977) | 79,252 (11,124) |
| Completeness (%) | 99.9 (100.0) | 98.0 (95.3) |
| Rmerge (%) | 8.6 (32.3) | 6.9 (37.2) |
| Multiplicity | 3.2 (3.1) | 7.0 (6.5) |
| I/σ(I) | 9.2 (3.3) | 15.8 (4.4) |
| Wilson B-factor (Å2) | 17.72 | 17.50 |
| Rcryst (%) | 17.2 (22.1) | 17.5 (24.1) |
| Rfree; free R test set size (%) | 22.1 (32.0); 5.0 | 20.1 (24.1); 5.0 |
| Atoms/molecules in refinement | ||
| Protein atoms | 7,688 | 3,895 |
| Sulfate ions | 7 | 9 |
| Ethylene glycol molecules | 20 | 8 |
| Carbon dioxide molecules | 3 | 1 |
| Water molecules | 449 | 543 |
| Average B factor (Å2) | 22.63 | 21.55 |
| rmsd Bond lengths (Å) | 0.025 | 0.011 |
| rmsd Bond angles (°) | 2.009 | 1.342 |
| rmsd Planar groups(Å) | 0.011 | 0.007 |
| rmsd Chiral centers (Å3) | 0.164 | 0.095 |
| esd on atomic positions (Å) | 0.095 | 0.048 |
| Ramachandran favored (%) | 95.3 | 97.1 |
| Ramachandran allowed (%) | 4.7 | 2.9 |
Data in parentheses refer to the highest resolution shell. esd, estimated standard deviations; rmsd, root mean square deviation.
Fig. 2.
Overall structures of the OXA-10 laboratory variants with the engineered β5–β6 loop. The residues of the β5–β6 loop in these laboratory variants are shown as sticks, and the corresponding 2Fc-Fo Fourier difference map (contoured at 1.5 σ) is shown in magenta. PDB codes are as in Fig. 1A. (A) Stereoview of the superimposed structures of OXA-10 (gray), its β5–β6 loop laboratory variant OXA-10loop24 (purple), and OXA-24 (yellow). The tertiary structure of the OXA-10loop24 variant is similar to that of the native OXA-10, except in the region of the β5–β6 loop, which now adopts a conformation very similar to that observed in the enzyme from which it originates (OXA-24). (B) Stereoview of the superimposed structures of OXA-10 (gray), its β5–β6 loop laboratory variant OXA-10loop48 (green), and OXA-48 (blue). Similarly to the OXA-10loop24 variant, the tertiary structure of the OXA-10loop48 variant is similar to that of the native OXA-10, except in the region of the β5–β6 loop, which also adopts the peculiar conformation observed in OXA-48.
This observation shows that the conformation of the loop is maintained despite the important sequence heterogeneity, reinforcing the idea that such conformation is determined by the loop length but also by the presence of a conserved PxxG motif, which is found in the carboxyl-terminal part of the β5–β6 loop (residues 217–220 in the OXA-48 numbering). Indeed, by comparing all of the available class D β-lactamase amino acid sequences (http://www.ncbi.nlm.nih.gov/), it emerged that this motif appears in all of the enzymes with carbapenemase properties, but never occurs in other OXA-type enzymes.
Concluding Remarks.
β-Lactamases provided Bacteria with an extraordinarily versatile tool that contributed and still contributes to the development of resistance to successive generations of β-lactam antibiotics. Considering the worldwide dissemination of bacterial isolates exhibiting resistance to most available antibacterial drugs, the development of new β-lactamase inhibitors would be desirable. From this perspective, the understanding of the structure-function dependencies of clinically-relevant β-lactamases, like the class D carbapenemases investigated here, is of primary importance.
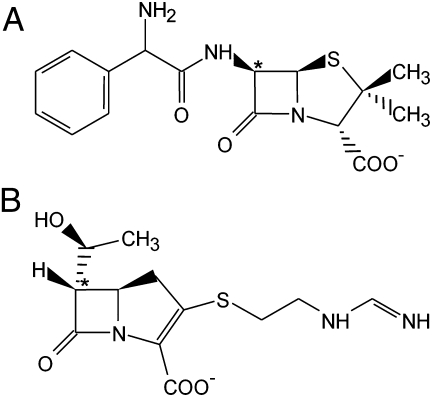
In this work, we successfully evolved a narrow-spectrum class D β-lactamase into a carbapenemase using a rational structure-based approach. The hydrolytic properties of OXA-10–engineered laboratory variants, in which the β5–β6 loop found in various clinically relevant class D carbapenemases (OXA-23, OXA-24, and OXA-48) was inserted in the structurally equivalent position in OXA-10, were significantly altered as these variants acquired significant imipenem-hydrolyzing activity. The data presented here clearly demonstrate the critical role of the residues of the β5–β6 loop in determining the behavior of the enzyme toward carbapenem substrates, although not in affecting its ability to hydrolyze other substrates (like penicillins or cephalosporins). Interestingly, this finding should be paralleled with the distinctive transconformation shown by carbapenem compounds (Fig. 3), whose hydroxyethyl moiety is located on the α-face of the β-lactam ring and is interacting with the side chains of residues in the β5–β6 loop.
Fig. 3.
The structure of two representative β-lactams. (A) Ampicillin, a penicillin compound with the substitutents of the β-lactam ring in a cis conformation (also characteristic of cephalosporins and monobactams) and (B) imipenem, a carbapenem compound, with a 6α-hydroxyethyl substituent that determines a trans conformation of the molecule (the C-6 atom is indicated by an asterisk).
Beyond their clinical relevance, β-lactamases also represent interesting model enzymes to investigate protein structure function and molecular evolution (15, 16). From an evolutionary standpoint, modifications of the loops close to the enzyme active site, which could possibly arise from homologous recombination events, have been recognized as a factor contributing to the evolution or diversification of enzyme function within a family of structurally related proteins (16). The data presented here are in agreement with such a model and represent an additional example of the structural plasticity exhibited by a β-lactamase, in this case the class D enzyme OXA-10. Indeed, this protein tolerated the substitution of an entire active site loop connecting two conserved secondary structures without significantly altering its tertiary structure. Moreover, this substitution was specifically responsible for a dramatic broadening of the enzyme substrate profile, now including carbapenems, without determining a trade-off of the enzyme original properties, thus further highlighting the importance of active site loops in the functional properties of class D β-lactamases.
Finally, this work also represents an interesting example of rational in vitro evolution of a protein substrate profile, following a molecular dynamics study that provided relevant information to hypothesize a catalytic mechanism (i.e., the role of the β5–β6 loop in promoting the rotation of the 6α-hydroxyethyl moiety of the acylated substrate, allowing for the correct activation of the deacylation water molecule) (7), which is now clearly supported by experimental data. Moreover, this mechanism seems to be adopted by all three class D carbapenemases investigated in this study and might well extend to other class D enzymes with similar properties (e.g., the acquired OXA-58 enzyme), thus providing previously unexplored insights potentially useful for the design of new β-lactamase inhibitors.
Materials and Methods
Molecular Techniques.
An OXA-10-producing E. coli laboratory strain was obtained following PCR amplification of the blaOXA-10 ORF with specific primers (Table 5) with the Expand High Fidelity PCR system (Roche Biochemicals), as previously described (17). The reaction conditions used were the following: an initial denaturation step at 96 °C for 2 min; 30 cycles of denaturation at 94 °C for 1 min, annealing at 48 °C for 1 min, extension at 72 °C for 1 min and 30 s, and a final extension step at 72 °C for 20 min. The 760-bp amplification product was subcloned in the EcoRI and BamHI restriction sites of pLB-II vector (18), to obtain the pLBII-OXA-10 plasmid. The OXA-10 laboratory variants were constructed by overlapping PCR with partially overlapping primers (Table 5), as previously described (19), using the pLBII-OXA10 plasmid as the template. The resulting mutated β-lactamase genes were sequenced to confirm the absence of any unwanted mutations.
Table 5.
Primers used in this study
| Primer | Sequence (5′–3′) |
| OXA-10-EXP/f* | CCGAATTCATCATGAAAACATTTGCCGCAT |
| OXA-10-EXP/r* | CCGGATCCTTAGCCACCAATGATGCCC |
| OXA-10loop23/f† | ATTAAACCGCAGGTGGGCTGGTGGGTTGGGTGGGTTG |
| OXA-10loop23/r† | TTTAATATCCATCGCCCAACCAGTTTTTGAATGCACTAG |
| OXA-10loop24/f† | GTTACTCCACAGGTAGGTTGGTGGGTTGGGTGGGTTG |
| OXA-10loop24/r† | AGTAACACCCATTCCCCAACCAGTTTTTGAATGCACTAG |
| OXA-10loop48/f† | ATTGAACCGAAAATTGGCTGGTGGGTTGGGTGGGTTG |
| OXA-10loop48/r† | TTCAATGCGGGTGCTATAACCAGTTTTTGAATGCACTAGAT |
*Primers used for cloning the blaOXA-10 ORF.
†Primers used to generate the various blaOXA-10 mutants.
In Vitro Antimicrobial Susceptibility Testing and Evaluation of Carbapenemase Activity in the Bacterial Host.
The in vitro antimicrobial susceptibility of E. coli DH5α carrying pLBII-OXA10 and other vectors was determined by the microdilution broth method, as recommended by the Clinical Laboratory Standards Institute (20), using supplemented Mueller-Hinton broth (Difco Laboratories) with an inoculum size of 104 CFU per well. Results were recorded after 18 h of incubation at 37 °C.
Microbiological Detection of Carbapenemase Activity in OXA-10 Variants (Carbapenemase Plate Assay).
An overnight culture of the various strains (grown in Mueller-Hinton broth at 37 °C) was added (final cell density, A600 = 0.5) to a 10 μg/mL (3.3 μM) imipenem solution and incubated at 30 °C for at least 4 h. Ten-microliter aliquots of the antibiotic solution were spotted down every hour onto a seeded top agar lawn of B. subtilis ATCC6633 on Mueller-Hinton plate (supplemented with colistin 2 μg/mL to prevent the growth of the E. coli strain). The diameter of the growth inhibition zone was measured after an incubation for 18 h at 37 °C.
Protein Production and Purification.
OXA-10 and its variants were produced from 1-L cultures of E. coli DH5α carrying plasmid pLBII-OXA10 or pLBII-OXA10loop24, and from culture of E. coli BL21(DE3) carrying plasmid pET-24-OXA10 loop48 in ZYP-5052 medium (21) at 30 °C for 24 h (for OXA-10 and OXA-10loop48) and 48 h (for OXA-10loop24). Cells were harvested by centrifugation (10,000 × g, 30 min, 4 °C), resuspended, and lysed by sonication. Cellular debris was removed by centrifugation (12,000 × g, 40 min, 4 °C). The proteins were purified in two or three chromatography steps. The first step was a cation exchange on a Hi-Trap SP-High Performance (prepacked column GE Healthcare) for both wild-type and mutants at pH 6.0 and pH 7.5, respectively; the second step was for the wild-type anion exchange (performed with a Mono Q, pH 7.6) and for the mutant cation exchange (performed with a Mono S, pH 7.5). Protein elution was performed using a linear gradient of K2SO4 (up to 0.5 M) instead of NaCl, to prevent inhibition of the enzyme by chloride ions. When required, the β-lactam–containing fractions were pooled and loaded on a XK 16/70 column packed with 120 mL of Superdex75 prep grade gel (GE Healthcare) using buffer with 20 mM Tris H2SO4 plus 0.3 M K2SO4, which was also used for protein elution (10).
Protein Analysis Techniques and Enzyme Kinetics.
The presence of β-lactamase in crude cell extracts and during all of the purification steps was assayed spectrophotometrically by monitoring the hydrolysis of 1 mM ampicillin at 235 nM. Enzyme purity and authenticity were assayed by SDS-PAGE and peptide mass fingerprint MALDI-TOF mass spectrometry (following proteins digestion by trypsin) (22). Kinetic parameters for the hydrolysis of β-lactam substrates of purified OXA-10 and the variants thereof were determined at 30 °C in using buffer with 20 mM Tris H2SO4 plus 0.3 M K2SO4 (10) supplemented with Na2CO3 50 mM using a Cary 100 UV-visible light spectrophotometer (Varian), as previously described (17). The enzyme concentration in the various assays ranged 1 to 500 nM.
Crystallization, X-Ray Diffraction, and Model Refinement.
Crystals of OXA-10 variants (OXA-10loop24 and OXA-10loop48) were obtained in a sitting-drop set-up (24-well Cryschem microplates; Hampton Research) in the following conditions: 0.1 M Tris pH 8.0, 2.8 M (NH4)2SO4 or 0.1 M Bicine pH 9.0, 2.9 M (NH4)2SO4, for OXA-10loop24 and OXA-10loop48, respectively. The drop consisted of 2 μL of 10 mg/mL of protein and 2 μL of the precipitant solution. Single crystals of β-lactamase were observed after several days of incubation at room temperature. Diffraction data were collected on the ID14-1 and ID23-2 beamlines at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France, at 100 K on crystals cryoprotected by 20% to 30% glycerol added to the respective crystallization solutions. The data were processed using the program MOSFLM and scaled using the program SCALA (23). The data collection parameters and data reduction statistics are reported in Table 4. The structure of the OXA-10 variants was solved by molecular replacement using standard Patterson search techniques, as implemented in the software MOLREP (24). The structure of the wild-type OXA-10 (PDB code, 1K4F) with all water molecules and ions omitted was used as the search model. The refinement was carried out by using REFMAC5 (25). Between the refinement cycles, the model was subjected to manual rebuild using COOT (26). The same program was used to model the various ligands (sulfate and ethylene glycol). Water molecules were added by using the standard procedure within the ARP/wARP suite (27) and checked by visual inspection. The stereochemical quality of both refined models was assessed using the program PROCHECK (28). Figures were made using softwares PyMol (http://www.pymol.org/) or CCP4mg (29). The coordinates and structure factors of OXA-10loop24 and OXA-10loop48 have been deposited at the Protein Data Bank under codes 3QNB and 3QNC, respectively.
Supplementary Material
Acknowledgments
We thank L. Bini and A. Armini for performing the peptide mass fingerprint analysis; the European Synchrotron Radiation Facility (Grenoble, France) for having provided access to the ID14-1 and ID23-2 beamlines; and the European Synchrotron Radiation Facility staff for excellent assistance. This work was funded in part by a grant from the Italian Ministero dell'Istruzione, Università e Ricerca (Contract No. 2005061894_004).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3QNB and 3QNC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110530108/-/DCSupplemental.
References
- 1.Goossens H, Ferech M, Vander Stichele R, Elseviers M, ESAC Project Group Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet. 2005;365:579–587. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- 2.Rice LB. The clinical consequences of antimicrobial resistance. Curr Opin Microbiol. 2009;12:476–481. doi: 10.1016/j.mib.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Livermore DM. Has the era of untreatable infections arrived? J Antimicrob Chemother. 2009;64(Suppl 1):i29–i36. doi: 10.1093/jac/dkp255. [DOI] [PubMed] [Google Scholar]
- 4.Queenan AM, Bush K. Carbapenemases: The versatile β-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilavsky E, Schwaber MJ, Carmeli Y. How to stem the tide of carbapenemase-producing enterobacteriaceae?: Proactive versus reactive strategies. Curr Opin Infect Dis. 2010;23:327–331. doi: 10.1097/QCO.0b013e32833b3571. [DOI] [PubMed] [Google Scholar]
- 6.Santillana E, Beceiro A, Bou G, Romero A. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc Natl Acad Sci USA. 2007;104:5354–5359. doi: 10.1073/pnas.0607557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Docquier JD, et al. Crystal structure of the OXA-48 β-lactamase reveals mechanistic diversity among class D carbapenemases. Chem Biol. 2009;16:540–547. doi: 10.1016/j.chembiol.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Danel F, Hall LM, Gur D, Livermore DM. OXA-16, a further extended-spectrum variant of OXA-10 β-lactamase, from two Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother. 1998;42:3117–3122. doi: 10.1128/aac.42.12.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun T, Nukaga M, Mayama K, Braswell EH, Knox JR. Comparison of β-lactamases of classes A and D: 1.5-A crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci. 2003;12:82–91. doi: 10.1110/ps.0224303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paetzel M, et al. Crystal structure of the class D β-lactamase OXA-10. Nat Struct Biol. 2000;7:918–925. doi: 10.1038/79688. [DOI] [PubMed] [Google Scholar]
- 11.Maveyraud L, et al. Insights into class D β-lactamases are revealed by the crystal structure of the OXA10 enzyme from Pseudomonas aeruginosa. Structure. 2000;8:1289–1298. doi: 10.1016/s0969-2126(00)00534-7. [DOI] [PubMed] [Google Scholar]
- 12.Pernot L, et al. Crystal structures of the class D β-lactamase OXA-13 in the native form and in complex with meropenem. J Mol Biol. 2001;310:859–874. doi: 10.1006/jmbi.2001.4805. [DOI] [PubMed] [Google Scholar]
- 13.Docquier JD, et al. Crystal structure of the narrow-spectrum OXA-46 class D β-lactamase: Relationship between active-site lysine carbamylation and inhibition by polycarboxylates. Antimicrob Agents Chemother. 2010;54:2167–2174. doi: 10.1128/AAC.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumura N, Minami S, Watanabe Y, Iyobe S, Mitsuhashi S. Role of permeability in the activities of β-lactams against gram-negative bacteria which produce a group 3 β-lactamase. Antimicrob Agents Chemother. 1999;43:2084–2086. doi: 10.1128/aac.43.8.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol. 2001;8:238–242. doi: 10.1038/84981. [DOI] [PubMed] [Google Scholar]
- 16.Tawfik DS. Biochemistry. Loop grafting and the origins of enzyme species. Science. 2006;311:475–476. doi: 10.1126/science.1123883. [DOI] [PubMed] [Google Scholar]
- 17.Docquier JD, et al. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J Antimicrob Chemother. 2003;51:257–266. doi: 10.1093/jac/dkg067. [DOI] [PubMed] [Google Scholar]
- 18.Borgianni L, et al. Mutational analysis of VIM-2 reveals an essential determinant for metallo-β-lactamase stability and folding. Antimicrob Agents Chemother. 2010;54:3197–3204. doi: 10.1128/AAC.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clinical Laboratory Standards Institute . Wayne, PA: Clinical Laboratory Standards Institute; 2006. Methods for dilution antimicrobial susceptibility tests for Bacteria that grow aerobically, approved standard. 7th edition. CLSI document M7-A7; p. 26. [Google Scholar]
- 21.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Lovato L, et al. Transketolase and 2′,3′-cyclic-nucleotide 3′-phosphodiesterase type I isoforms are specifically recognized by IgG autoantibodies in multiple sclerosis patients. Mol Cell Proteomics. 2008;7:2337–2349. doi: 10.1074/mcp.M700277-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Evans PR. SCALA, continuous scaling program. Joint CCP4 and ESF-EACBM Newsletter. 1997;33:22–24. [Google Scholar]
- 24.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 25.Vagin AA, et al. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Cohen SX, et al. ARP/wARP and molecular replacement: The next generation. Acta Crystallogr D Biol Crystallogr. 2008;64:49–60. doi: 10.1107/S0907444907047580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laskowski RA, Macarthur MW, Moss DS, Thornton JE. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 29.Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M. The CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2002;58:1955–1957. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.