Abstract
Recent studies of ancient genomes have suggested that gene flow from archaic hominin groups to the ancestors of modern humans occurred on two separate occasions during the modern human expansion out of Africa. At the same time, decreasing levels of human genetic diversity have been found at increasing distance from Africa as a consequence of human expansion out of Africa. We analyzed the signal of archaic ancestry in modern human populations, and we investigated how serial founder models of human expansion affect the signal of archaic ancestry using simulations. For descendants of an archaic admixture event, we show that genetic drift coupled with ascertainment bias for common alleles can cause artificial but largely predictable differences in similarity to archaic genomes. In genotype data from non-Africans, this effect results in a biased genetic similarity to Neandertals with increasing distance from Africa. However, in addition to the previously reported gene flow between Neandertals and non-Africans as well as gene flow between an archaic human population from Siberia (“Denisovans”) and Oceanians, we found a significant affinity between East Asians, particularly Southeast Asians, and the Denisova genome—a pattern that is not expected under a model of solely Neandertal admixture in the ancestry of East Asians. These results suggest admixture between Denisovans or a Denisova-related population and the ancestors of East Asians, and that the history of anatomically modern and archaic humans might be more complex than previously proposed.
Keywords: human origins, ancient DNA
Widespread evidence from genetics, linguistics, fossils, and archeology suggests that a wave of migration of anatomically modern humans out from Africa occurred within the last ∼100,000 years (1–8). Until recently, evidence from genetic data has been inconclusive (9) with regard to the possibility of gene flow from the archaic human populations that already resided in Eurasia at the time of the out of Africa migration into the expanding population of anatomically modern humans, with some studies favoring some degree of admixture or shared ancestry (10–14) and others concluding either that admixture is unsupported or unnecessary to explain the data at hand (15–22).
Recent analyses of large-scale ancient genomic sequence data provided the most rigorously tested genetic evidence for admixture so far, suggesting that a fraction of the ancestry of all modern humans of recent non-African ancestry traces back to Neandertals (23) and that a related archaic group, Denisovans, contributed an additional fraction of the ancestry of humans living in Oceania today (24). However, neither fine-scale geographic patterns of archaic ancestry nor the impact of many demographic events—such as founder events causing genetic drift—on archaic ancestry signals have been extensively studied. In addition, a model positing Neandertal-related gene flow into the ancestors of non-Africans—potentially occurring in the Middle East (23)—is supported by the possible Late Pleistocene overlap of Neandertals and early modern humans in the Eastern Mediterranean (4, 6, 25). Similarly, the suggestion of archaic Asian ancestry in Oceania is partly supported by some morphological interpretations of the fossil record (4, 26). However, similar, and arguably more suggestive (4, 6), morphological evidence for admixture with archaic populations has been found in early modern human remains from East Asia (4, 6, 27–30) and Europe (4, 31–33). Thus, it is possible that additional genomic signs of archaic admixture remain undetected because of inadequate sampling of ancient and/or contemporary human genetic variation (23–25).
Although genome sequence data only exist for a small number of individuals representing a handful of populations (8), genome-wide SNP genotype data (34, 35) have been collected for a large number of populations from around the world, including urban, rural, and indigenous groups (36, 37). However, inference using genotype data is complicated by ascertainment bias; the bias that arises from discovering SNPs in sequence data from a limited number of individuals resulting in enrichment of common alleles, particularly in the populations from which the discovery panel was constructed (38, 39). To determine fine-scale patterns of archaic admixture in the large collection of populations that have been SNP genotyped, we need to understand the impact of ascertainment bias on signals of archaic admixture.
In this study, we analyzed patterns of genetic variation in modern humans in the light of the two archaic genomes using genotype data from a diverse set of extant populations, and we found a signal of Denisova admixture in contemporary East Asian populations. We also studied the effect of ascertainment bias under serial founder models of human expansion to show that the signal of Denisova admixture in contemporary East Asians is opposite to the expectation under a model of solely Neandertal admixture with the ancestral population of non-Africans followed by greater genetic drift in East Asia than in Europe (40).
Results
To investigate the distribution of the signal of archaic human ancestry in a diverse and worldwide set of populations, we extracted the Neandertal variant and the Denisova variant from the Neandertal (23) and Denisova (24) genomes at 40,656 loci overlapping with genotypes from 1,568 globally distributed extant humans (34, 35, 41, 42) and the chimpanzee genome (43) using largely the same filters for base quality, mapping quality, and postmortem degradation as previous studies (24). From each extant individual, we used one randomly sampled allele at each SNP to mimic the data from the archaic individuals, and we included one SNP from each pair of SNPs in high linkage disequilibrium (r2 > 0.2), resulting in 38,848 SNPs.
Principal Component Analysis of Archaic Ancestry.
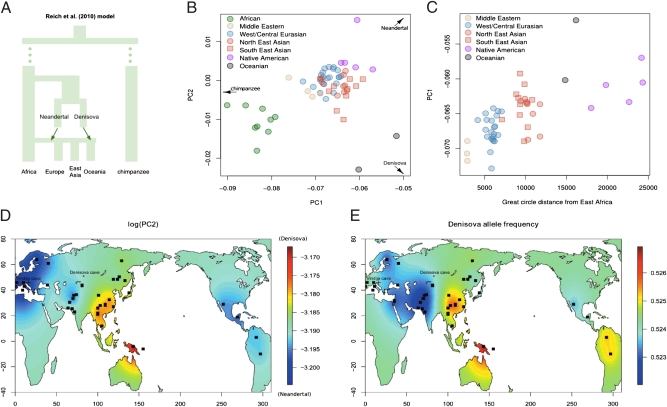
We performed principal component analysis (PCA) (44) by defining the first two principal components (PCs) using the Denisova, the Neandertal, and the chimpanzee and projected extant humans on the resulting axes of variation (24). This setup resulted in PC1 describing general genetic similarity to archaic humans (represented by both the Neandertal and Denisova genomes) and PC2 contrasting genetic similarity between Neandertal and Denisova. Under the assumption of a common shared history between Neandertal and Denisova (24) as well as no admixture between archaic populations and the ancestors of extant human populations since the diversification of modern humans (Fig. 1A), extant individuals are expected to be homogeneously distributed between archaic human and chimpanzee variations (24) (SI Materials and Methods).
Fig. 1.
Modern human genetic variation projected on axes of variation defined by chimpanzee, Denisova, and Neandertal. (A) A model of hominin evolutionary history suggested in the work in ref. 24 with putative admixture events indicated by arrows. (B) Population means for each of 62 populations. (C) PC1 in individuals from Eurasia and America as a function of distance from East Africa (kilometers). (D) Interpolated spatial distribution of PC2 [transformed for visualization as (−1) × log10 (x + C), where x is the PC loading and C = 0.04231] in individuals from Eurasia, Oceania, and America. (E) Interpolated spatial distribution of the frequency of Denisova alleles at SNPs where Denisova is different from chimpanzee and Neandertal. Sample localities are indicated with rectangles.
We recovered the previously reported (24) pattern—that extant human variation is largely organized in three clusters corresponding to Africans, Oceanians, and other non-Africans, respectively (Fig. 1B and Fig. S1). In the worldwide collection of extant populations, we used Procrustes superimposition (45) to compare PC1 and PC2 with geographic coordinates (longitude and latitude) for each population and found that sampling location was mirrored, to some extent, by archaic ancestry (individuals: Procrustes correlation = 0.127, P << 10−6; population means: Procrustes correlation = 0.309, P < 0.01). This pattern was expected given a model with two episodes of archaic gene flow involving non-Africans and Oceanians (Fig. 2B) (23, 24). However, PC1 and PC2 were also correlated with geography in a region comprising Eurasia and the Americas (individuals: Procrustes correlation = 0.104, P < 10−4; population means: Procrustes correlation = 0.335, P = 0.017), which seemed incompatible with previously suggested admixture scenarios postulating that archaic ancestry is homogeneous in people of European, Asian, and Native American descent (23, 24).
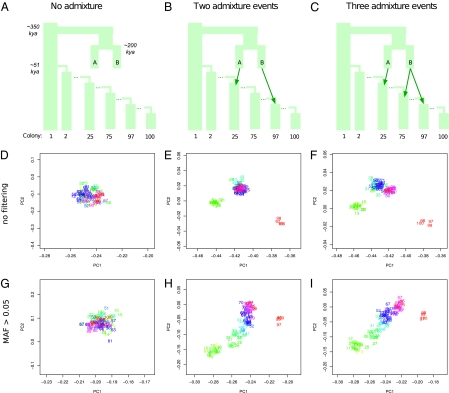
Fig. 2.
PCA of archaic ancestry under a serial founder model with (or without) archaic admixture. PCA was performed by projecting hypothetical extant humans onto the variation of a hypothetical chimpanzee and two hypothetical archaic hominins that shared a recent history with modern humans ∼400 kya. (A) A model with no admixture between colonizing and archaic populations. (B) A model with two separate admixture events: from archaic population A into colony 25 (forward in time, 2.5% of colony 25 is replaced) and from archaic population B into colony 97 (forward in time, 5% of colony 97 is replaced). This model is similar to the model suggested in ref. 24 (Fig. 1A). (C) A model with the two admixture events in B and an additional admixture event from archaic population B into colony 75 (forward in time, 1% of colony 75 is replaced). D–F show PCA results from models A, B, and C, respectively. G–I show PCA results for models A, B and C, respectively, but with SNPs with minor allele frequency (MAF) < 5% excluded. In D–I, we display the mean PC loading of 10 samples from each colony numbered and colored according to distance from the founding population. The model in C produced the qualitatively most similar result compared with the empirical data in Fig. 1B.
By comparing the geographic locations of individuals from different regions with the archaic ancestry signal (PC1 values), East Asian (two-sided t test: P < 10−6) and Native American (P < 10−8) populations were found to be more similar to archaic hominins compared with European and Central/South Asian populations (Fig. 1B and Table S1). In addition, the archaic ancestry signal was positively correlated with distance from East Africa (individuals: Spearman's rank correlation coefficient rs = 0.189, P < 10−9; population means: rs = 0.600, P < 10−4) (Fig. 1C). Because it is well-known that PCs of modern human genetic variation capture geography to some extent (35, 46), we also computed PCs using modern human genetic variation in Eurasia (SI Results) and found that the top two PCs of differentiation between Europe and East Asia were correlated with the archaic ancestry signal (PC1: rs = 0.138, P < 10−5; PC2: rs = 0.090, P = 0.002). This result suggests that, if separated along the major axes of differentiation between Europe and East Asia, individuals on the eastern end of the spectrum tend to be more similar to archaic human genomes. In contrast, after correction for multiple tests, we found no correlations between the archaic ancestry signal and intraregional PCs within Europe, and we did not find patterns of variation in archaic ancestry within Africa and America that could not be explained by more recent admixture with non-African populations (SI Results and Table S2).
To disentangle the signal of Denisova admixture from the signal of Neanderthal admixture, we investigated the distribution of PC2, which separated Denisovans and Neandertals, and found that proximity to Neandertal increases with distance from Africa (individuals: rs = 0.078, P = 0.010). However, East Asians were an exception to this trend in that they were significantly closer to Denisovans compared with Europeans (P = 0.003), all West/Central Eurasians (P = 0.006), and Americans (P < 10−4). In a spatial interpolation of log-transformed PC2 values, the affinity to the Denisova genome seemed to be strongest in Southern China and Southeast Asia (Fig. 1D), which is noteworthy considering the supposed East Eurasian distribution of Denisovans (24). To investigate this geographical pattern using an alternative approach, we identified SNPs in chimpanzees and Neandertals that shared the ancestral allele, and Denisova had the derived allele; we computed the frequency of the derived Denisova allele in global modern human populations. Using the same method of spatial interpolation, we found that East Asian populations, particularly Southeast Asian populations, had, on average, a greater frequency of the derived Denisova allele compared with other populations (except for Oceanians) (Fig. 1E). For example, although the greatest Denisova allele frequency was found in Papuans (53.5%), Yizu from Southern China had a greater frequency of the Denisova allele than Melanesians from Bougainville (53.0% vs. 52.9%) (Table S3).
Simulating the Effect of Ascertainment Bias and Genetic Drift.
Although the above described spatial patterns of variation in archaic ancestry among extant humans suggest that the distribution of archaic ancestry is more complex than previously suggested, it is also well-known that population differentiation, genetic diversity, and frequencies of derived alleles correlate with distance from Africa (1, 3, 34, 35), which is in line with anatomically modern human expansion out from Africa (3, 21). To investigate the impact of demography on signals of archaic admixture, we simulated a model of human expansion out of Africa, which is similar to the approach used in ref. 21. Briefly, the model comprises serial founder events with successive bottlenecks initiated ∼51,000 years ago (kya) and leaves 100 descendant populations (colonies). We also model two archaic hominin populations that may (or may not) contribute genetic material to the extant populations (Materials and Methods and Fig. 2 A–C). Based on simulated data from the model, we projected individuals from the colony populations onto PCs defined by a hypothetical chimpanzee and one sample from each archaic population.
Assuming no admixture with archaic populations, we obtain no distinct clustering of different colony populations (Fig. 2 A, D, and G). In contrast, if we assume two separate admixture events involving each archaic population, akin to the conclusions of the works in refs. 23 and 24, we observe similar clustering patterns as in the empirical data but without regional intracluster patterns (Fig. 2 B and E). However, if we filter out rare alleles to mimic some aspects of ascertainment bias affecting SNP-chip data, there is a strong correlation between PC1/PC2 and colony number among the hypothetical Eurasian colonies—the colonies that have only been involved in one episode of admixture (Fig. 2H). Thus, this model predicts increasing affinity to the hypothetical Neandertal with increasing distance from the founder population, which is caused by the combination of ascertainment bias and genetic drift. This observation is in line with the correlations of the signal of archaic ancestry and distance from Africa that we reported above but in stark contrast to the observation that East Asians are significantly closer to Denisova relative to Neandertal. Interestingly, when we add a third archaic admixture event representing a Denisova-related contribution to the ancestral population of East Asians, we obtain a pattern that is qualitatively more similar to the empirical data (Fig. 2 C, F, and I).
Formal Test for Archaic Admixture.
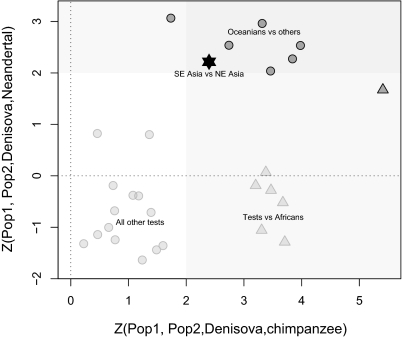
To test the hypothesis of Denisovan ancestry in East Asians using an alternative approach, we performed 4-population tests (23, 24, 40, 47, 48) on diploid genotype data from different regions. Note that using population allele frequencies is confounded by demographic effects (48) but may also provide more power and a more diverse set of samples compared with using low-coverage shotgun sequences from a smaller set of individuals (23, 24). To investigate fine-scale geographical patterns suggested by the PC analyses (Fig. 1 D and E), we divided the East Asian populations in our dataset into two groups, South and North of Beijing (with the North subgroup including Beijing and Japan). We then tested the hypothesis of no gene flow in the population topology (Denisova, chimpanzee), (Northeast Asians, Southeast Asians) and found that allele frequencies in the Southeastern Asian group were significantly more similar to the Denisova data (D = 0.55 ± 0.23% SE, Z = 2.40) (Table S4). In addition, we found that allele frequencies in the Southeastern Asian group were significantly more similar to the Denisova genome compared with the Neandertal genome (D = 0.66 ± 0.30%, Z = 2.22). This differentiation is not predicted by the effects of ascertainment bias, which was observed in our simulations, because ascertainment bias is expected to artificially increase similarity to Neandertal under the model suggested by previous studies (23, 24). Indeed, the only other pairwise regional comparisons for which we see a strong pattern of similarity to Denisova compared with both chimpanzee and Neandertal include Oceanians (Fig. 3 and Table S4).
Fig. 3.
Results of 4-population tests suggest Denisova-related ancestry in Southeast (SE) Asia. Z scores for the D statistic in all pairwise comparisons between Africa, Middle East, Central/South Asia, Southeast Asia, Northeast (NE) Asia, Oceania, and America are displayed. The configuration of each pairwise comparison that gives a positive value of D in the test (Pop1, Pop2, Denisova, chimpanzee) was chosen to ease visualization. Except for Oceanians and the comparison between SE Asia and NE Asia, populations that show high affinity to Denisova compared with chimpanzee tend to also show a higher affinity to Neandertal compared with Denisova (negative values on the y axis). Comparisons with Africans are shown by triangles. The area corresponding to significant deviations from 0 (|Z| > 2) is shaded, with the overlap representing significant deviations for both tests (Table S4).
The reason why an affinity to Denisova is not as clearly seen in 4-population tests between Southeast Asians and other populations (except Northeast Asians) (Fig. 3 and Table S4) could be the joint effect of ascertainment bias and genetic drift, which is indicated by our simulations. For example, a test of the topology [archaic population A, archaic population B, (colonies 50–70), (colonies 97–100)] fails to detect a fraction of 5% ancestry contributed by archaic population B to colonies 97–100 in simulation model ii (Fig. 2B), with filtering for minor allele frequency > 5%, and in fact, it suggests a skew toward colonies 97–100 being more similar to archaic population A (D = 1.0 ± 3.7%, Z = 0.27). In general, a signal of archaic ancestry is detected only if tests are performed between populations in which the magnitude of genetic drift has been similar. For example, if colonies 50–70 are substituted with colonies 90–96 in the test above, the test statistic significantly deviates from zero, and the admixture is detected (D = −7.0 ± 2.7%, Z = −2.6), although colonies 50–70 have exactly the same true fraction of archaic ancestry as colonies 90–96. However, the admixture is detected in both cases (Z < −8) if the data are unascertained (no minor allele frequency filtering).
Comparison with Complete Genomes.
To circumvent the effects of ascertainment bias, we identified polymorphisms between complete genomes sequenced to high coverage from seven individuals of Korean, Chinese, European, and African ancestry (49–52) and retained positions that overlapped with data from Neandertal and Denisova. We then tested the hypothesis of no gene flow in the population topology (Denisova, Neandertal), [East Asian, (European or African)] using haploid sets of SNPs from one individual from each population (23, 24, 48). Similar to previous analysis using this approach (23, 24), we did not observe any significant deviations from the null hypothesis in tests between Europeans and East Asians (Table S5). However, we note that this approach offers less power compared to using multiple individuals from each population (SEs were 3.4–4.4 times larger than in the population-based test between Southeast and Northeast Asia) (23, 24), and neither of the two complete East Asian genomes were from Southeast Asia, the region where we observe the strongest signal of Denisova ancestry.
Discussion
We have shown that complex signals of archaic ancestry arise in analyses of human genetic variation. Specifically, we find that the joint effect of ascertainment bias and genetic drift results in artificial differences between populations that have exactly the same admixture history. Although we have investigated these patterns in the context of a serial founder model, our conclusions generalize to related models (53–55) where populations have experienced different magnitudes of genetic drift since their diversification (40) but not necessarily because of founder events. One possible reason for this effect could be that a large part of the detectable signal for recent admixture is comprised by rare alleles. Because genetic drift increases the variance in allele frequencies among loci, the chance that a variant introduced by admixture increases in frequency above the discovery threshold might be greater in populations that have experienced stronger genetic drift. Although this explanation is compatible with the patterns in our simulations (Fig. 2), more complex (population biased) ascertainment schemes might have additional effects, but these are not expected to increase the rate of false positive tests for admixture (48).
Although the observed pattern of increased similarity to Neandertals with increasing distance from Africa is confounded by SNP ascertainment bias, the greater affinity to Denisova in East Asian (and Oceanian) populations, particularly Southeast Asian populations, is contrary to what is expected under a model of solely Neandertal-related gene flow into the ancestral population of non-Africans. It remains possible that this observation is influenced by population-biased SNP discovery (39, 48) and/or differences between sequencing and mapping methods used for the two archaic genomes (24), but we note that many of the tests for admixture and estimated fractions of archaic ancestry computed from sequencing data in refs. 23 and 24 were skewed (nonsignificantly) to East Asians compared with other Eurasian individuals. Moreover, although it is possible that contamination of the Denisovan genome sequence by DNA from modern day Southeast Asians could give rise to a similar affinity between Southeast Asians and Denisova as reported here, the very low inferred contamination rates in the Denisova data suggest that this explanation is unlikely (24).
An alternative explanation for the observed pattern would be that the signal is caused by gene flow from the ancestors of Oceanians (and/or East Asians) into the ancestors of the Denisovan individual. Such gene flow would be expected to cause a general affinity between Denisova and anatomically modern human populations correlated with relatedness to the source population of admixture. For example, if ancestors of the extant Oceanian population contributed genetic material to the ancestry of the Denisovan individual, it could be expected that Southeast Asians would be genetically more similar to Denisova than other human populations followed by Northeast Asians and Americans. However, differences in the fraction of derived alleles shared with Africans indicate that at least some of the gene flow was into the ancestors of Oceanians (24). Moreover, the age (>50,000 B.P.) and location (Altai, Siberia) of the Denisovan individual (24) suggest that gene flow from the ancestors of extant Oceanians into the descent of the Denisovan individual is a less parsimonious hypothesis than a small fraction of Denisovan-related ancestry in extant East Asian populations, especially because Denisova ancestry has been found in Oceania (24).
Although this Denisova ancestry in Southeast Asians could possibly have been introduced by gene flow from indigenous Oceanian populations after the introduction of Denisova-related genetic variants in Oceanians, evidence for such large-scale migration has not been found (56, 57). Moreover, this gene flow would have had to be substantial enough not to dilute the Denisova ancestry present in Oceanians beyond detection. Assuming ∼5% Denisova-related ancestry in Oceanians (24), any fraction of Denisova-related ancestry in Southeast Asia would require a ∼20 times as large contribution from Oceanian populations to be explained solely by modern human gene flow.
Quantitative estimation of the precise fraction of Denisova-related ancestry in Southeast Asian populations based on genotype data are unfortunately sensitive to ascertainment bias and genetic drift, and such estimates will require genome sequence data that are currently unavailable. However, both the PCA results (Fig. 1B) and the approximately six times lower absolute values of the D statistic in tests between Northeast Asians and Southeast Asians compared with tests between Northeast Asians and Oceanians (Table S4) indicate a relatively low fraction of Denisova-related ancestry. Thus, the fraction is likely to be smaller than both the ∼5% fraction of Denisova-related ancestry present in Oceanians and the ∼2.5% fraction of Neandertal ancestry present in non-Africans (23, 24), perhaps around 1%.
The lack of evidence of Denisova ancestry in other Eurasian populations indicates that this genetic material was introduced into the ancestral population of Southeast Asians after the time of divergence from Europeans, a date that has been estimated to 23–45 kya (58, 59) but could also have occurred considerably earlier (27, 28, 30). The apparent absence of Denisova ancestry in Native Americans in our study could be influenced by the biased affinity to the Neandertal genome that is expected because of ascertainment bias and genetic drift, but analyses of unascertained low-coverage shotgun sequence data from a single Native American individual resulted in a similar conclusion (24). If absent in America, the Denisova component must have appeared in the ancestors of East Asians after their divergence from Native Americans, which has been dated to ∼14–30 kya (58, 60, 61).
An alternative model for explaining the affinity between Neandertals and extant non-Africans postulates a structured ancestral African population rather than a temporally restricted admixture event (23). Ancient population structure in Africa (62, 63) could, in principle, also explain the Denisova ancestry in Oceanians and East Asians, but it is becoming increasingly difficult to imagine a structure model that can fully explain the complex pattern of archaic ancestry in non-Africans without invoking any restricted admixture events with archaic humans. Instead, we suggest that direct gene flow from archaic populations is the most likely explanation for the shared genetic ancestry between East Asian populations and the Denisova genome, which is in line with some previous findings based on fossils (4, 27–30) and genetic data from extant East Asians alone (10, 14). Whether this contact was separate from the contact with the ancestors of Oceanians or a population ancestral to both East Asians and Oceanians (and later diluted in East Asia by gene flow from other populations) is not clear. One possibility, suggested by the presence of highly divergent mitochondrial DNA lineages in two Denisova individuals (24, 64), is gene flow from a third as of yet unsampled archaic population into both Denisovans and the ancestors of East Asians. Regardless, the possibility of intracontinental variation in archaic ancestry highlights the importance of complete sequencing of genomes from diverse human populations for obtaining a detailed picture of human origins and demographic history.
Materials and Methods
Data Acquisition and Processing.
We obtained haploid autosomal variants from the Neandertal (23) and Denisova (24) genomes and removed bases with quality <40 and from within 5 and 1 bp from the 5′ end of the sequence reads from Neandertal and Denisova, respectively (24). We filtered reads with mapping quality <90 (Neandertal) and <37 (Denisova) (24) and randomly chose a single read from positions covered by multiple reads. We obtained phased HapMap 3 genotypes from 630 individuals (41, 42) and phased genotypes for 938 globally distributed individuals from the Human Genome Diversity Project (35). We intersected this dataset with chimpanzee (43), Neandertal, and Denisova genotypes, resulting in 228,984 overlapping SNPs. To avoid the effect of postmortem nucleotide misincorporations in analyses that included ancient genomes, we followed the information in refs. 23 and 24 by removing all transition SNPs (C/T and G/A), resulting in 40,656 SNPs.
From the seven complete genome sequences, we identified SNPs using genotypes called by the 1,000 Genomes Project (49). We excluded hypermutable CpG sites and transition substitutions and retained SNPs for which there was both Neandertal and Denisova genome data, using the same genotyping criteria for the ancient genomes as described above.
PC Analysis.
For PC analyses, we used a dataset created by randomly sampling one allele from each individual at each position to allow comparison with the single-pass ancient data. PCA was performed with EIGENSOFT 4.0 (44) on a dataset where one SNP from each pair of SNPs with r2 > 0.2 had been excluded. In the dataset that included only transversion SNPs, the number of SNPs used was 38,848, but the full dataset comprised 183,166 SNPs. We first performed PCA using chimpanzee, Denisova, and Neandertal, and thereafter, projected all other samples on the resulting components. In the transversion analysis, (PC1, PC2) loadings were chimpanzee (−0.81, 0.072), Denisova (0.34, −0.74), and Neandertal: (0.47, 0.67).
For Procrustes analyses, two-sided t tests, and Pearson correlation analyses on the PCA results, we excluded Middle Easterners and Balochi because of possible gene flow from Africa. Procrustes analysis was performed using the vegan R package (65), and significance was assessed with 1,000,000 permutations. All tests were performed on raw PC scores unless explicitly stated otherwise.
Simulations.
We used the computer program ms (66) to simulate a serial founder model as in ref. 21, with the main modification being inclusion of two archaic human populations A and B (hypothetical Neandertal and Denisova, respectively). We assumed that the two archaic populations diverged from each other 200 kya and had a divergence time of the hypothetical Neandertal/Denisova ancestral population and modern humans of 350 kya (18, 23, 24). Outgroup (hypothetical chimpanzee) population divergence was assumed to be 4 million years (y). We assumed a generation time of 25 y. We sampled 10 haploid samples from each of 100 extant populations. The age was set to ∼50 kya for both the archaic samples by moving their lineages to an isolated population at that specific time point. This movement prevents the lineages from coalescing with other lineages in the simulation, and any private mutations do not contribute to any tests for admixture under an infinite sites mutation model. We simulated three scenarios: (i) no admixture with archaic populations, (ii) admixture fraction from archaic population A of 2.5% in colony 25 and admixture fraction of 5% from archaic population B in colony 97, and (iii) admixture fractions as in scenario ii with the addition of a 1% admixture fraction from archaic population B in colony 75. We based our analyses on two different datasets, each consisting of 38,848 independent SNPs. In the first dataset, only singleton SNPs were excluded (because they are not informative in our analyses). In the second dataset, SNPs with minor allele frequency > 5% were excluded (to investigate the effect of ascertainment bias).
4-Population Tests.
4-population tests on allele frequencies (23, 24, 40, 47, 48) were implemented as in ref. 24 and performed using diploid genotype data from Africa, Europe, Oceania, America, the Middle East, Southeast Asia, and Northeast Asia. We computed SEs using a block jackknife, dropping 1 of 114 contiguous blocks with 600 SNPs in each block. Tests on complete genome sequence data (23, 24, 48) used a randomly sampled haploid copy from each individual, and SEs were computed using a block jackknife over contiguous blocks with 200 informative SNPs (ABBA or ABAB) in each block. Tests on the simulated data were performed by computing a block jackknife SE over 129 contiguous windows of 600 SNPs each. We used computed SEs to obtain Z scores for the test statistic D and interpreted |Z| > 2.0 as a statistically significant deviation from zero.
Supplementary Material
Acknowledgments
We thank Noah Rosenberg, Michael Blum, Anders Götherström, Carina Schlebusch, Sohini Ramachandran, and two anonymous reviewers for valuable comments. Financial support was provided by the Swedish Research Council and the Lawski Foundation. Computations were performed on Swedish National Infrastructure for Computing (SNIC) and Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) resources (Project b2010050).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108181108/-/DCSupplemental.
References
- 1.Cann RL, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 2.Stringer C. Modern human origins: Progress and prospects. Philos Trans R Soc Lond B Biol Sci. 2002;357:563–579. doi: 10.1098/rstb.2001.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trinkaus E. Early modern humans. Annu Rev Anthropol. 2005;34:207–230. [Google Scholar]
- 5.Mellars P. Going east: New genetic and archaeological perspectives on the modern human colonization of Eurasia. Science. 2006;313:796–800. doi: 10.1126/science.1128402. [DOI] [PubMed] [Google Scholar]
- 6.Klein RG. Out of Africa and the evolution of human behavior. Evol Anthropol. 2008;17:267–281. [Google Scholar]
- 7.Atkinson QD. Phonemic diversity supports a serial founder effect model of language expansion from Africa. Science. 2011;332:346–349. doi: 10.1126/science.1199295. [DOI] [PubMed] [Google Scholar]
- 8.Stoneking M, Krause J. Learning about human population history from ancient and modern genomes. Nat Rev Genet. 2011;12:603–614. doi: 10.1038/nrg3029. [DOI] [PubMed] [Google Scholar]
- 9.Nordborg M. On the probability of Neanderthal ancestry. Am J Hum Genet. 1998;63:1237–1240. doi: 10.1086/302052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrigan D, Mobasher Z, Severson T, Wilder JA, Hammer MF. Evidence for archaic Asian ancestry on the human X chromosome. Mol Biol Evol. 2005;22:189–192. doi: 10.1093/molbev/msi013. [DOI] [PubMed] [Google Scholar]
- 11.Garrigan D, Hammer MF. Reconstructing human origins in the genomic era. Nat Rev Genet. 2006;7:669–680. doi: 10.1038/nrg1941. [DOI] [PubMed] [Google Scholar]
- 12.Green RE, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 13.Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wall JD, Lohmueller KE, Plagnol V. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Mol Biol Evol. 2009;26:1823–1827. doi: 10.1093/molbev/msp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krings M, et al. Neandertal DNA sequences and the origin of modern humans. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 16.Currat M, Excoffier L. Modern humans did not admix with Neanderthals during their range expansion into Europe. PLoS Biol. 2004;2:e421. doi: 10.1371/journal.pbio.0020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serre D, et al. No evidence of neandertal mtDNA contribution to early modern humans. Plos Biol. 2004;2:E57. doi: 10.1371/journal.pbio.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noonan JP, et al. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fagundes NJR, et al. Statistical evaluation of alternative models of human evolution. Proc Natl Acad Sci USA. 2007;104:17614–17619. doi: 10.1073/pnas.0708280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wall JD, Kim SK. Inconsistencies in Neanderthal genomic DNA sequences. PLoS Genet. 2007;3:1862–1866. doi: 10.1371/journal.pgen.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeGiorgio M, Jakobsson M, Rosenberg NA. Out of Africa: Modern human origins special feature: Explaining worldwide patterns of human genetic variation using a coalescent-based serial founder model of migration outward from Africa. Proc Natl Acad Sci USA. 2009;106:16057–16062. doi: 10.1073/pnas.0903341106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blum MGB, Jakobsson M. Deep divergences of human gene trees and models of human origins. Mol Biol Evol. 2011;28:889–898. doi: 10.1093/molbev/msq265. [DOI] [PubMed] [Google Scholar]
- 23.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodgson JA, Bergey CM, Disotell TR. Neandertal genome: The ins and outs of African genetic diversity. Curr Biol. 2010;20:R517–R519. doi: 10.1016/j.cub.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Wolpoff MH, Hawks J, Frayer DW, Hunley K. Modern human ancestry at the peripheries: A test of the replacement theory. Science. 2001;291:293–297. doi: 10.1126/science.291.5502.293. [DOI] [PubMed] [Google Scholar]
- 27.Etler DA. The fossil evidence for human evolution in Asia. Annu Rev Anthropol. 1996;25:275–301. [Google Scholar]
- 28.Wu X. On the origin of modern humans in China. Quatern Int. 2004;117:131–140. [Google Scholar]
- 29.Shang H, Tong H, Zhang S, Chen F, Trinkaus E. An early modern human from Tianyuan Cave, Zhoukoudian, China. Proc Natl Acad Sci USA. 2007;104:6573–6578. doi: 10.1073/pnas.0702169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, et al. Human remains from Zhirendong, South China, and modern human emergence in East Asia. Proc Natl Acad Sci USA. 2010;107:19201–19206. doi: 10.1073/pnas.1014386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duarte C, et al. The early Upper Paleolithic human skeleton from the Abrigo do Lagar Velho (Portugal) and modern human emergence in Iberia. Proc Natl Acad Sci USA. 1999;96:7604–7609. doi: 10.1073/pnas.96.13.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trinkaus E, et al. An early modern human from the Peştera cu Oase, Romania. Proc Natl Acad Sci USA. 2003;100:11231–11236. doi: 10.1073/pnas.2035108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trinkaus E. European early modern humans and the fate of the Neandertals. Proc Natl Acad Sci USA. 2007;104:7367–7372. doi: 10.1073/pnas.0702214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 35.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 36.Cann HM, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 37.Novembre J, Ramachandran S. Perspectives on human population structure at the cusp of the sequencing era. Annu Rev Genomics Hum Genet. 2011;12:245–274. doi: 10.1146/annurev-genom-090810-183123. [DOI] [PubMed] [Google Scholar]
- 38.Kuhner MK, Beerli P, Yamato J, Felsenstein J. Usefulness of single nucleotide polymorphism data for estimating population parameters. Genetics. 2000;156:439–447. doi: 10.1093/genetics/156.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albrechtsen A, Nielsen FC, Nielsen R. Ascertainment biases in SNP chips affect measures of population divergence. Mol Biol Evol. 2010;27:2534–2547. doi: 10.1093/molbev/msq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altshuler DM, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Surakka I, et al. Founder population-specific HapMap panel increases power in GWA studies through improved imputation accuracy and CNV tagging. Genome Res. 2010;20:1344–1351. doi: 10.1101/gr.106534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 44.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang C, et al. Comparing spatial maps of human population-genetic variation using Procrustes analysis. Stat Appl Genet Mol Biol. 2010;9:13. doi: 10.2202/1544-6115.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahn S-M, et al. The first Korean genome sequence and analysis: Full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenberg NA, et al. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hunley KL, Healy ME, Long JC. The global pattern of gene identity variation reveals a history of long-range migrations, bottlenecks, and local mate exchange: Implications for biological race. Am J Phys Anthropol. 2009;139:35–46. doi: 10.1002/ajpa.20932. [DOI] [PubMed] [Google Scholar]
- 55.Handley LJ, Manica A, Goudet J, Balloux F. Going the distance: Human population genetics in a clinal world. Trends Genet. 2007;23:432–439. doi: 10.1016/j.tig.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 56.Friedlaender JS, et al. The genetic structure of Pacific Islanders. PLoS Genet. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wollstein A, et al. Demographic history of Oceania inferred from genome-wide data. Curr Biol. 2010;20:1983–1992. doi: 10.1016/j.cub.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 58.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gravel S, et al. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci USA. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dillehay TD. Monte Verde: A Late Pleistocene Settlement in Chile. Washington, DC: Smithsonian Institution Press; 1997. [DOI] [PubMed] [Google Scholar]
- 61.Gilbert MTP, et al. DNA from pre-Clovis human coprolites in Oregon, North America. Science. 2008;320:786–789. doi: 10.1126/science.1154116. [DOI] [PubMed] [Google Scholar]
- 62.Harding RM, McVean G. A structured ancestral population for the evolution of modern humans. Curr Opin Genet Dev. 2004;14:667–674. doi: 10.1016/j.gde.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 63.Gunz P, et al. Early modern human diversity suggests subdivided population structure and a complex out-of-Africa scenario. Proc Natl Acad Sci USA. 2009;106:6094–6098. doi: 10.1073/pnas.0808160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krause J, et al. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 2010;464:894–897. doi: 10.1038/nature08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–930. [Google Scholar]
- 66.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.