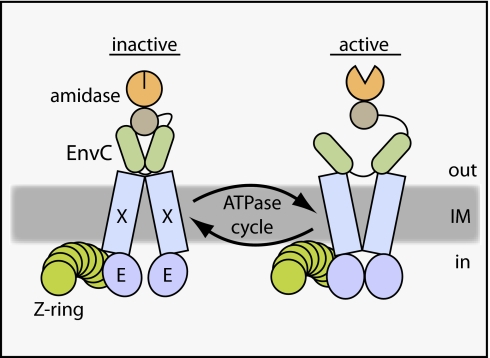
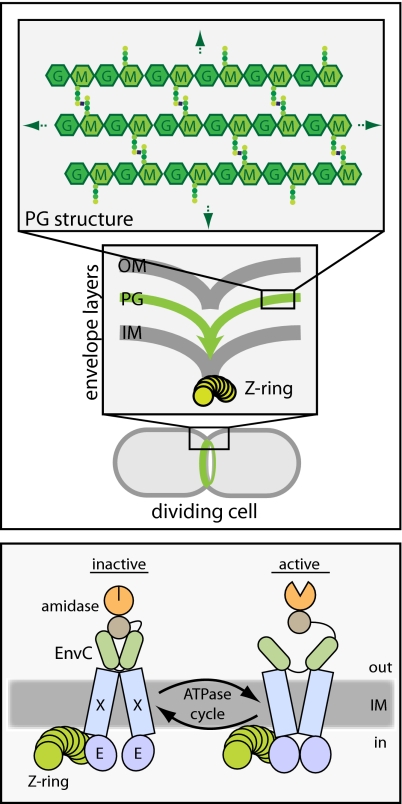
Abstract
ATP-binding cassette transporters are ubiquitous membrane protein complexes that move substrates across membranes. They do so using ATP-induced conformational changes in their nucleotide-binding domains to alter the conformation of the transport cavity formed by their transmembrane domains. In Escherichia coli, an ATP-binding cassette transporter-like complex composed of FtsE (nucleotide-binding domain) and FtsX (transmembrane domain) has long been known to be important for cytokinesis, but its role in the process has remained mysterious. Here we identify FtsEX as a regulator of cell-wall hydrolysis at the division site. Cell-wall material synthesized by the division machinery is shared initially by daughter cells and must be split by hydrolytic enzymes called “amidases” to drive daughter-cell separation. We recently showed that the amidases require activation at the cytokinetic ring by proteins with LytM domains, of which EnvC is the most critical. In this report, we demonstrate that FtsEX directly recruits EnvC to the septum via an interaction between EnvC and a periplasmic loop of FtsX. Importantly, we also show that FtsEX variants predicted to be ATPase defective still recruit EnvC to the septum but fail to promote cell separation. Our results thus suggest that amidase activation via EnvC in the periplasm is regulated by conformational changes in the FtsEX complex mediated by ATP hydrolysis in the cytoplasm. Since FtsE has been reported to interact with the tubulin-like FtsZ protein, our model provides a potential mechanism for coupling amidase activity with the contraction of the FtsZ cytoskeletal ring.
Keywords: morphogenesis, peptidoglycan, cell division, cell envelope
Cytokinesis in Escherichia coli and other bacteria is mediated by a ring-shaped multiprotein machine called the “septal ring” or “divisome” (1). Assembly of this machine begins with the polymerization of the tubulin-like FtsZ protein into a ring-like structure just underneath the cell membrane at the prospective site of division (2). Several FtsZ-binding proteins have been identified in E. coli (FtsA, ZipA, ZapA, and ZapC). Along with the ZapA-binding protein ZapB, they appear to play partially redundant roles in the formation and stabilization of the Z-ring structure (3–10). Once formed, the Z-ring promotes septal-ring assembly by facilitating the recruitment of the remaining essential and nonessential division proteins to the division site according to a mostly linear dependency pathway (1).
Because the functions of many of its components are ill-defined, the mechanism(s) by which the septal ring promotes cell constriction remain largely mysterious (1). One of the most enigmatic division factors has been the ATP-binding cassette (ABC)-transporter–like complex formed by FtsE and FtsX (FtsEX) (11). ABC transporters are integral membrane protein complexes that use the energy of ATP hydrolysis to transport substrates across membranes (12). They typically are composed of two polytopic transmembrane domains (TMDs) and a pair of cytosolic nucleotide-binding domains (NBDs). Structural studies of complete ABC-transporters indicate that these complexes undergo remarkable conformational changes in response to nucleotide binding and hydrolysis (12). The NBD subunits interconvert between an open and a closed conformation at the membrane surface during an ATP hydrolysis cycle (12). These conformational changes, in turn, are transmitted to the TMD subunits to promote transitions between outward-facing and inward-facing conformations of the central cavity formed by the TMDs (12). Transport thus is promoted by alternating access of the substrate-binding site to opposing sides of the membrane.
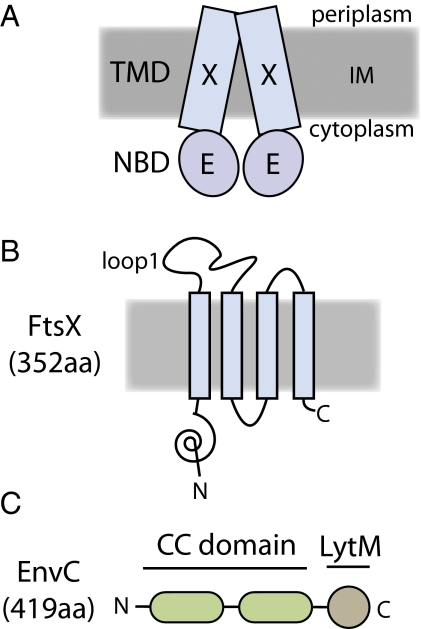
The role of the FtsEX complex in cell division has remained unclear for some time (13, 14). FtsE is the NBD component of the complex, and FtsX is the TMD component (15) (Fig. 1 A and B). Both factors localize to the septal ring and are conditionally essential for cell division (11). In medium of low osmotic strength, such as LB without added NaCl, cells lacking FtsEX display a lethal division defect (11, 15). They form smooth filaments that assemble Z-rings, but these structures are compromised because they fail to recruit FtsK and other downstream division factors (11). This phenotype along with the observation that FtsE and FtsX interact with several different division proteins (16, 17) has led to the idea that one important function of FtsEX is to stabilize the septal-ring structure (18–20). Consistent with this idea, the division defect of FtsEX− cells can be suppressed partially by the overproduction of other division proteins (FtsZ, FtsN, or FtsP) (18). Increasing the osmolarity of the medium and lowering the growth temperature also restores division function to FtsEX− mutants (11, 15, 18, 19). The mechanisms by which high osmolarity or division protein overproduction bypass the FtsEX requirement for cell division are not known. However, these growth conditions presumably allow essential division proteins downstream of FtsEX in the recruitment hierarchy to assemble at the division site in the absence of the complex. Indeed, FtsN has been shown to localize to sites of constriction in FtsEX− cells grown in LB with 1% NaCl (11). Over the years, many functions for FtsEX have been proposed, most of them assuming that it must transport a substrate (20 and references therein). In this report, we present evidence that FtsEX may not be a transporter at all but instead is an important regulator of cell-wall turnover at the division site.
Fig. 1.
Domain structure of FtsEX and EnvC. (A) Diagram of the FtsEX ABC system. FtsX (X) is the TMD component, and FtsE (E) is the ATPase component (15). The complex is in the inner membrane (IM) with FtsE located on the cytoplasmic face of the membrane. (B) Membrane topology of FtsX as determined by Weiss and coworkers (20). The loop1 domain is composed of residues 93–223. Membrane orientation is the same as in A. (C) Domain structure of EnvC. EnvC is a periplasmic protein. It possesses an N-terminal signal sequence (residues 1–34), two relatively long segments predicted to form coiled coils (CC domain, residues 35–277), and a C-terminal LytM domain (residues 278–419) needed for amidase activation.
Most bacteria surround themselves with a polysaccharide cell-wall matrix called “peptidoglycan” (PG) (21). This meshwork is essential for cellular integrity and is composed of glycan strands connected to one another by crosslinks between attached peptide moieties (Author Summary, Fig. P1) (21). During cytokinesis in Gram-negative bacteria, septal PG is synthesized by the divisome (1). This material is thought to be shared initially by the developing daughter cells and must be split to facilitate outer membrane constriction and daughter-cell separation (1) (Author Summary, Fig. P1). Septal PG splitting is mediated by the periplasmic PG amidases, AmiA, AmiB, and AmiC (22). Amidases are PG hydrolases that break crosslinks in the PG meshwork by cleaving bonds that link stem peptides to the glycan strands. Mutants lacking amidase activity complete inner membrane constriction and fusion. However, they fail to split septal PG and form long chains of cells connected by shared layers of PG and a partially constricted outer membrane layer (22, 23).
The amidases must be controlled tightly to prevent them from creating lesions in the cell wall that can result in cell lysis. Part of this regulation appears to rely on the fact that the PG amidases alone are weakly active enzymes (24). To hydrolyze PG efficiently, they require activation by EnvC and NlpD (24), divisome-associated proteins with LytM domains (Pfam, Peptidase_M23) (25, 26). EnvC specifically activates AmiA and AmiB, whereas NlpD specifically activates AmiC (24). Accordingly, mutants lacking both EnvC and NlpD have a chaining phenotype that resembles a triple-amidase mutant (25). A major unresolved question has been how the LytM factors themselves are controlled so that amidase activity at the septum is coordinated properly with other activities of the septal ring, such as membrane invagination and septal PG synthesis.
Using a genetic screen designed to identify factors involved in cell-wall assembly (27), we found that mutations in ftsEX and envC are synthetically lethal with loss-of-function mutations in a common set of genes. This result suggested that FtsEX and EnvC might participate in the same biochemical pathway. Indeed, when grown under permissive conditions, FtsEX− cells phenocopy the cell-separation defect displayed by EnvC− cells. We further show that EnvC requires FtsEX for its recruitment to the division site and that EnvC interacts directly with the large periplasmic loop domain of FtsX. Importantly, we also show that FtsEX variants predicted to be ATPase defective still recruit EnvC to the septum but fail to promote cell separation. Our results thus suggest that FtsX regulates amidase activation by EnvC in the periplasm through conformational changes induced by FtsE-mediated ATP hydrolysis on the opposite side of the membrane. Because FtsE has been reported to interact with FtsZ (16), this finding provides a potential mechanism for directly coupling septal PG hydrolysis with the contraction of the Z-ring during cell constriction. Interestingly, the accompanying report by Sham and coworkers (28) similarly connects FtsEX with cell separation in the Gram-positive pathogen Streptococcus pneumoniae, indicating that the regulation of cell-wall turnover is likely to be a broadly conserved function for FtsEX.
Results
ftsEX and envC Mutants Share Common Genetic Interactions.
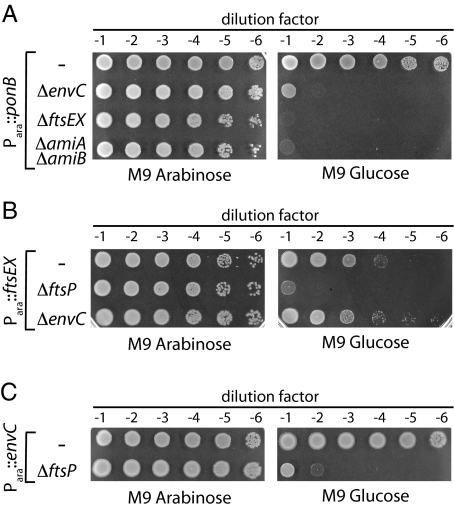
To identify factors important for cell-wall biogenesis, we recently performed a genetic screen for mutations synthetically lethal with the inactivation of the PG synthase PBP1b (slb mutants) (27). In addition to mutants in the lpoA gene coding for a lipoprotein cofactor required for the activity of PBP1a (27, 29), we isolated mutants with transposon insertions in envC and ftsEX. The ftsEX mutant was isolated in a screen performed at room temperature, a condition that we find suppresses the lethal effects of FtsEX inactivation. To confirm the synthetic lethality of the mutant combinations, ΔenvC and ΔftsEX alleles were transduced into strain MM11 (Para::ponB) harboring the low-copy plasmid pTB63 (ftsQAZ), which expresses the ftsQAZ operon from native promoters and suppresses the FtsEX− growth defect. In MM11, the native ponB promoter was replaced with Para such that ponB expression is arabinose-dependent (27). Depletion of PBP1b in the absence of FtsEX or EnvC was confirmed to be lethal (Fig. 2A). The terminal phenotype in both cases was cell lysis. Since EnvC is needed to activate the amidases AmiA and AmiB at the developing septum, we tested whether the combined inactivation of AmiA and AmiB also was lethal upon PBP1b depletion. This indeed proved to be the case (Fig. 2A). We do not currently know why the inactivation of FtsEX, EnvC, or AmiA/AmiB renders PBP1b essential for growth. Nevertheless, the observed phenotypes suggested that FtsEX may function in the process of septal PG splitting with EnvC and the amidases.
Fig. 2.
Shared synthetic lethal phenotypes of envC and ftsEX mutants. (A) Cells of MM11/pTB63 (Para::ponB/ftsQAZ) and its ΔenvC, ΔftsEX, or ΔamiA ΔamiB derivatives were grown overnight in M9 arabinose medium supplemented with 5 μg/mL Tet (Tet5) at 37 °C. Following normalization for cell density (OD600 = 2), the resulting cultures were serially diluted (10−1 to 10−6), and 5 μL of each dilution was spotted on the indicated medium. Plates were incubated overnight at 37 °C and photographed. (B) Cells of TU191(attλTU188) [ΔftsEX (Para::ftsEX)] and its ΔenvC or ΔftsP derivatives were processed as in A except growth was in the absence of Tet. (C) Cells of TB140(attλTD25) [ΔenvC (Para::envC)] and its ΔftsP derivative were grown and processed as in B.
The loss of FtsEX function was shown previously to be synthetically lethal with the deletion of ftsP (sufI) (18). We confirmed this result and found that the depletion of EnvC also was synthetically lethal with ΔftsP (Fig. 2 B and C), thus further connecting the functions of EnvC and FtsEX. As expected for proteins functioning in the same pathway, loss of EnvC function was not lethal upon FtsEX depletion (Fig. 2B).
FtsEX Is Required For Daughter-Cell Separation.
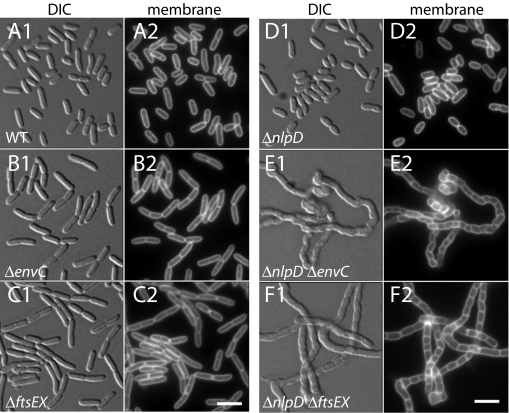
While performing the FtsEX depletion experiments in Fig. 2B, we found that growth on minimal agar at 37 °C partially suppressed the FtsEX− growth defect. We assume this suppression is caused by the higher osmolarity of the medium relative to standard LB (0.5% NaCl) and the slower overall growth rate of the cells. The suppression in liquid minimal medium was even more pronounced, especially when cells were grown at 30 °C. Here, FtsEX− cells displayed a much milder division defect resembling that typically observed for EnvC− cells (25) (see below). A similar division defect was observed for ΔenvC and ΔftsEX mutants harboring an ftsQAZ-overproducing plasmid (pTB63) when we grew them in LB (1% NaCl) (Fig. 3 A–C). It thus appeared that under conditions that suppressed the septal-ring stability defect of FtsEX− cells, a potential role for the FtsEX complex in EnvC-mediated cell separation was revealed. Interestingly, we note that, although not as severe as the FtsEX− division defect, EnvC− cells display a lethal filamentation phenotype when grown in LB without added NaCl at 42 °C (30–32). This finding suggests that at least part of the constriction defect of FtsEX− cells may be the result of EnvC inactivation.
Fig. 3.
FtsEX− and EnvC− cells have similar division defects. Cells of TB28 (WT) (A), KP4 (ΔenvC) (B), KP5 (ΔftsEX) (C), TB145 (ΔnlpD) (D), KP6 (ΔnlpD ΔenvC) (E), and KP7 (ΔnlpD ΔftsEX) (F) harboring pTB63 (ftsQAZ) were grown overnight in LB-Tet5 1.0% NaCl (A–C) or 1.5% NaCl (D–F) at 30 °C. Cultures were diluted 1:200 into the same medium and grown at 30 °C to an OD600 of 0.6–0.8. Cells then were incubated with the fixable membrane stain FM1-43FX (5 μg/mL) for 10 min and were fixed. Fixed cells were visualized on 2% agarose pads using differential interference contrast microscopy (DIC) (A1–F1) and GFP (A2–F2) optics. (Scale bars, 4 microns.)
We previously showed that cells lacking EnvC become dependent on NlpD for cell separation (25). Cells lacking EnvC and NlpD appear to be completely defective in cell separation and form very long chains that resemble triple-amidase mutants (25) (Fig. 3 D–F). We reasoned that if FtsEX truly is required for EnvC-mediated cell separation, the simultaneous inactivation of FtsEX and NlpD also should result in a severe chaining phenotype, as indeed proved to be the case (Fig. 3 D–F). We therefore conclude that FtsEX is required for EnvC to promote septal PG splitting.
EnvC Is Stable and Released into the Periplasm in the Absence of FtsEX.
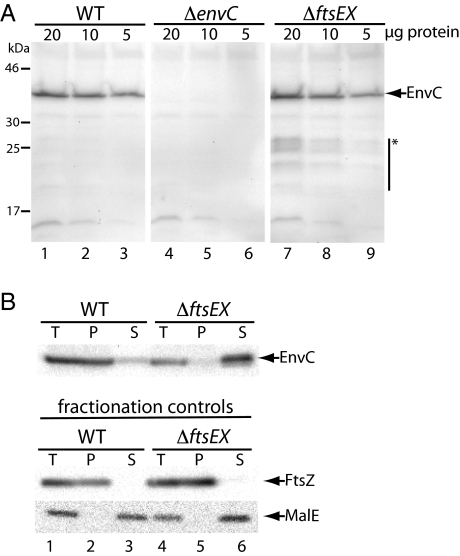
A potential reason for the FtsEX–EnvC connection is that EnvC requires FtsEX for its stable accumulation. To investigate this possibility, we compared EnvC levels in total cell extracts prepared from WT versus ΔftsEX cells (Fig. 4A). Immunoblotting with affinity-purified anti-EnvC polyclonal antibodies revealed that similar amounts of EnvC accumulate whether or not cells produce FtsEX (Fig. 4A, compare lanes 1–3 and 7–9). However, we did observe low concentrations of smaller immunoreactive species in the ΔftsEX extract that were not seen in the WT or ΔenvC extracts, indicating that a small portion of EnvC probably is processed proteolytically in the absence of FtsEX (Fig. 4A). This low level of processing is unlikely to explain the EnvC− phenotype of ΔftsEX cells.
Fig. 4.
Change in EnvC subcellular localization in the absence of FtsEX. (A) Cells of TB28 (WT), KP4 (ΔenvC), or KP5 (ΔftsEX) harboring pTB63 (ftsQAZ) were grown overnight in LB (1.0% NaCl)-Tet5 at 30 °C. Cultures were diluted 1:100 into LB-Tet5 and grown at 30 °C to an OD600 of 0.66–0.72. Cell extracts then were prepared, and the indicated total protein amounts were subjected to immunoblotting using affinity-purified EnvC antibodies. Arrow indicates position of FLEnvC bands, and the bar with the asterisk highlights what appears to be a small amount of EnvC degradation products in extracts from the ΔftsEX mutant. Positions of the molecular weight markers are indicated on the left. (B) Cultures of the strains in A were grown as above to an OD600 of 0.39–0.46. One aliquot of cells was used to prepare a total-cell extract. The remaining cells were converted to spheroplasts and pelleted by centrifugation. The resulting pellet (P) and supernatant (S) fractions along with the total-cell extract (T) were analyzed by SDS-PAGE and immunoblotting for EnvC, FtsZ, and MalE as indicated. FtsZ and MalE served as markers for the cytoplasm/spheroplast membranes and periplasm, respectively.
Because FtsEX is related to ABC transporters, one possible way in which it might promote EnvC function is by facilitating its export to the periplasm. To investigate this possibility, we fractionated cells to determine the subcellular localization of EnvC in the presence or absence of FtsEX. WT and ΔftsEX cells were suspended in buffer containing sucrose and converted to spheroplasts by the addition of EDTA and lysozyme. In WT cells, EnvC pelleted with the spheroplasts rather than remaining in the soluble periplasmic fraction like the MalE control (Fig. 4B, lanes 1–3). In contrast, EnvC was found almost exclusively in the periplasmic fraction from FtsEX− cells. The FtsZ and MalE fractionation controls indicated that this change in EnvC localization was not caused by altered fractionation properties of the FtsEX− cells (Fig. 4B, lanes 4–6). Thus, FtsEX is not required for the export of EnvC to the periplasm. This result is consistent with previous reports suggesting that EnvC is a Sec substrate (32) and that its signal sequence can be replaced functionally by alternative signal peptides for Tat- or Sec-mediated transport (24, 26). Furthermore, the fractionation results suggest that EnvC remains associated with the outer face of the inner membrane of WT cells, possibly through an interaction with FtsX, and that it is released into the periplasm in the absence of this interaction (see below).
FtsEX Is Required for the Recruitment of EnvC to the Septal Ring.
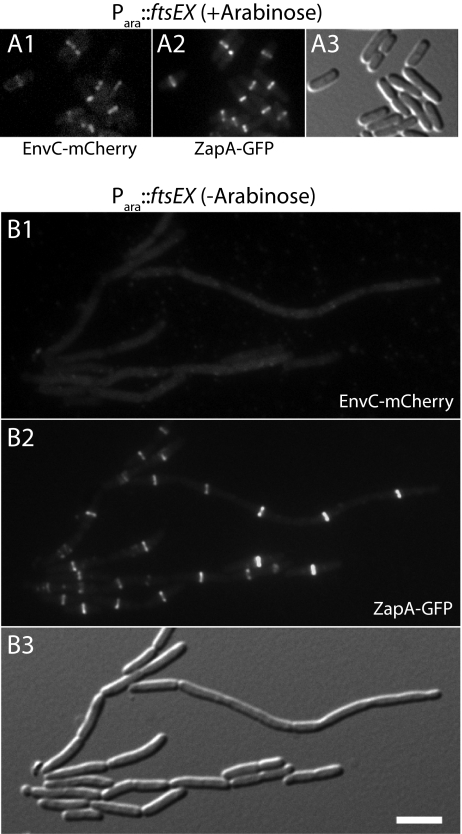
In addition to subcellular fractionations, we also investigated the role of FtsEX in the recruitment of EnvC to the septal ring. To do so, we constructed an FtsEX depletion strain NP69(attλTU188)(attHKTB316) [ΔftsEX ΔenvC zapA-gfp (Para::ftsEX)(Plac::envC-mCherry)]. In this strain, the native ftsEX locus was deleted, and a second copy of ftsEX under arabinose promoter (Para) control was integrated at the λ att site. The strain also encodes zapA-gfp at the native zapA locus and expresses envC-mCherry from an expression cassette integrated at the HK022 att site. When grown in M9 maltose medium containing a small amount of arabinose (0.01%), cells of NP69(attλTU188)(attHKTB316) divided normally and displayed bands of both EnvC-mCherry and ZapA-GFP at the division sites of most cells (Fig. 5A). Conversely, when the same cells were grown in M9 maltose medium containing a small amount of glucose (0.01%) to repress Para::ftsEX, they displayed the heterogeneous cell constriction and separation phenotype typical of FtsEX− and EnvC− cells (Fig. 5B). Although most cells appeared capable of cell division, many were elongated and appeared to have difficulty completing cell separation. The Z-ring marker, ZapA-GFP, formed rings/bands in these elongated cells at fairly regular intervals, but EnvC-mCherry failed to be recruited to these structures (Fig. 5B). Immunoblot analysis indicated that this localization defect was not caused by excessive degradation of the EnvC-mCherry fusion (SI Appendix, Fig. S1). We thus conclude that FtsEX is required for the recruitment of EnvC to the septal ring.
Fig. 5.
FtsEX is required for the recruitment of EnvC to the division site. Cells of NP69(attλTU188)(attHKTB316) [ΔftsEX ΔenvC zapA-gfp (Para::ftsEX)(Plac::envC-mCherry)] were grown overnight in M9 maltose supplemented with 0.01% arabinose. Cells were washed twice with and resuspended in an equal volume of M9 medium without added sugar. They then were diluted 1:100 into M9 maltose, and growth was continued at 30 °C with the addition of either 0.01% arabinose (A) or 0.01% glucose (B). When the cultures reached an OD600 of 0.4–0.6, cells were visualized on 2% agarose pads using mCherry (Panel 1), GFP (Panel 2), or DIC (Panel 3) optics. (Scale bar, 4 microns.) Note that a peripheral EnvC-mCherry signal was not observed in B as might be expected for a periplasmic protein, likely because too little fusion protein is present to raise the peripheral signal above background.
Large Periplasmic Loop of FtsX Interacts Directly with EnvC.
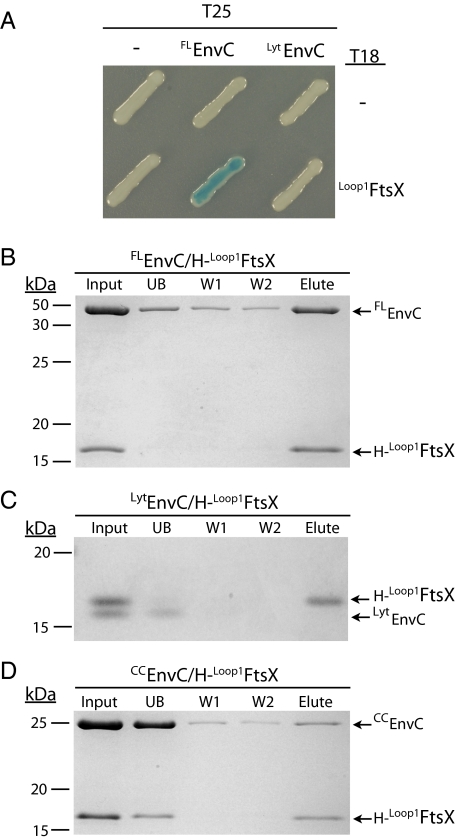
Our results thus far suggested that FtsEX may interact directly with EnvC to recruit it to the division site. The large periplasmic loop of FtsX (residues 93–223, Loop1FtsX) (Fig. 1B) seemed a likely candidate for mediating the interaction with EnvC. We therefore tested the potential interaction between full-length, mature EnvC (FLEnvC) and Loop1FtsX using a bacterial two-hybrid (BACTH) assay based on the reconstitution of adenylate cyclase activity from the fragments T18 and T25 (33). FLEnvC-T25 showed a strong interaction signal when paired with a Loop1FtsX-T18 fusion (Fig. 6A). EnvC contains three identifiable domains: a signal peptide (residues 1–34), a coiled-coil (CC) domain (residues 35–271), and a LytM domain (residues 318–413) (Fig. 1C) (26, 31, 32). An interaction signal with Loop1FtsX-T18 was not detected when an EnvC truncation lacking the CC domain (LytEnvC) was fused to T25 in place of FLEnvC (Fig. 6A). This result suggests that the EnvC–FtsX interaction is mediated by contacts between Loop1FtsX and the CC domain of EnvC (CCEnvC). However, for reasons that are not clear, T25 fusions to CCEnvC did not show an interaction signal when coexpressed with Loop1FtsX-T18.
Fig. 6.
EnvC interacts directly with the large periplasmic loop of FtsX. (A) Plasmid pairs encoding the indicated FLEnvC-T25 or LytEnvC-T25 and Loop1FtsX-T18 fusion proteins were cotransformed into BTH101 (cya-99). Individual colonies were patched on M9-glucose supplemented with Amp, Kan, X-Gal, and 1 mM IPTG. Plates were incubated at room temperature and photographed after 72 h. For this particular BACTH assay, interacting partners bring together T18 and T25 to reconstitute adenylate cyclase activity. This activity is detected using lacZ induction as a reporter. (B–D) Purified H-Loop1FtsX was incubated with FLEnvC (B), LytEnvC (C), or CCEnvC (D) for 100 min at room temperature in binding buffer [20 mM Tris-HCl (pH 7.4), 150 mM NaCl]. Ni-NTA magnetic agarose beads (Qiagen) then were added to each reaction and were incubated further for 120 min at 4 °C with rotation. The magnetic beads were captured with a magnet and were washed twice with binding buffer containing 50 mM imidazole. Proteins retained on the resin were eluted with sample buffer containing EDTA (100 mM). Proteins in the initial reaction (Input), initial supernatant (UB), wash supernatants (W1 and W2), and eluate (Elute) were separated on a 15% Tris-Tricine polyacrylamide gel and stained with Coomassie Brilliant Blue. All proteins were present in the initial binding reaction at a concentration of 4 μM. Positions of molecular weight markers (numbers in kDa) are given to the left of each gel. Control reactions indicated that none of the purified EnvC derivatives could be pulled up with H-GFP.
To investigate further the interaction between FtsX and EnvC, we purified untagged versions of FLEnvC, LytEnvC, and CCEnvC as well as a 10× His (H)-tagged version of Loop1FtsX (H-Loop1FtsX). We then tested for interactions using pull-up assays with magnetic Ni-NTA beads. When FLEnvC was incubated with H-Loop1FtsX, it was found in the eluate from Ni-NTA beads following two wash steps (Fig. 6B). Consistent with the BACTH results, when LytEnvC was incubated with H-Loop1FtsX (Fig. 6C), LytEnvC was found primarily in the unbound fraction. When we incubated CCEnvC with H-Loop1FtsX, only about half of the H-Loop1FtsX was retained by the beads. It was accompanied in the eluate by a small fraction (about 10%) of the total CCEnvC, indicating that the two domains interact weakly. The remaining H-Loop1FtsX was found in the unbound fraction with the majority of CCEnvC. Because H-Loop1FtsX bound efficiently to the beads in the presence of FLEnvC and LytEnvC, the addition of CCEnvC appears to affect the accessibility of the H-tag on H-Loop1FtsX adversely. This effect may be the result of CCEnvC inducing some sort of conformational change in H-Loop1FtsX, but further investigation is required to test this possibility.
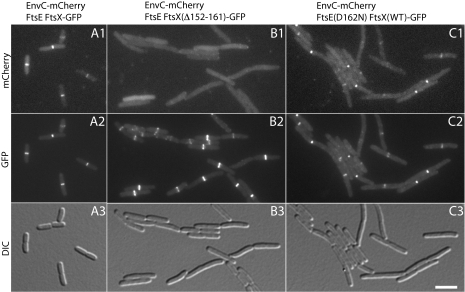
To determine the physiological relevance of the EnvC–Loop1FtsX interaction, we investigated the effect of deleting various portions of the FtsX loop domain on the recruitment of EnvC to the division site. To do so, we a generated a construct expressing the ftsEX operon under control of Plac with the ftsX reading frame fused to the coding sequence for GFP. The expression construct thus produces untagged FtsE as well as FtsX-GFP. For convenience, we will refer to the fusion as FtsEX-GFP. In addition to the WT fusion, we also generated several variants deleted for various portions of the Loop1 domain of FtsX. When produced as the only source of FtsEX in strain DY18(attλTD80)(attHKDY156) [ΔftsEX ΔenvC (Plac::envC-mCherry)(Plac::ftsEX-gfp)], the FtsEX-GFP fusion corrected the ΔftsEX division phenotype, localized to the division site, and was functional for the recruitment of EnvC-mCherry to the septal ring (Fig. 7A). All the Loop1-deletion derivatives of FtsX-GFP that we tested (Δ152–161, Δ146–165, Δ137–176, and Δ109–213) localized to the division site (Fig. 7B and SI Appendix, Fig. S2), indicating that the Loop1 domain is not an important localization determinant for FtsX. However, even the derivative with the smallest deletion (Δ152–161) failed to recruit EnvC to the septum, and cells expressing these derivatives displayed an EnvC−-like division defect (Fig. 7B and SI Appendix, Fig. S2). We conclude that the Loop1–EnvC interaction we detected in the BACTH and in vitro assays is required for the recruitment of EnvC to the septal ring.
Fig. 7.
EnvC localization in cells producing FtsEX variants. Cells of DY18(attλTD80) [ΔftsEX ΔenvC (Plac::envC-mCherry)] harboring the integrated expression constructs attHKDY156 (Plac::ftsEX-GFP) (A), attHKDY161 (Plac::ftsEXΔ152-161-GFP) (B), or attHKDY167 [Plac::ftsE(D162N)X-GFP] (C) were grown overnight at 30 °C in LB (1% NaCl). They then were diluted 1:100 into M9 maltose supplemented with 500 μM IPTG. When cells reached an OD600 of 0.42–0.54, they were visualized as described in the legend for Fig. 5.
ATPase Activity of FtsE Is Likely Required for Amidase Activation by EnvC.
Weiss and coworkers (20) recently showed that FtsE residues in and around the Walker A motif (GxxGxGKS/T, where x is any residue) and Walker B motif (φφφφD, where φ is a hydrophobic residue) predicted to be important for ATP hydrolysis are required for FtsEX to function in cell division (20). Interestingly, however, these residues were not needed for the stability of FtsE or the recruitment of either FtsE or FtsX to the division site. Also, unlike ftsEX-null mutants, division proteins downstream of FtsEX in the localization hierarchy were recruited to the septal ring in the presence of these predicted ATPase-defective variants when mutant cells were grown under nonpermissive conditions (20). Thus, by all indications, complete septal rings were formed in the filamentous cells producing the FtsE mutants, but the rings were unable to promote cell constriction without ATP hydrolysis by FtsEX (20).
Our results thus far indicate that FtsEX directly recruits EnvC to the septal ring. Given the findings of Arends et al. (20), we also wondered if the ATPase activity of FtsE might be required to stimulate amidase activation by EnvC. We therefore generated ftsE(K41M), ftsE(D162N), and ftsE(E163Q) derivatives of the Plac::ftsEX-gfp construct described above. The encoded FtsE variants (FtsE*) in these constructs are similar to those studied previously by Arends et al. (20), except that more conservative amino acid changes were made. The K41M substitution in the Walker A motif of FtsE is predicted to reduce affinity for ATP greatly (34). The Walker B residue, D162, is predicted to coordinate Mg2+ in the active site (34). Substitutions at this residue in other NBDs abrogate ATP-binding (34). An acidic residue, E163, in FtsE typically follows the Walker B motif in ABC transporters. The role of this residue in the ATPase catalytic mechanism is controversial (34). NBDs with Glu-to-Gln substitutions at this position still bind ATP, but, depending on the particular transporter, may or may not retain residual ATPase activity (34).
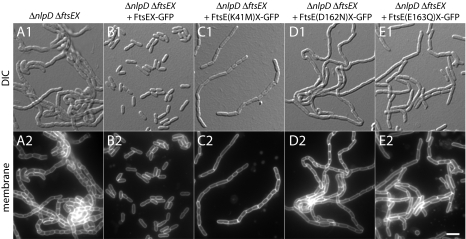
The ability of the FtsE* variants to promote cell separation was assessed first by producing them from the Plac::ftsEX-gfp construct in ΔftsEX ΔnlpD cells. As expected, a construct producing WT FtsE completely reversed the severe chaining phenotype of the FtsEX− NlpD− mutants (Fig. 8 and SI Appendix, Table S1). Constructs producing the FtsE* variants, on the other hand, all failed to restore normal cell separation (Fig. 8 and SI Appendix, Table S1). However, the mutants did not behave identically. Although FtsE(D162N) appeared to be completely defective for cell separation, the FtsE(K41M) and FtsE(E163Q) variants appeared to retain partial function (SI Appendix, Table S1). Based on results from the related LolCDE system, FtsE(K41M) is likely to retain weak affinity for ATP (35). This weak affinity may allow enough ATPase activity to reduce the length of the chains observed in ΔftsEX ΔnlpD cells. For FtsE(E163Q), its ability to shorten ΔftsEX ΔnlpD chains may indicate either that ATP-binding alone is sufficient for low-level cell-separation activity or that the E/Q substitution in FtsEX does not abolish ATPase activity completely. Measurement of the ATPase activity of FtsE and the FtsE* variants is required to distinguish between these possibilities, but this measurement has not been possible because of problems with FtsE solubility when it was overproduced. Nevertheless, the (partial) loss-of-function phenotypes displayed by the FtsE* variants clearly highlight a role for key ATP-binding site residues in FtsE for it to promote proper cell separation. Importantly, FtsX-GFP remained capable of recruiting to septal rings in cells producing the FtsE* variants, and the efficiency with which EnvC-mCherry was recruited to the FtsX-containing rings was largely unaffected by the FtsE lesions (Fig. 7C and SI Appendix, Fig. S3 and Table S2). Furthermore, based on the results of Arends et al. (20) with similar mutants, we assume that the FtsE* variants described here also are recruited normally to the septal ring. Thus, FtsE*X–EnvC complexes likely do form at the division site in cells expressing the ftsE* alleles but fail to function or function poorly in the septal PG splitting process. We therefore infer that the ATPase activity of the FtsEX complex plays an important role in promoting amidase activation by EnvC at the septum.
Fig. 8.
FtsE ATP-binding site lesions result in a cell-separation defect. Cells of KP7/pTB63 (ΔftsEX ΔnlpD/ftsQAZ) without an integrated expression cassette (A) or harboring attHKDY156 (Plac::ftsEX-GFP) (B), attHKDY166 [Plac::ftsE(K41M)X-GFP] (C), attHKDY167 [Plac::ftsE(D162N)X-GFP] (D), or attHKDY168 [Plac::ftsE(E163Q)X-GFP] (E) were grown and visualized as described in the legend to Fig. 3, except that the medium contained 500 μM IPTG.
Discussion
The role of the FtsEX ABC system in cytokinesis has long remained mysterious. In this study, we genetically and physically connected FtsEX with the process of septal PG splitting. Our results indicate that the complex is directly responsible for the recruitment of the amidase activator EnvC to the cytokinetic ring. Importantly, we also showed that variants of the FtsEX complex predicted to have inactive ATPase subunits still recruited EnvC to the septum but failed to induce septal PG splitting. Thus, in addition to connecting EnvC to the septal ring, FtsEX appears to use its ATPase activity to promote amidase activation. An attractive possibility is that it does so using ATPase-induced conformational changes similar to those observed in other ABC systems (12) to regulate EnvC activity in the periplasm allosterically (Fig. 9).
Fig. 9.
Model for FtsEX function in regulating PG hydrolase activity at the division site. Shown is a schematic diagram of a putative FtsEX-EnvC-amidase complex at the Z-ring. We propose that conformational changes in FtsEX induced by FtsE-mediated ATP hydrolysis are transmitted to EnvC to control its ability to activate the amidases so that they can cleave the septal PG (not drawn). The model is not meant to reflect actual interaction stoichiometries, because they have yet to be determined. In addition, it is not yet clear if the amidases remain in complex with EnvC as drawn or if this interaction is also regulated. See text for details.
Consistent with this model, sequence analysis groups FtsEX with a subclass of ABC systems mainly comprising substrate binding protein (SBP)-dependent importers such as the maltose transporter (MalFGK2) (34). The structure of MalFGK2 in complex with maltose-binding protein (MalE) indicates that ATP-driven conformational changes in the transporter can alter the conformation of periplasmic MalE (36). In this case, MalE is converted from its closed, maltose-bound conformation to an open conformation that releases maltose into the outward-facing cavity of the transporter for subsequent import (36). We therefore propose that EnvC may be an SBP analog for FtsEX and envision that the conformation of EnvC is modulated similarly by the ABC system so that it interconverts between an “on” and “off” state during an ATPase cycle (Fig. 9). Such a model is appealing for several reasons. Most significantly, it would provide a means for converting septal PG hydrolysis into a discrete process with a fixed number of PG bonds being broken per ATP hydrolyzed. Requiring the activation of each cleavage event would afford the septal ring exquisite control over PG hydrolysis, which is highly desirable given the inherent risks involved in promoting localized PG degradation. Additionally, because FtsE interacts with FtsZ in the cytoplasm, the ATPase activity of FtsE could be coupled directly to Z-ring dynamics. Thus, the FtsEX complex could serve as a molecular governor to coordinate properly the rate of septal PG hydrolysis with the contraction of the Z-ring. Finally, in addition to connecting the Z-ring with septal PG hydrolysis, interactions of FtsX with other transmembrane components of the divisome may help couple the activity of the integral membrane PG synthases with the cell-separation amidases. For example, the FtsEX complex may promote EnvC-activated amidase activity only when it is engaged with an active PG synthetic complex.
Control of EnvC activity by FtsEX is likely mediated in part by the observed interaction between the CC domain of EnvC and Loop1FtsX. Consistent with this possibility, the CC domain of EnvC was shown previously to be important for EnvC regulation (24). An EnvC truncation lacking the CC domain (LytEnvC) failed to be recruited to the division site and inappropriately activated AmiA and AmiB to induce cell lysis (24). Because cell lysis is not triggered when FLEnvC is displaced from the septum by CCEnvC overproduction (24) or by the deletion of ftsEX, proper regulation of amidase activation appears to entail more than just controlling EnvC localization. Because FLEnvC activates the amidases just as well as LytEnvC in vitro (24), we suspect either that there is something about the physiochemical environment of the periplasm that promotes the direct inhibition of FLEnvC activity by the CC domain itself or that inhibition is mediated by an additional factor that associates with the CC domain. According to our model for EnvC regulation by FtsEX, it is this auto- or trans-inhibition of EnvC that is cycled on and off in response to the ATPase activity of FtsEX (Fig. 9).
Although we favor a model in which FtsEX serves as a transmembrane allosteric regulator of EnvC, scenarios in which FtsEX transports a molecule needed for EnvC activity are difficult to exclude. Arguing against a transport function is the fact that FtsEX is most similar to the LolCDE ABC system. Rather than catalyzing the transport of a substrate across a membrane, LolCDE facilitates the transfer of lipoproteins from the outer leaflet of the inner membrane to the periplasmic LolA carrier protein for their ultimate insertion into the outer membrane (37). The individual TMD components of both FtsEX and LolCDE have only four transmembrane helices, unlike most ABC transporters, which typically have at least six (20, 34, 38). Moreover, the membrane-spanning helices of FtsX do not contain any charged amino acids as might be expected for a factor that transports an electrolyte (20). Based on these observations, Weiss and coworkers (20) also have proposed that FtsEX may not be a transporter. Rather, they hypothesized that, as we propose here, FtsE uses ATP hydrolysis to drive a periplasmic activity through conformational changes in FtsX (20). Reconstitution of EnvC regulation by FtsEX is required to demonstrate an allosteric control mechanism definitively. So far, our attempts at reconstitution have been unsuccessful. Addition of purified Loop1FtsX or full-length FtsX to mixtures of EnvC and AmiB did not affect amidase activation using our standard reaction conditions (24). This result suggests that the full FtsEX complex may be needed to observe regulation in vitro. Unfortunately, efforts to purify the complete FtsEX complex have been hampered by the insolubility of FtsE when it is overproduced. This hurdle must be overcome before additional attempts at reconstitution can be pursued.
A role for FtsEX in the process of septal PG splitting appears to be conserved. In an accompanying report Sham et al. (28) directly connect the essential cell-separation factor PcsB (39) with FtsEX in S. pneumoniae. Although their sequences are largely unrelated, the domain structures of PcsB and EnvC are strikingly similar. Like EnvC, PcsB has an N-terminal region predicted to form coiled coils and a C-terminal PG hydrolase-like CHAP domain (39). Currently however, it is not known whether PcsB directly degrades PG or if, analogous to EnvC, it is an activator of other PG hydrolases. Nevertheless, the results of Sham et al. (28) indicate that PcsB activity is likely governed by FtsEX to control properly the process of septal PG splitting in S. pneumoniae. Interestingly, FtsEX does not appear to be involved in cell division in Bacillus subtilis (40). We suspect this difference has to do with the different septal geometries of these organisms. Gram-negative bacteria like E. coli and ovococci like S. pneumoniae appear to couple septal PG splitting with the invagination of their cytoplasmic membrane to give their predivisional cells a constricted appearance (1, 41). This mode of division likely requires regulators of PG hydrolysis such as FtsEX to be associated with the septal ring. B. subtilis and many other Gram-positive bacteria, on the other hand, first construct a flat septum, the splitting of which appears to be uncoupled from membrane constriction and fission (42). Septal PG splitting in these cells therefore is likely to have regulatory requirements that differ from those of constricting cells. The function of FtsEX is in cells with a flat septal morphology is not clear, but it may involve the regulation of PG hydrolysis needed for other aspects of cell-wall growth and remodeling.
In conclusion, we have identified a role for the FtsEX ABC system in the regulation of septal PG hydrolysis by the amidases AmiA and AmiB and the LytM factor EnvC. A second pathway for septal PG splitting involving AmiC and the LytM factor NlpD also is operative in E. coli (24). How this system is regulated is not known currently. However, because NlpD is an outer- membrane lipoprotein (43) rather than a periplasmic protein like EnvC, its activity is likely to be modulated by a distinct mechanism. In summary, this report provides evidence for the transmembrane regulation of enzymatic activity by an ABC system. Given the diversity and ubiquity of ABC systems in nature (34), it is likely that the ATP-driven conformational changes in these membrane complexes have been adapted to regulate a variety of biological processes.
Experimental Procedures
Media, Bacterial Strains, and Plasmids.
Cells were grown in LB (1% tryptone, 0.5% yeast extract, 0.5–1.5% NaCl as indicated) or minimal M9 medium (44) supplemented with 0.2% casamino acids and 0.2% sugar (glucose, maltose, or arabinose as indicated). Unless otherwise indicated, antibiotics were used at 5, 10, 15, 20, or 50 μg/mL for tetracycline (Tet), chloramphenicol (Cam), ampicillin (Amp), kanamycin (Kan), or spectinomycin (Spec), respectively.
The bacterial strains used in this study are listed in SI Appendix, Table S3. All strains used in the reported experiments are derivatives of MG1655. Plasmids used in this study are listed in SI Appendix, Table S4. Vectors with R6K origins are all derivatives of the CRIM plasmids developed by Haldimann and Wanner (45). They were maintained in the cloning strain DH5α(λpir) where they replicate as plasmids or were integrated into phage attachment sites (HK022 or λ) using the helper vectors pTB102 (46) or pInt-ts (45), respectively, as described previously (45). See SI Appendix for Details of plasmid and strain construction.
Synthetic Lethal Screen.
The screen for mutants with a Slb phenotype was performed as previously described (27). Briefly, TU122/pTU110 (ΔlacIZYA ΔponB/Plac::ponB lacZ ) was mutagenized with the EzTn-Kan2 transposome (Epicentre) as previously described (26). Mutants were selected for Kan resistance at room temperature, yielding a library of 75,000 independent transposon insertions. This mutant library was plated on LB (0.5% NaCl) agar supplemented with 50 μM IPTG and X-Gal (40 μg/mL) at 30 °C and room temperature to identify mutants with a Slb phenotype. In addition to the transposon insertions in ponA and lpoA described previously (27), we also isolated mutants with insertions in envC (between codons 74 and 75) and ftsX (within codon 59).
Fluorescence Microscopy and Protein Methods.
Fluorescence microscopy was performed as described previously (25). Specific growth conditions used for each experiment are given in figure legends. Cell fixation and membrane staining was performed as described previously (25). Protein purification, cell fractionation, and immunoblotting methods are described in SI Appendix. The BACTH assay and Ni-NTA pull-up assays are described in the legend to Fig. 6.
Supplementary Material
Acknowledgments
We thank members of the laboratory for discussions, support, and critical reading of the manuscript. We also thank Malcolm Winkler and co-workers for communicating their independent discovery prior to publication that FtsEX in S. pneumoniae interacts with the cell-separation factor PcsB. This work was supported by the Massachusetts Life Science Center, the Burroughs Wellcome Fund, and by National Institutes of Health Grant R01 AI083365. T.G.B. holds a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 18209.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107780108/-/DCSupplemental.
References
- 1.de Boer PAJ. Advances in understanding E. coli cell fission. Curr Opin Microbiol. 2010;13:730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 3.Dajkovic A, Pichoff S, Lutkenhaus J, Wirtz D. Cross-linking FtsZ polymers into coherent Z rings. Mol Microbiol. 2010;78:651–668. doi: 10.1111/j.1365-2958.2010.07352.x. [DOI] [PubMed] [Google Scholar]
- 4.Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 6.Hale CA, et al. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J Bacteriol. 2011;193:1393–1404. doi: 10.1128/JB.01245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J Bacteriol. 2011;193:1405–1413. doi: 10.1128/JB.01258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galli E, Gerdes K. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol Microbiol. 2010;76:1514–1526. doi: 10.1111/j.1365-2958.2010.07183.x. [DOI] [PubMed] [Google Scholar]
- 10.Ebersbach G, Galli E, Møller-Jensen J, Löwe J, Gerdes K. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol Microbiol. 2008;68:720–735. doi: 10.1111/j.1365-2958.2008.06190.x. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt KL, et al. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J Bacteriol. 2004;186:785–793. doi: 10.1128/JB.186.3.785-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rees DC, Johnson E, Lewinson O. ABC transporters: The power to change. Nat Rev Mol Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricard M, Hirota Y. Process of cellular division in Escherichia coli: Physiological study on thermosensitive mutants defective in cell division. J Bacteriol. 1973;116:314–322. doi: 10.1128/jb.116.1.314-322.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill DR, Hatfull GF, Salmond GP. A new cell division operon in Escherichia coli. Mol Gen Genet. 1986;205:134–145. doi: 10.1007/BF02428043. [DOI] [PubMed] [Google Scholar]
- 15.de Leeuw E, et al. Molecular characterization of Escherichia coli FtsE and FtsX. Mol Microbiol. 1999;31:983–993. doi: 10.1046/j.1365-2958.1999.01245.x. [DOI] [PubMed] [Google Scholar]
- 16.Corbin BD, Wang Y, Beuria TK, Margolin W. Interaction between cell division proteins FtsE and FtsZ. J Bacteriol. 2007;189:3026–3035. doi: 10.1128/JB.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy M. Role of FtsEX in cell division of Escherichia coli: Viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J Bacteriol. 2007;189:98–108. doi: 10.1128/JB.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samaluru H, SaiSree L, Reddy M. Role of SufI (FtsP) in cell division of Escherichia coli: Evidence for its involvement in stabilizing the assembly of the divisome. J Bacteriol. 2007;189:8044–8052. doi: 10.1128/JB.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arends SJ, Kustusch RJ, Weiss DS. ATP-binding site lesions in FtsE impair cell division. J Bacteriol. 2009;191:3772–3784. doi: 10.1128/JB.00179-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 22.Heidrich C, et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- 23.Priyadarshini R, de Pedro MA, Young KD. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J Bacteriol. 2007;189:5334–5347. doi: 10.1128/JB.00415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 2010;29:1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernhardt TG, de Boer PAJ. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol. 2004;52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paradis-Bleau C, et al. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sham L, Barendt SM, Kopecky KE, Winkler ME. The essential CHAP-domain protein PcsB interacts with the essential cell division protein FtsX in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 2011;108:E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Typas A, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodolakis A, Thomas P, Starka J. Morphological mutants of Escherichia coli. Isolation and ultrastructure of a chain-forming envC mutant. J Gen Microbiol. 1973;75:409–416. doi: 10.1099/00221287-75-2-409. [DOI] [PubMed] [Google Scholar]
- 31.Ichimura T, Yamazoe M, Maeda M, Wada C, Hiraga S. Proteolytic activity of YibP protein in Escherichia coli. J Bacteriol. 2002;184:2595–2602. doi: 10.1128/JB.184.10.2595-2602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hara H, et al. Identification and characterization of the Escherichia coli envC gene encoding a periplasmic coiled-coil protein with putative peptidase activity. FEMS Microbiol Lett. 2002;212:229–236. doi: 10.1111/j.1574-6968.2002.tb11271.x. [DOI] [PubMed] [Google Scholar]
- 33.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito Y, Kanamaru K, Taniguchi N, Miyamoto S, Tokuda H. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins. Mol Microbiol. 2006;62:1064–1075. doi: 10.1111/j.1365-2958.2006.05378.x. [DOI] [PubMed] [Google Scholar]
- 36.Khare D, Oldham ML, Orelle C, Davidson AL, Chen J. Alternating access in maltose transporter mediated by rigid-body rotations. Mol Cell. 2009;33:528–536. doi: 10.1016/j.molcel.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yakushi T, Masuda K, Narita S, Matsuyama S, Tokuda H. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat Cell Biol. 2000;2:212–218. doi: 10.1038/35008635. [DOI] [PubMed] [Google Scholar]
- 38.Daley DO, et al. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 39.Ng W-L, Kazmierczak KM, Winkler ME. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol Microbiol. 2004;53:1161–1175. doi: 10.1111/j.1365-2958.2004.04196.x. [DOI] [PubMed] [Google Scholar]
- 40.Garti-Levi S, Hazan R, Kain J, Fujita M, Ben-Yehuda S. The FtsEX ABC transporter directs cellular differentiation in Bacillus subtilis. Mol Microbiol. 2008;69:1018–1028. doi: 10.1111/j.1365-2958.2008.06340.x. [DOI] [PubMed] [Google Scholar]
- 41.Higgins ML, Shockman GD. Model for cell wall growth of Streptococcus faecalis. J Bacteriol. 1970;101:643–648. doi: 10.1128/jb.101.2.643-648.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burdett ID. Electron microscope study of the rod-to-coccus shape change in a temperature-sensitive rod- mutant of Bacillus subtilis. J Bacteriol. 1979;137:1395–1405. doi: 10.1128/jb.137.3.1395-1405.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ichikawa JK, Li C, Fu J, Clarke S. A gene at 59 minutes on the Escherichia coli chromosome encodes a lipoprotein with unusual amino acid repeat sequences. J Bacteriol. 1994;176:1630–1638. doi: 10.1128/jb.176.6.1630-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 45.Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernhardt TG, de Boer PAJ. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]